Effects of Solvent Evaporation Methods and Short-Term Room Temperature Storage on High-Coverage Cellular Metabolome Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cell Culture
2.3. Sample Preparation
2.4. Chemical Isotope Labeling
2.5. LC-MS Analysis
2.6. Data Processing, Metabolite Identification, and Statistical Analysis
3. Results and Discussion
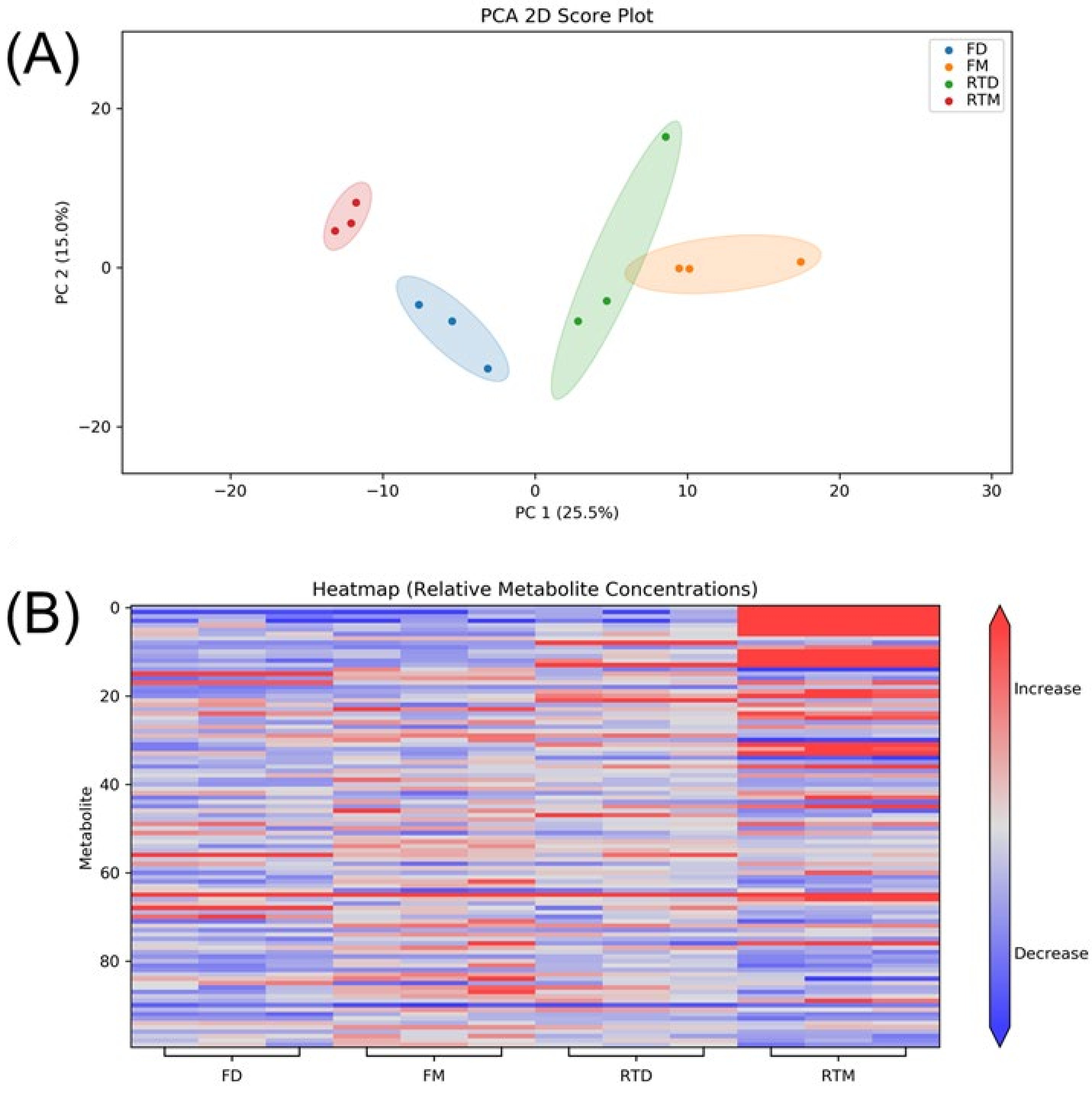
3.1. Cell Line
3.2. Metabolite Identification
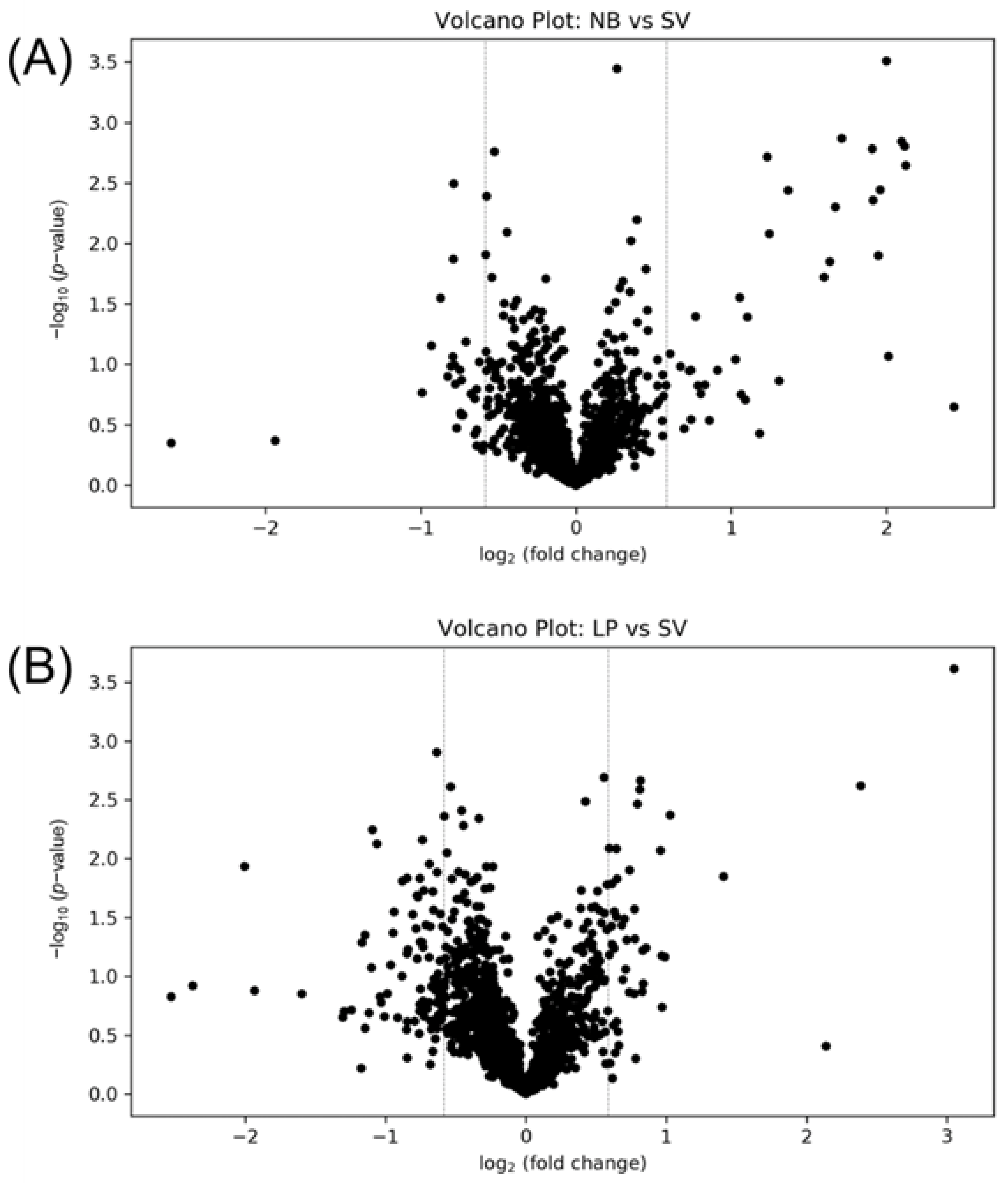
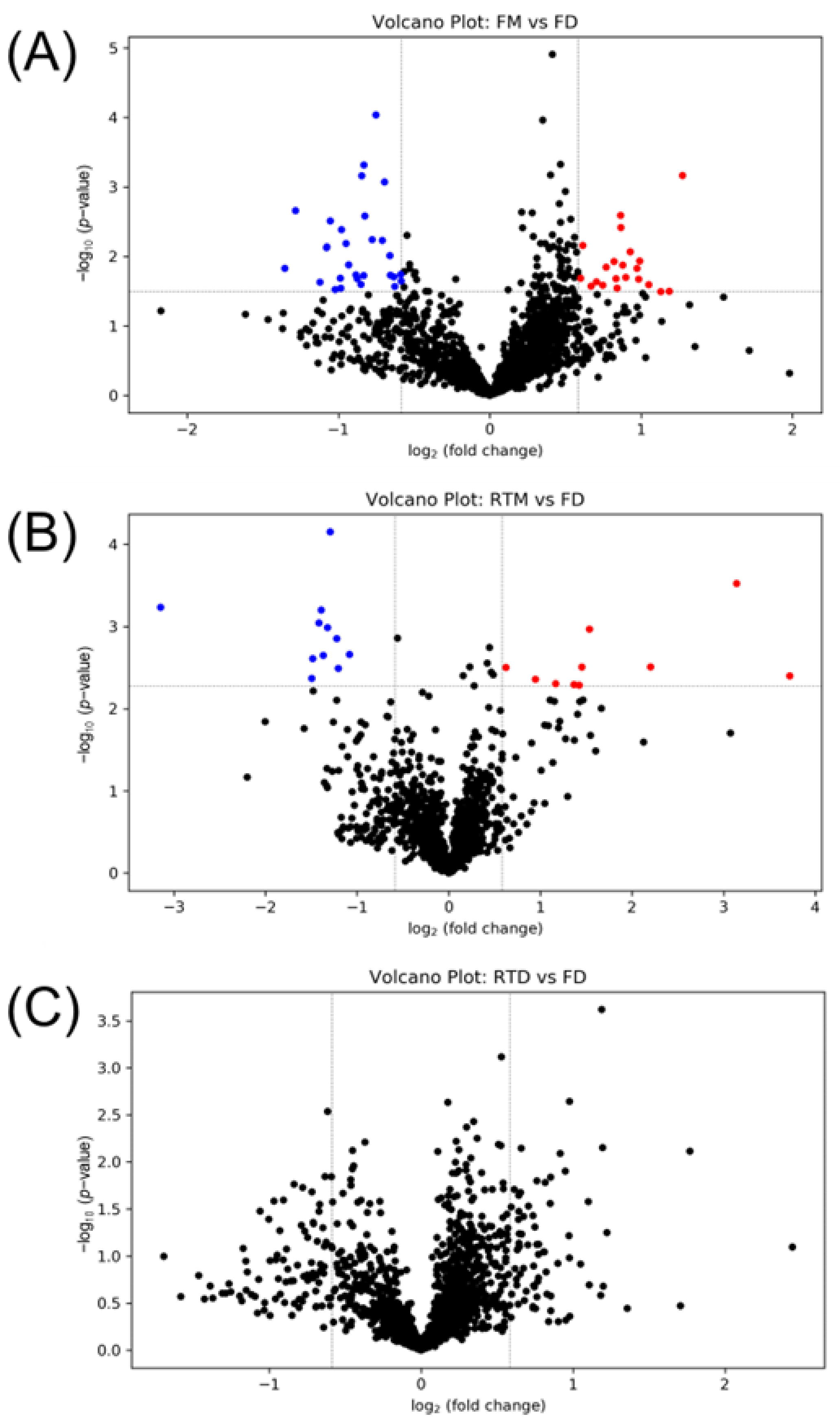
3.3. Solvent Evaporation Equipment Selection
3.4. Storage Condition
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Muschet, C.; Möller, G.; Prehn, C.; de Angelis, M.H.; Adamski, J.; Tokarz, J. Removing the Bottlenecks of Cell Culture Metabolomics: Fast Normalization Procedure, Correlation of Metabolites to Cell Number, and Impact of the Cell Harvesting Method. Metabolomics 2016, 12, 151. [Google Scholar] [CrossRef]
- Domenick, T.M.; Gill, E.L.; Vedam-Mai, V.; Yost, R.A. Mass Spectrometry-Based Cellular Metabolomics: Current Approaches, Applications, and Future Directions. Anal. Chem. 2021, 93, 546–566. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, L. Metabolomics of Small Numbers of Cells: Metabolomic Profiling of 100, 1000, and 10,000 Human Breast Cancer Cells. Anal. Chem. 2017, 89, 11664–11671. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Gu, X.; Li, L. Development of a Simple and Efficient Method of Harvesting and Lysing Adherent Mammalian Cells for Chemical Isotope Labeling LC-MS-Based Cellular Metabolomics. Anal. Chim. Acta 2018, 1037, 97–106. [Google Scholar] [CrossRef]
- Yuan, B.-F.; Zhu, Q.-F.; Guo, N.; Zheng, S.-J.; Wang, Y.-L.; Wang, J.; Xu, J.; Liu, S.-J.; He, K.; Hu, T.; et al. Comprehensive Profiling of Fecal Metabolome of Mice by Integrated Chemical Isotope Labeling-Mass Spectrometry Analysis. Anal. Chem. 2018, 90, 3512–3520. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhou, X.; Yue, M.; Zhu, S.; Liu, Q.; Zhao, X.-E. Advances and Perspectives in Chemical Isotope Labeling-Based Mass Spectrometry Methods for Metabolome and Exposome Analysis. TrAC Trends Anal. Chem. 2023, 162, 117022. [Google Scholar] [CrossRef]
- Jia, S.; Xu, T.; Huan, T.; Chong, M.; Liu, M.; Fang, W.; Fang, M. Chemical Isotope Labeling Exposome (CIL-EXPOSOME): One High-Throughput Platform for Human Urinary Global Exposome Characterization. Environ. Sci. Technol. 2019, 53, 5445–5453. [Google Scholar] [CrossRef] [PubMed]
- Bruheim, P.; Kvitvang, H.F.N.; Villas-Boas, S.G. Stable Isotope Coded Derivatizing Reagents as Internal Standards in Metabolite Profiling. J. Chromatogr. A 2013, 1296, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Powers, R. The Current State of Drug Discovery and a Potential Role for NMR Metabolomics. J. Med. Chem. 2014, 57, 5860–5870. [Google Scholar] [CrossRef] [PubMed]
- Tounta, V.; Liu, Y.; Cheyne, A.; Larrouy-Maumus, G. Metabolomics in Infectious Diseases and Drug Discovery. Mol. Omics 2021, 17, 376–393. [Google Scholar] [CrossRef] [PubMed]
- Hasunuma, T.; Sanda, T.; Yamada, R.; Yoshimura, K.; Ishii, J.; Kondo, A. Metabolic Pathway Engineering Based on Metabolomics Confers Acetic and Formic Acid Tolerance to a Recombinant Xylose-Fermenting Strain of Saccharomyces Cerevisiae. Microb. Cell Fact. 2011, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Araújo, A.M.; Carvalho, F.; de Pinho, P.; Carvalho, M. Toxicometabolomics: Small Molecules to Answer Big Toxicological Questions. Metabolites 2021, 11, 692. [Google Scholar] [CrossRef] [PubMed]
- León, Z.; García-Cañaveras, J.C.; Donato, M.T.; Lahoz, A. Mammalian Cell Metabolomics: Experimental Design and Sample Preparation. Electrophoresis 2013, 34, 2762–2775. [Google Scholar] [CrossRef] [PubMed]
- Madji Hounoum, B.; Blasco, H.; Emond, P.; Mavel, S. Liquid Chromatography–High-Resolution Mass Spectrometry-Based Cell Metabolomics: Experimental Design, Recommendations, and Applications. TrAC Trends Anal. Chem. 2016, 75, 118–128. [Google Scholar] [CrossRef]
- Lorenz, M.A.; Burant, C.F.; Kennedy, R.T. Reducing Time and Increasing Sensitivity in Sample Preparation for Adherent Mammalian Cell Metabolomics. Anal. Chem. 2011, 83, 3406–3414. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Xu, H.; Qiu, S.; Wang, X. Cell Metabolomics. OMICS 2013, 17, 495–501. [Google Scholar] [CrossRef]
- Teng, Q.; Huang, W.; Collette, T.W.; Ekman, D.R.; Tan, C. A Direct Cell Quenching Method for Cell-Culture Based Metabolomics. Metabolomics 2009, 5, 199–208. [Google Scholar] [CrossRef]
- Yuan, W.; Zhang, J.; Li, S.; Edwards, J.L. Amine Metabolomics of Hyperglycemic Endothelial Cells Using Capillary LC–MS with Isobaric Tagging. J. Proteome Res. 2011, 10, 5242–5250. [Google Scholar] [CrossRef] [PubMed]

| Speed * | Equipment Cost | Consumable Cost | |
|---|---|---|---|
| Nitrogen Blowdown Evaporator | ~an hour | ~USD 2000 | ~USD 300 per week (nitrogen gas from high pressure cylinder) or ~USD 0 (nitrogen gas from nitrogen generator) |
| SpeedVac Concentrator | ~an hour | ~USD 20,000 | N/A |
| Lyophilizer | ~one day | ~ USD 50,000 | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, X.; Li, L. Effects of Solvent Evaporation Methods and Short-Term Room Temperature Storage on High-Coverage Cellular Metabolome Analysis. Metabolites 2023, 13, 1052. https://doi.org/10.3390/metabo13101052
Luo X, Li L. Effects of Solvent Evaporation Methods and Short-Term Room Temperature Storage on High-Coverage Cellular Metabolome Analysis. Metabolites. 2023; 13(10):1052. https://doi.org/10.3390/metabo13101052
Chicago/Turabian StyleLuo, Xian, and Liang Li. 2023. "Effects of Solvent Evaporation Methods and Short-Term Room Temperature Storage on High-Coverage Cellular Metabolome Analysis" Metabolites 13, no. 10: 1052. https://doi.org/10.3390/metabo13101052
APA StyleLuo, X., & Li, L. (2023). Effects of Solvent Evaporation Methods and Short-Term Room Temperature Storage on High-Coverage Cellular Metabolome Analysis. Metabolites, 13(10), 1052. https://doi.org/10.3390/metabo13101052