UPLC-QTOF-MS Based Comparison of Rotundic Acid Metabolic Profiles in Normal and NAFLD Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Material and Chemicals
2.2. Animals Experiments
2.2.1. Modeling of NAFLD Rats
2.2.2. Pathological Staining
2.2.3. Biochemical Detection
2.3. Preparations of Biological Samples
- Plasma
- Urine
- Feces
- Liver tissues
2.4. RA Metabolism In Vitro
2.5. UPLC-QTOF-MS Conditions
2.6. Data Analysis
3. Results
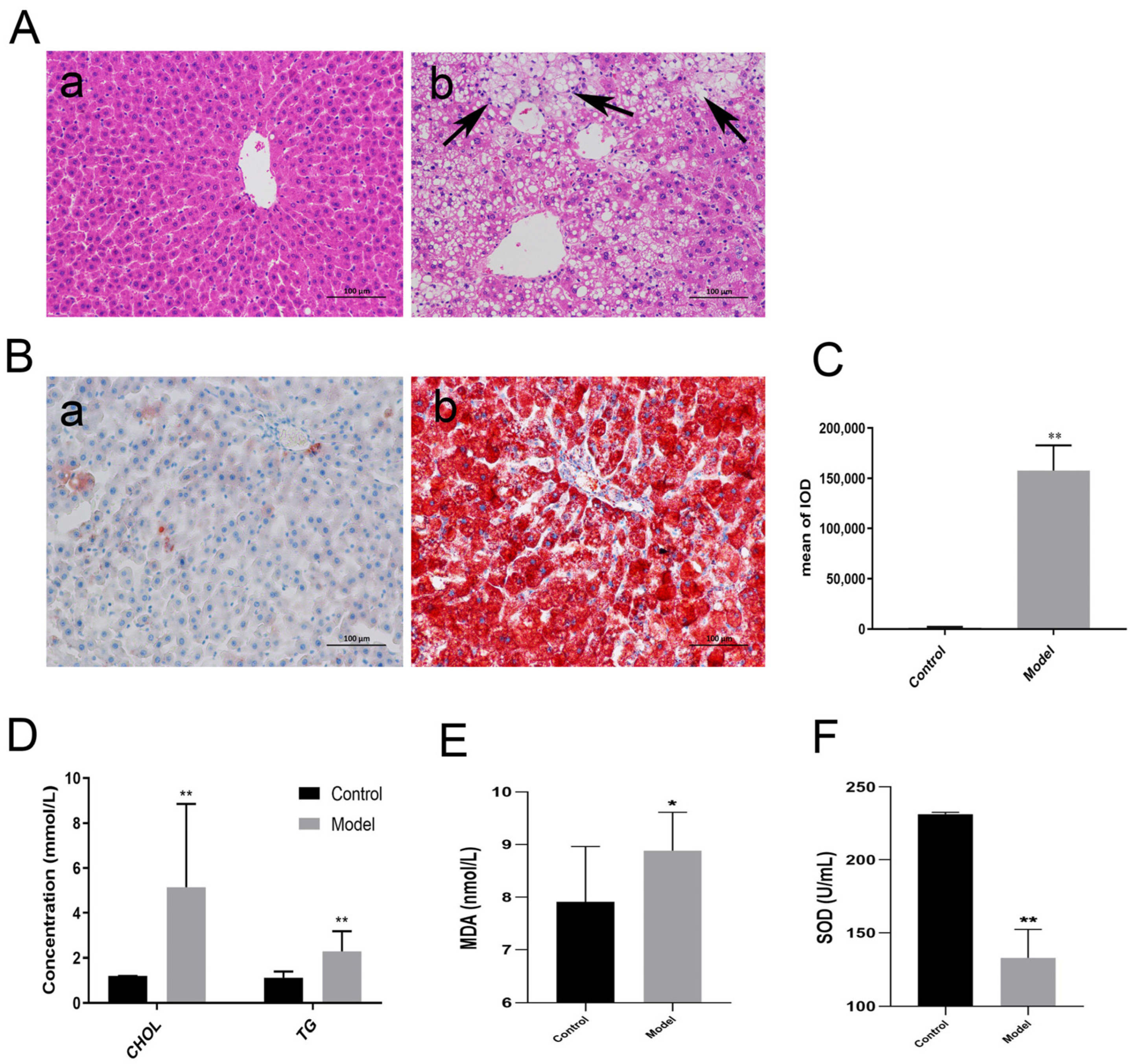
3.1. Establishment of NAFLD Model
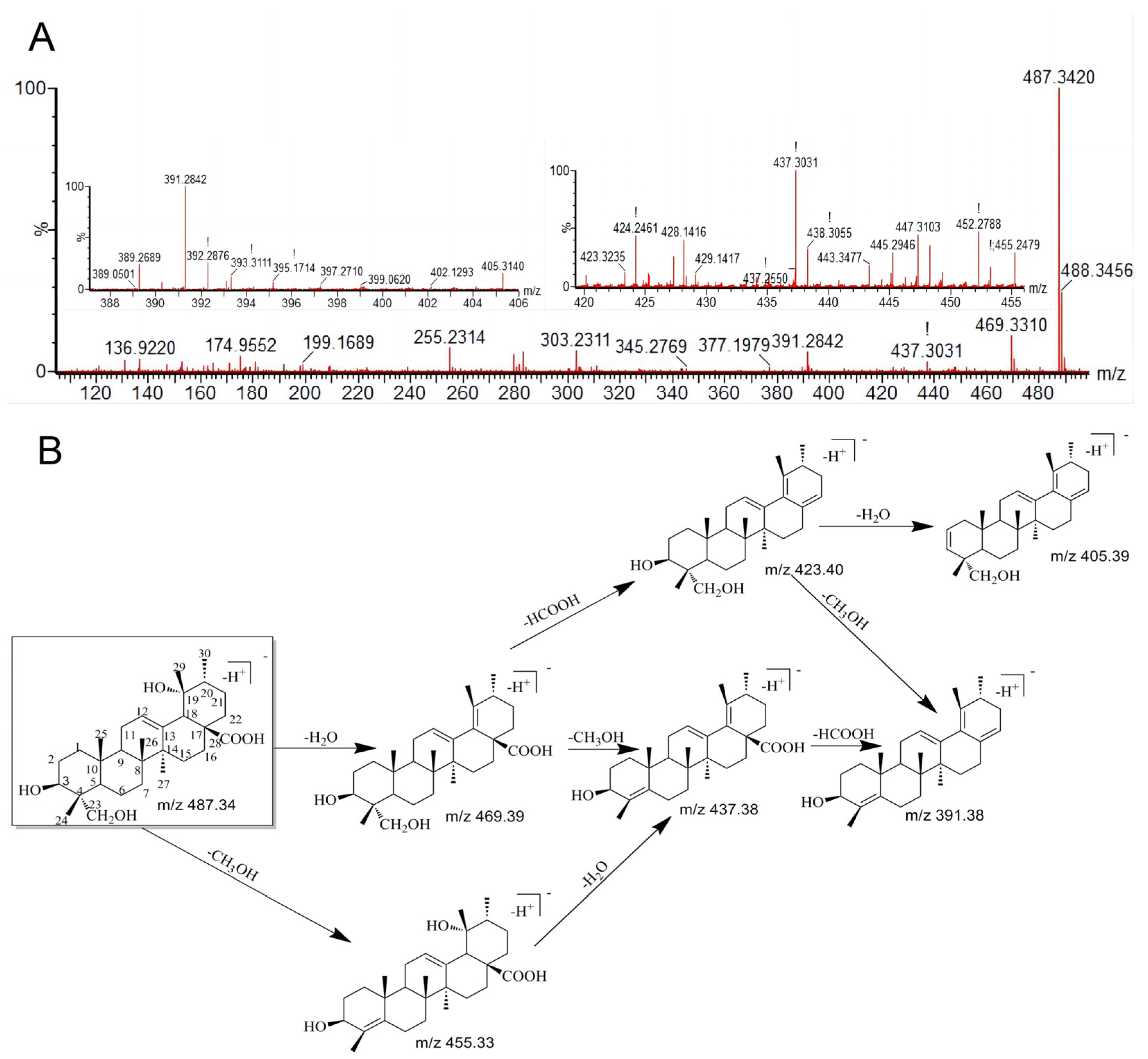
3.2. The Characteristic Fragmentation of RA
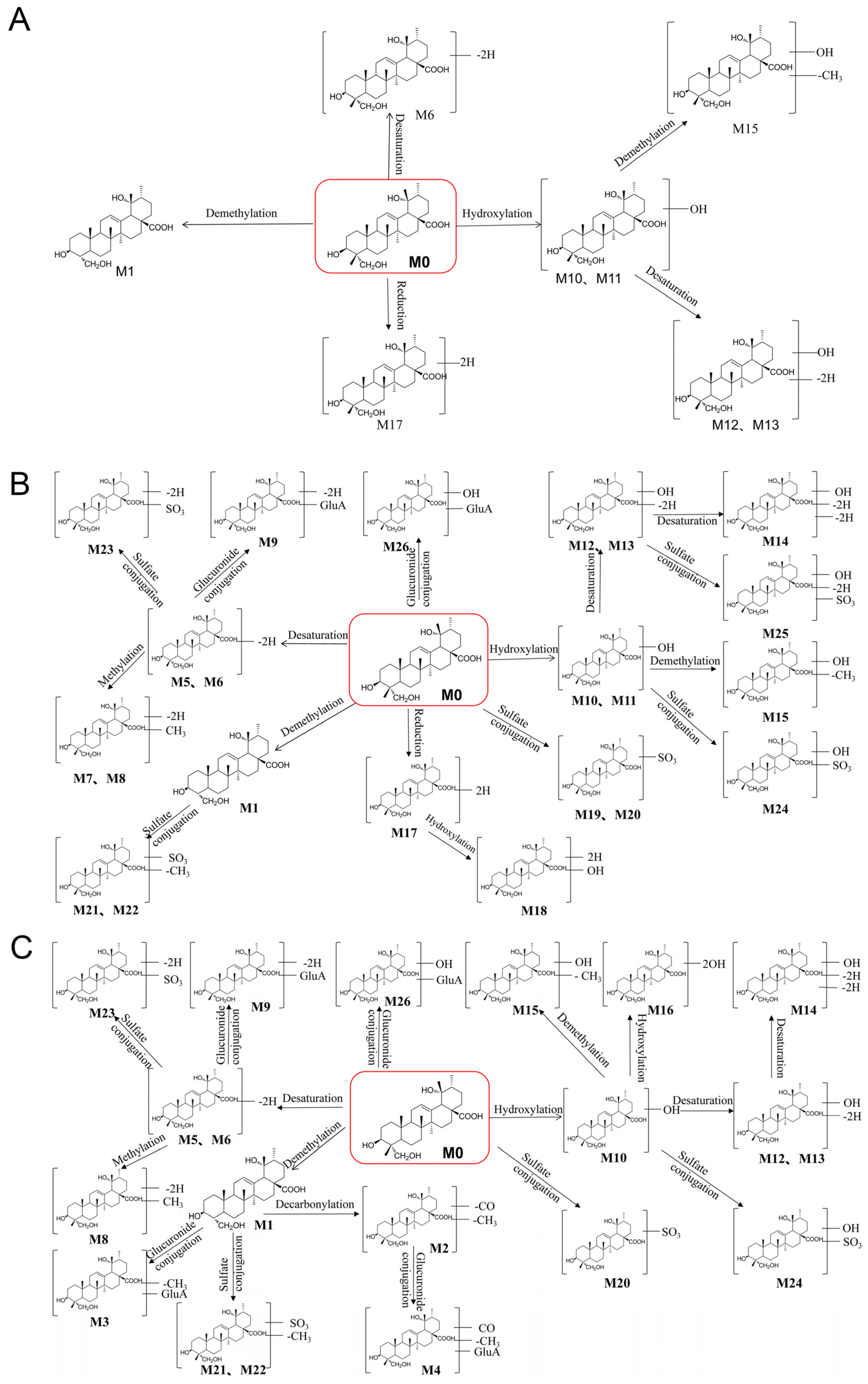
3.3. Identification of the Metabolites of RA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, H.; Wang, Z.J.; Wu, Y.L.; Jiang, H.T.; Zhou, F.; Xie, X.H.; Wang, R.L.; Hua, C. Untargeted Metabonomics Reveals Intervention Effects of Chicory Polysaccharide in a Rat Model of Non-Alcoholic Fatty Liver Disease. Int. J. Biol. Macromol. 2019, 128, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Farzanegi, P.; Dana, A.; Ebrahimpoor, Z.; Asadi, M.; Azarbayjani, M.A. Mechanisms of Beneficial Effects of Exercise Training on Non-Alcoholic Fatty Liver Disease (NAFLD): Roles of Oxidative Stress and Inflammation. Eur. J. Sport Sci. 2019, 19, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Shulman, G.I. Ectopic Fat in Insulin Resistance, Dyslipidemia, and Cardiometabolic Disease. N. Engl. J. Med. 2014, 371, 2237–2238. [Google Scholar] [CrossRef]
- Chen, K.; Ma, J.B.; Jia, X.Y.; Ai, W.; Ma, Z.R.; Pan, Q.W. Advancing the Understanding of NAFLD to Hepatocellular Carcinoma Development: From Experimental Models to Humans. Biochim. Biophys. Acta-Rev. Cancer 2019, 1871, 117–125. [Google Scholar] [CrossRef]
- Leoni, S.; Tovoli, F.; Napoli, L.; Serio, I.; Ferri, S.; Bolondi, L. Current Guidelines for the Management of Non-Alcoholic Fatty Liver Disease: A Systematic Review with Comparative Analysis. World J. Gastroenterol. 2018, 24, 3361–3373. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Zhou, L.; Xiong, T.Q.; Zhou, J.S.; Li, Q.G.; Tan, Q.L.; Zhao, Z.X.; Jin, J. Antiplatelet Aggregation Triterpene Saponins from the Barks of Ilex Rotunda. Fitoterapia 2015, 101, 19–26. [Google Scholar] [CrossRef]
- Liu, W.J.; Peng, Y.Y.; Chen, H.; Liu, X.F.; Liang, J.Y.; Sun, J.B. Triterpenoid Saponins with Potential Cytotoxic Activities from the Root Bark of Ilex Rotunda Thunb. Chem. Biodivers. 2017, 14, e1600209. [Google Scholar] [CrossRef]
- Wang, Z.F.; Sun, W.Y.; Yu, D.H.; Zhao, Y.; Xu, H.M.; He, Y.F.; Li, H.J. Rotundic Acid Enhances the Impact of Radiological Toxicity on MCF-7 Cells through the ATM/P53 Pathway. Int. J. Oncol. 2018, 53, 2269–2277. [Google Scholar] [CrossRef]
- He, Y.F.; Nan, M.L.; Sun, J.M.; Meng, Z.J.; Li, W.; Zhang, M. Design, Synthesis and Cytotoxicity of Cell Death Mechanism of Rotundic Acid Derivatives. Bioorg. Med. Chem. Lett. 2013, 23, 2543–2547. [Google Scholar] [CrossRef]
- He, Y.F.; Nan, M.L.; Sun, J.M.; Meng, Z.J.; Yue, F.G.; Zhao, Q.C.; Yang, X.H.; Wang, H. Synthesis, Characterization and Cytotoxicity of New Rotundic Acid Derivatives. Molecules 2012, 17, 1278–1291. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.M.; Hung, Y.C.; Hu, L.H.; Lee, Y.J.; Yin, M.C. Anti-Diabetic Effects of Madecassic Acid and Rotundic Acid. Nutrients 2015, 7, 10065–10075. [Google Scholar] [CrossRef] [PubMed]
- Roy, G.; Guan, S.; Liu, H.X.; Zhang, L. Rotundic Acid Induces DNA Damage and Cell Death in Hepatocellular Carcinoma Through AKT/MTOR and MAPK Pathways. Front. Oncol. 2019, 9, 545. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, Q.; He, Y.; Nan, M. Use of Rotundic Acid in Preparing Medicine, e.g., Tablet for Regulating Blood Fat. CN101849950-A, 6 October 2010. [Google Scholar]
- Li, G.; Zhao, Q.; He, Y.; Nan, M.; Wang, L.; Zhao, Y. Preparation of Medicine Composition, e.g., for Preventing and Treating Alcoholic Liver Injury, by Crushing Cortex Ilicis Rotundae, Extracting with Ethanol, Filtering, Passing through Processed Macroporous Resin Column, Eluting, and Drying. CN102058632-A, 30 May 2012. [Google Scholar]
- Liu, H.J.; Cao, S.T.; Wen, B.Y.; Han, X.; Li, Y.; Li, S.; Li, J.; Zhang, L. Rotundic Acid Ameliorates Non-Alcoholic Steatohepatitis via SREBP-1c/SCD1 Signaling Pathway and Modulating Gut Microbiota. Int. Immunopharmacol. 2021, 99, 108065. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Zou, Y.K.; Jia, B.J.; Wu, L.; Yang, Z.C.; Yuan, F.; Zhang, L. Pharmacokinetic and Metabolic Studies of Vortioxetine in Rats Using Ultra High Performance Liquid Chromatography with Tandem Mass Spectrometry. J. Sep. Sci. 2018, 41, 4469–4479. [Google Scholar] [CrossRef] [PubMed]
- Song, G.S.; Jin, M.M.; Du, Y.F.; Cao, L.; Xu, H.J. UPLC-QTOF-MS/MS Based Screening and Identification of the Metabolites in Rat Bile after Oral Administration of Imperatorin. J. Chromatogr. B-Anal. Technol. Biomed. Life Sci. 2016, 1022, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, B.; Cao, D.; Zhou, L.; Wang, Q.; Rong, L.; Zhou, X.H.; Jin, J.; Zhao, Z.X. Identification of Rotundic Acid Metabolites after Oral Administration to Rats and Comparison with the Biotransformation by Syncephalastrum Racemosum AS 3.264. J. Pharm. Biomed. Anal. 2018, 150, 406–412. [Google Scholar] [CrossRef]
- Shang, H.H.; Wang, Z.; Ma, H.; Sun, Y.H.; Ci, X.Y.; Gu, Y.; Liu, C.X.; Si, D.Y. Influence of Verapamil on the Pharmacokinetics of Rotundic Acid in Rats and Its Potential Mechanism. Pharm. Biol. 2021, 59, 200–208. [Google Scholar] [CrossRef]
- Shang, H.H.; Dai, X.H.; Li, M.; Kai, Y.Y.; Liu, Z.R.; Wang, M.; Li, Q.S.; Gu, Y.; Liu, C.X.; Si, D.Y. Absolute Bioavailability, Dose Proportionality, and Tissue Distribution of Rotundic Acid in Rats Based on Validated LC-QqQ-MS/MS Method. J. Pharm. Anal. 2022, 12, 278–286. [Google Scholar] [CrossRef]
- Yang, Z.; Zhu, M.Z.; Zhang, Y.B.; Wen, B.B.; An, H.M.; Ou, X.C.; Xiong, Y.F.; Lin, H.Y.; Liu, Z.H.; Huang, J.A. Coadministration of Epigallocatechin-3-Gallate (EGCG) and Caffeine in Low Dose Ameliorates Obesity and Nonalcoholic Fatty Liver Disease in Obese Rats. Phyther. Res. 2019, 33, 1019–1026. [Google Scholar] [CrossRef]
- Zhao, H.; Luo, Y.E.; Zhang, Y.A.; Chen, X.X.; Wang, H.; Guo, D.Y.; Wu, Z.X. Effects of Bacillus Subtilis on Hepatic Lipid Metabolism and Oxidative Stress Response in Grass Carp (Ctenopharyngodon Idellus) Fed a High-Fat Diet. Mar. Life Sci. Technol. 2020, 2, 50–59. [Google Scholar] [CrossRef]
- Yang, Q.C.; Wu, W.H.; Han, F.M.; Chen, Y. Identification of In-Vivo and in-Vitro Metabolites of Palmatine by Liquid Chromatography-Tandem Mass Spectrometry. J. Pharm. Pharmacol. 2009, 61, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.B.; Bai, L.; Li, X.R.; Xiong, J.; Xu, P.X.; Xue, M. Structural Analysis of Metabolites of Asiatic Acid and Its Analogue Madecassic Acid in Zebrafish Using LC/IT-MSn. Molecules 2015, 20, 3001–3019. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Li, H.; Ruan, Q.F.; Tong, Y.; Liu, Z.Q.; Xuan, S.X.; Jin, J.; Zhao, Z.X. Rapid Profiling and Pharmacokinetic Studies of Multiple Potential Bioactive Triterpenoids in Rat Plasma Using UPLC/Q-TOF-MS/MS after Oral Administration of Ilicis Rotundae Cortex Extract. Fitoterapia 2018, 129, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Li, H.; Ruan, Q.F.; Xue, Y.Y.; Cao, D.; Zhou, X.H.; Jiang, S.Q.; Yi, T.; Jin, J.; Zhao, Z.X. A Facile and Selective Approach to the Qualitative and Quantitative Analysis of Triterpenoids and Phenylpropanoids by UPLC/Q-TOF-MS/MS for the Quality Control of Ilex Rotunda. J. Pharm. Biomed. Anal. 2018, 157, 44–58. [Google Scholar] [CrossRef]
- Cao, D.; Wang, Q.; Jin, J.; Qiu, M.S.; Zhou, L.; Zhou, X.H.; Li, H.; Zhao, Z.X. Simultaneous Qualitative and Quantitative Analyses of Triterpenoids in Ilex Pubescens by Ultra-High-Performance Liquid Chromatography Coupled with Quadrupole Time-of-Flight Mass Spectrometry. Phytochem. Anal. 2018, 29, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Z.; Jiang, Y.M.; Wu, J.J.; Zheng, C.Q.; Ran, X.R.; Li, D.S.; Huang, M.; Bi, H.C. Metabolic Mapping of Schisandra Sphenanthera Extract and Its Active Lignans Using a Metabolomic Approach Based on Ultra High Performance Liquid Chromatography with High-Resolution Mass Spectrometry. J. Sep. Sci. 2017, 40, 574–586. [Google Scholar] [CrossRef]
- Cobbina, E.; Akhlaghi, F. Non-Alcoholic Fatty Liver Disease (NAFLD)—Pathogenesis, Classification, and Effect on Drug Metabolizing Enzymes and Transporters. Drug Metab. Rev. 2017, 49, 197–211. [Google Scholar] [CrossRef]
- Xie, C.C.; Halegoua-DeMarzio, D. Role of Probiotics in Non-Alcoholic Fatty Liver Disease: Does Gut Microbiota Matter? Nutrients 2019, 11, 2837. [Google Scholar] [CrossRef]
- Ji, Y.; Yin, Y.; Li, Z.R.; Zhang, W.Z. Gut Microbiota-Derived Components and Metabolites in the Progression of Non-Alcoholic Fatty Liver Disease (NAFLD). Nutrients 2019, 11, 1712. [Google Scholar] [CrossRef] [PubMed]
- Aron-Wisnewsky, J.; Vigliotti, C.; Witjes, J.; Le, P.; Holleboom, A.G.; Verheij, J.; Nieuwdorp, M.; Clement, K. Gut Microbiota and Human NAFLD: Disentangling Microbial Signatures from Metabolic Disorders. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 279–297. [Google Scholar] [CrossRef] [PubMed]
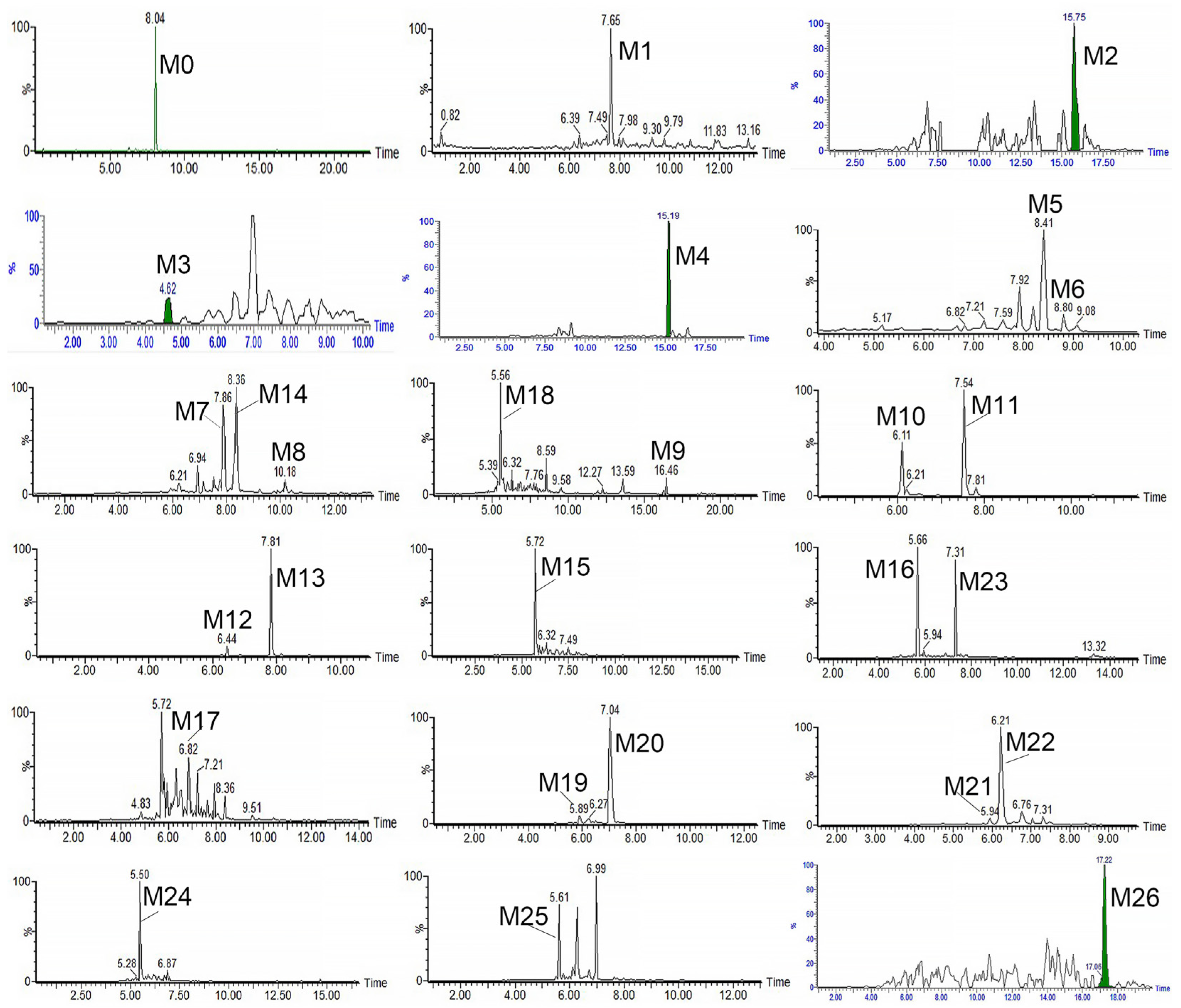
| NO. | tR (Min) | [M-H]− | Formula | Metabolite Description | MS/MS Fragment | Normal Rat | NAFLD Rat | LM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cal. | Exp. | F | P | U | L | F | P | U | L | ||||||
| 0 | 8.04 | 487.3423 | 487.3420 | C30H48O5 | Prototype | 469.3310, 455.2479, 437.3031, 423.3235, 405.3140, 393.3111, 391.2842 | + | + | + | + | + | + | + | + | + |
| 1 | 7.65 | 473.3267 | 473.3260 | C29H46O5 | Demethylation | 455.2859, 423.2159, 409.2301, 405.2641, 391.2842 | + | + | + | + | + | + | + | − | + |
| 2 | 15.75 | 445.3317 | 445.3348 | C28H46O4 | Demethylation + Decarbonylation | 427.35, 395.35, 391.34 | − | − | − | − | − | − | + | + | − |
| 3 | 4.62 | 649.3587 | 649.3563 | C35H54O11 | Demethylation + Glucuronide conjugation | 631.21, 599.20, 423.20 | − | − | − | − | + | + | + | − | − |
| 4 | 15.19 | 621.3638 | 621.3605 | C34H54O10 | Demethylation + Decarboxylation + Glucuronide conjugation | 603.43, 553.31, 445.39, 395.34 | − | − | − | − | − | + | − | + | − |
| 5 | 8.41 | 485.3267 | 485.3317 | C30H46O5 | Dehydrogenation | 467.3573, 455.3588, 437.3543, 411.3806, 389.3309 | + | + | + | + | + | − | + | + | − |
| 6 | 8.8 | 485.3267 | 485.3273 | C30H46O5 | Dehydrogenation | 467.3567, 405.3729, 389.3342 | + | + | − | − | + | + | + | − | + |
| 7 | 7.86 | 499.3423 | 499.3423 | C31H48O5 | Desaturation + Methylation | 485.3569, 481.3273, 469.3383, 419.3471, 437.3595 | + | + | + | − | − | − | − | − | − |
| 8 | 10.18 | 499.3423 | 499.3358 | C31H48O5 | Desaturation + Methylation | 481.3222, 439.3662 | + | − | − | + | + | − | − | + | − |
| 9 | 16.46 | 661.3587 | 661.3616 | C36H54O11 | Desaturation + Glucuronide conjugation | 643.39, 581.44, 409.24 | + | + | − | + | + | + | + | + | − |
| 10 | 6.11 | 503.3372 | 503.3366 | C30H48O6 | Hydroxylation | 485.3713, 453.3587, 407.3573, 391.3818 | + | − | + | − | + | + | − | + | + |
| 11 | 7.54 | 503.3372 | 503.3387 | C30H48O6 | Hydroxylation | 485.3718, 471.3510, 453.3677, 407.3552, 391.3692 | + | + | + | + | − | − | − | − | + |
| 12 | 6.44 | 501.3216 | 501.3211 | C30H46O6 | Hydroxylation + desaturation | 483.3675, 439.3691, 451.2440, 405.3373 | + | + | − | − | + | − | + | − | + |
| 13 | 7.81 | 501.3216 | 501.3580 | C30H46O6 | Hydroxylation + desaturation | 483.3567, 469.3797, 439.3788, 421.3737, 405.3845 | + | + | + | + | + | − | + | − | + |
| 14 | 8.36 | 499.3059 | 499.3107 | C30H44O6 | Hydroxylation + desaturation + Desaturation | 481.3325, 457.2943, 449.3637 | + | − | + | + | + | − | − | − | − |
| 15 | 5.72 | 489.3216 | 489.3261 | C29H46O6 | Hydroxylation + Demethylation | 471.3112, 439.2938, 421.2827 | + | − | + | + | + | + | + | + | + |
| 16 | 5.66 | 519.3321 | 519.3369 | C30H48O7 | 2 x Hydroxylation | 501.2282, 469.2262, 423.3068 | − | − | − | − | + | − | − | + | − |
| 17 | 6.82 | 489.358 | 489.3506 | C30H50O5 | Reduction | 471.2795, 407.3333 | + | − | − | + | − | − | − | − | + |
| 18 | 5.56 | 505.3529 | 505.3398 | C30H50O6 | Reduction + Hydroxylation | 487.3348, 423.3238, 391.2398 | + | − | + | − | − | − | − | − | − |
| 19 | 5.89 | 567.2991 | 567.2737 | C30H48O8S | Sulfate conjugation | 549.3577, 503.2623, 489.3547, 485.2561 | + | − | − | − | − | − | − | − | − |
| 20 | 7.04 | 567.2991 | 567.3094 | C30H48O8S | Sulfate conjugation | 523.3328, 487.3748, 453.2393, 407.3369 | + | + | + | − | + | − | − | + | − |
| 21 | 5.94 | 553.2835 | 553.3008 | C29H46O8S | Demethylation + Sulfate conjugation | 485.2623, 489.3580, 473.2652, 429.2460, 379.2833 | + | + | + | + | − | − | − | + | − |
| 22 | 6.21 | 553.2835 | 553.2967 | C29H46O8S | Demethylation + Sulfate conjugation | 503.3317, 489.3503, 485.2567, 473.3644, 429.2452, 379.3122 | + | + | − | + | − | + | − | + | − |
| 23 | 7.31 | 565.2835 | 565.2891 | C30H46O8S | Desaturation + Sulfate conjugation | 515.3238, 501.3277, 485.3311, 439.3272, 421.3105 | + | − | − | + | + | + | + | − | − |
| 24 | 5.5 | 583.294 | 583.2958 | C30H48O9S | Hydroxylation + Sulfate conjugation | 565.2947, 533.3004, 467.2841, 439.3002 | + | − | + | + | + | − | + | − | − |
| 25 | 5.61 | 581.2784 | 581.2805 | C30H46O9S | Hydroxylation + desaturation + Sulfate conjugation | 563.2995, 517.2929, 501.2476, 487.2725, 467.2502 | + | − | + | − | − | − | − | − | − |
| 26 | 17.22 | 663.3744 | 663.3741 | C36H56O11 | Glucuronide conjugation | 549.22, 487.22, 437.31 | + | + | + | − | + | − | + | + | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Xing, L.; Zou, Y.; Wang, Z.; Gou, Y.; Zhang, L.; Guan, S. UPLC-QTOF-MS Based Comparison of Rotundic Acid Metabolic Profiles in Normal and NAFLD Rats. Metabolites 2023, 13, 38. https://doi.org/10.3390/metabo13010038
Wu L, Xing L, Zou Y, Wang Z, Gou Y, Zhang L, Guan S. UPLC-QTOF-MS Based Comparison of Rotundic Acid Metabolic Profiles in Normal and NAFLD Rats. Metabolites. 2023; 13(1):38. https://doi.org/10.3390/metabo13010038
Chicago/Turabian StyleWu, Lvying, Lei Xing, Yake Zou, Zichen Wang, Yuanyuan Gou, Lei Zhang, and Su Guan. 2023. "UPLC-QTOF-MS Based Comparison of Rotundic Acid Metabolic Profiles in Normal and NAFLD Rats" Metabolites 13, no. 1: 38. https://doi.org/10.3390/metabo13010038
APA StyleWu, L., Xing, L., Zou, Y., Wang, Z., Gou, Y., Zhang, L., & Guan, S. (2023). UPLC-QTOF-MS Based Comparison of Rotundic Acid Metabolic Profiles in Normal and NAFLD Rats. Metabolites, 13(1), 38. https://doi.org/10.3390/metabo13010038





