Abstract
Although the inclusion of polyunsaturated fatty acids (PUFAs) in ruminants’ diets appears to be a well-documented strategy to enrich milk with PUFAs, several gene networks that regulate milk synthesis and mammary gland homeostasis could be impaired. The objective of this literature review is to assess the effects of nutritional strategies focused on enriching milk with PUFAs on gene networks regulating mammary gland function and lipogenesis, as well as the impact of feed additives and bioactive compounds with prominent antioxidant potential on immune-oxidative transcriptional profiling, as a part of mammary gland homeostasis and health. The findings support the conclusion that PUFAs’ inclusion in ruminants’ diets more strongly downregulate the stearoyl-CoA desaturase (SCD) gene compared to other key genes involved in de novo fatty acid synthesis in the mammary gland. Additionally, it was revealed that seed oils rich in linoleic and linolenic acids have no such strong impact on networks that regulate lipogenic homeostasis compared to marine oils rich in eicosapentaenoic and docosahexaenoic acids. Furthermore, ample evidence supports that cows and sheep are more prone to the suppression of lipogenesis pathways compared to goats under the impact of dietary marine PUFAs. On the other hand, the inclusion of feed additives and bioactive compounds with prominent antioxidant potential in ruminants’ diets can strengthen mammary gland immune-oxidative status. Considering that PUFA’s high propensity to oxidation can induce a cascade of pro-oxidant incidences, the simultaneous supplementation of antioxidant compounds and especially polyphenols may alleviate any side effects caused by PUFA overload in the mammary gland. In conclusion, future studies should deeply investigate the effects of PUFAs on mammary gland gene networks in an effort to holistically understand their impact on both milk fat depression syndrome and homeostatic disturbance.
Keywords:
antioxidant; immune system; gene networks; fat synthesis; marine lipids; milk; plant-derived lipids; PUFAs 1. Introduction
The fast pace of everyday life has rapidly shifted nutritional habits to foods with a high content of saturated fatty acids (SFAs) or sugars, thus increasing the risk for cardiovascular diseases [1], diabetes [2], and obesity [3]. Conversely, the influence of nutrition on health is summarized by Hippocrates’ quote “leave the drugs on the chemists’ pot, if you can heal a patient with food” [4]. In other words, the established idea that nutrition should just nourish people needs to be substituted with a more holistic approach that is not just focused on covering needs. Nowadays, nutraceuticals and functional foods with a high content of bioactive compounds, such as omega-3 polyunsaturated fatty acids (PUFAs) and antioxidants, have received considerable attention from both consumers and the industry.
In most developed countries, milk and dairy products are the major source of SFAs in the human diet. For this reason, there has been considerable interest in decreasing the proportions of SFAs in milk fat as a means of meeting national targets without requiring substantive changes in consumers’ eating habits. Several reviews have summarized the potential of dietary strategies on modifying the milk fat composition of lactating ruminants [5,6,7]. Amongst these strategies, dietary supplementation with unconventional feedstuffs rich in unsaturated fatty acids (UFA), such as soybean oil, linseed oil, sunflower oil, safflower oil, rapeseed, canola seeds, fish oil, and microalgae, have been well-documented in dairy ruminants, and their impact on milk fortification with PUFAs is now clear. However, scarce information exists about their impact on several other metabolic and cellular functions related to milk synthesis and the organisms’ homeostasis. In this context, a meta-transcriptomic analysis of the bovine mammary gland unveiled that more than 1000 genes are differentially expressed when Holstein cows are fed 5% linseed, and it argued that PUFA-rich diets downregulated genes involved in fatty acid/lipid synthesis and lipid metabolism pathways [8]. On the other hand, high levels of PUFAs in ruminants’ diet can also cause some detrimental effects, such as oxidative stress, and consequently, homeostatic imbalances in the mammary gland due to the high propensity of PUFAs to nonenzymatic and enzymatic oxidation [9]. Considering the aforementioned fact, it is acknowledged as being of utmost importance to investigate the effect of PUFA supplementation in ruminants’ diets on mammary gland gene networks related to milk synthesis and udder homeostasis, aiming to develop targeted and tailor-made strategies to produce functional dairy products fortified with PUFAs.
The advent of advanced lab techniques and software tools initiated the -omics technologies, such that Nutrigenomics, as a sub-field of nutritional study, refers to the examination of food and its constituents on the regulation of gene expression [10] and the subsequent effects on the metabolic level [11]. Understanding the interactions between a genome and nutrients may potentially promote customized diets for specific needs [12], such as the development of functional dairy products.
In this review, the intention is not to present the established knowledge of the field of Nutrigenomics in its entirety, but rather to place an emphasis on the mammary gland’s gene expression as related to fat synthesis under the implementation of dietary strategies focusing on the development of functional dairy products enriched with PUFAs and the potential of several dietary bioactive compounds to alleviate the oxidative balance and the homeostasis of the mammary gland.
2. The Mammary Gland: Gene Network and Milk Fat Synthesis
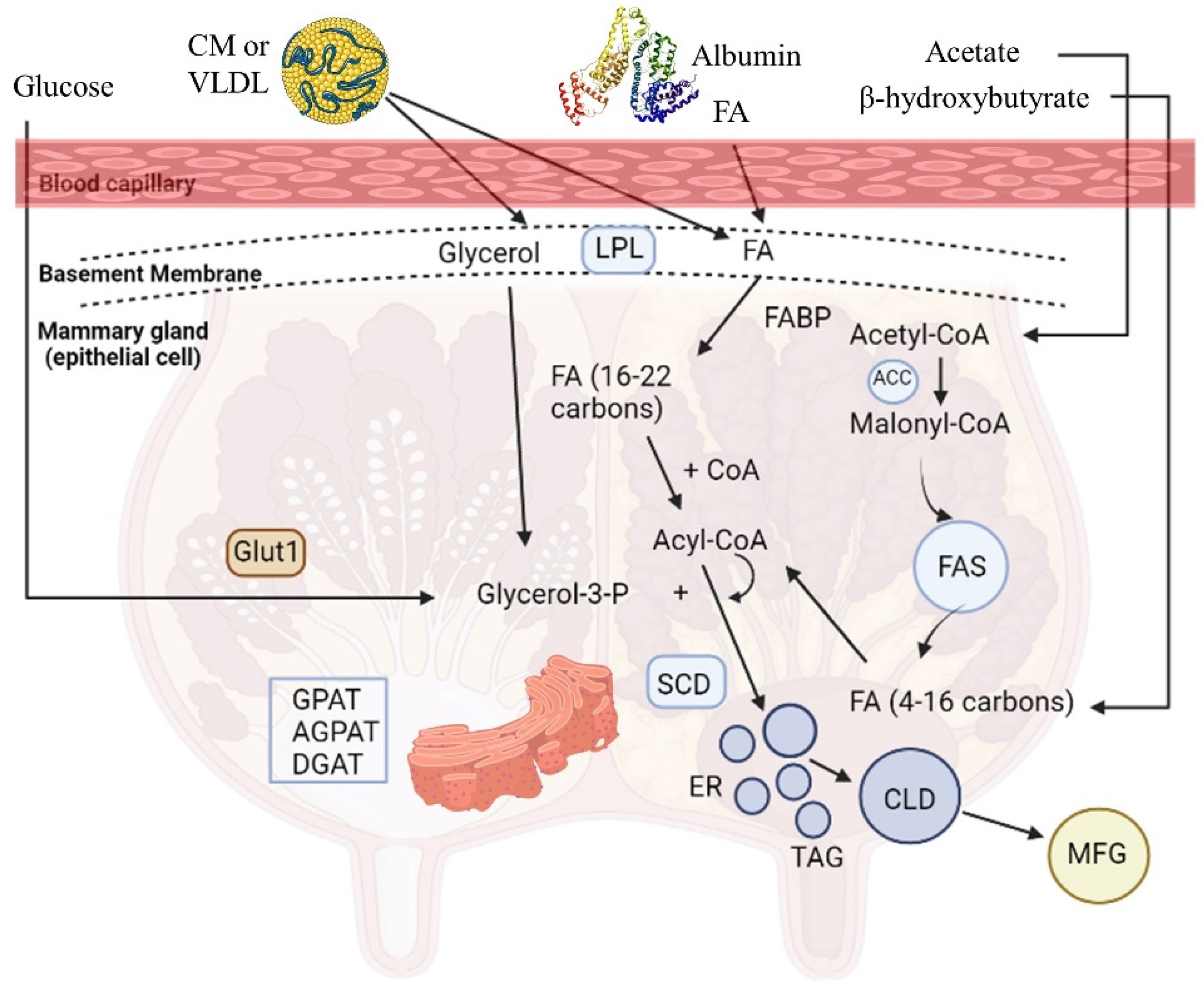
Milk is a highly nutritious product, crucial for the growth and health of neonates, which is synthesized by the mammary epithelial cells (MECs) and secreted by the mammary glands of mammals. The mammary gland synthesizes a variety of milk components, including proteins (casein 80% and whey protein 20%), carbohydrates (mainly lactose), coated lipid droplets, water, and ions [13]. Milk fat triglycerides (TAGs; 98% of total milk lipids) contain SFAs (C4:0-C18:0), palmitoleic (cis-9-16:1), oleic (cis-9-18:1), trans-18:1, and linoleic acid (cis-9, cis-12-18:2) [14]. Milk TAGs have a dual origin; one source is the de novo synthesis of fatty acids (FAs) produced by the MECs of the mammary gland through acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS) from circulating acetate and β-hydroxybutyrate (BHBA), a process which results in the formation of short- (SCFAs) and medium- (MCFAs) chain FAs (C4:0 to C16:0) that constitute almost half of the percentage of the milk FAs (Figure 1). The other source is FAs taken up by the mammary gland from blood circulation coming from the intestinal absorption of dietary and microbial FAs. Particularly, the long-chain FAs (LCFAs) (≥C18:0), along with 50% of C16:0 and a small proportion of C14:0, derive from blood plasma circulation released by lipoprotein lipase (LPL) from TAGs circulating in chylomicrons (CM) or very low-density lipoprotein (VLDL), or from the plasma NEFA bound to albumin [15] (Figure 1). Then, FAs are carried into the cells by FA translocase (CD36), FA transport protein (FATP), and FA binding protein (FABP) relative to the acyl-CoA synthetase or free FA receptors (FFARs). Subsequently, FAs are bound to FA binding proteins (e.g., FABP4) and subjected to different outcomes, including oxidation in mitochondria or esterification and storage in lipid droplets [8]. As for esterification, MCFAs are not esterified in cells which have an improper number of TAGs, while the storage of LCFAs does not have such a requirement [16].
De novo lipogenesis appears to be an important topic when the development of functional dairy products is concerned since there is ample evidence showing that PUFA supplementation in ruminants’ diets can severely downregulate these gene networks [8]. The three key enzymes that affect the de novo synthesis pathway of FAs are acyl-CoA synthetase short-chain family member 2 (ACSS2), ACC, and FAS. First, ACSS2 is the activator of acetic acid and BHBA in MECs, which are transformed to acetyl-CoA after connecting to pyruvate. The ACC enzyme carboxylates acetyl-CoA for the formation of malonyl-CoA, MCFAs, or LCFAs by FAS action in the cytosol [17] (Figure 1). Other enzymes, such as ATP-citrate lyase (ACL), FA elongase (ELOVL), and stearoyl-CoA desaturase (SCD), have a significant role in de novo synthesis [8]. ELOVL and SCD cooperate to synthesize MUFAs or PUFAs. FA desaturase enzymes (e.g., SCD) catalyze the conversion of SFAs to MUFAs or PUFAs, while FA elongases (ELOVL1 to ELOVL7) the inhibiting step of the LCFA elongation cycle in ruminants. SCD (Figure 1) is an enzyme from the endoplasmic reticulum (ER) enzyme, called a Δ9-desaturase because of a cis-9 double bond on the fatty acyl-CoA substrates, mostly from C14:0 to C19:0 [18]. SCD mostly uses stearic acid (C18:0) as a substrate for cis-9-18:1 synthesis, the main UFA found in milk triacylglycerol, implicated in the regulation of milk fat globules’ fluidity in the mammary gland [19]. Also, SCD can convert trans-vaccenic acid (C18:1 trans-11), an intermediate of biohydrogenation, to CLA cis-9, trans 11 [20].
As mentioned previously, the operation of TAGs is underscored to fully explain the mechanism of TAG synthesis in the smooth ER (Figure 1) and the coordination via subsequent reactions by the products of glycerol-3-phosphate acyltransferase (GPAT), lipin 1 (LPIN1), and diacylglycerol acyltransferase 1 (DGAT1) genes. The first step is the acylation of glycerol-3-phosphate (glycerol-3-P) for lysophosphatidic acid (LPA) formation, catalyzed by the GPAT enzyme. LPA acyltransferase (AGPAT) transfers a FA to LPA in order to produce phosphatidate (PA). The PA is converted to diacylglycerol (DAG) by the lipin, an ER enzyme, and lastly, from DAG to TAG by the DGAT enzyme [15,21]. TAGs in small lipid droplet formation are enveloped by the ER plasma membrane and escape from the MECs in larger lipid droplets through the apical membrane into the alveoli lumen and mammary gland duct, and they dissociate from the cell (Figure 1).
The detection of transcription factors (TFs) that perceive the presence of lipids, notably FA, along with the comprehension of FA interference in several pathways and functions in cells, has shed light on the biological roles of FAs [22]. Overall data suggests that SCD is a key gene for milk fat synthesis, as it is the one with the highest gene expression, while the TF of sterol regulatory element binding transcription factor 1 (SREBF1), as well as the collaborative action of proliferator activated receptor γ (PPARγ), PPARγ coactivator 1 alpha (PPARγC1A), and insulin-induced gene 1 (INSIG1), have a crucial role in this process [23]. Milk fat synthesis in ruminant MECs is summarized in Figure 1, while the key genes involved in this process are listed in Table 1.

Table 1.
Key genes involved in milk fat synthesis in ruminants [17].
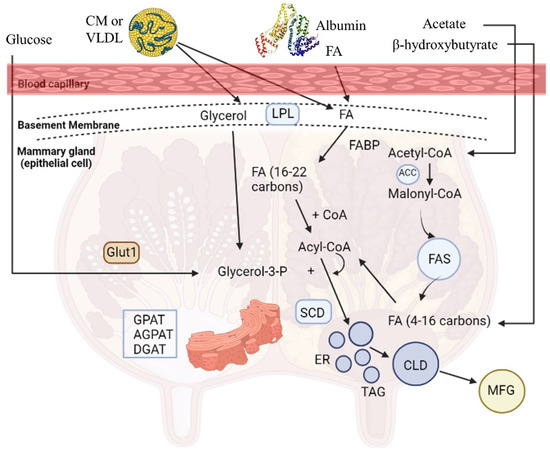
Figure 1.
Milk fat synthesis in the ruminant mammary epithelial cell [15]. Abbreviations: ACC = acetyl-CoA carboxylase; AGPAT = acyl glycerol phosphate acyl transferase; CD36 = fatty acid transferase (CD36); CLD = cytoplasmic lipid droplet; CoA = coenzyme A; CM = chylomicron; DGAT = diacylglycerol acyltransferase; ER = endoplasmic reticulum; FA = fatty acid; FABP = fatty acid binding protein; FAS = fatty acid synthase; Glut1 = glucose transporter1; GPAT = glycerol-3 phosphate acyltransferase; LPL = lipoprotein lipase; MFG = milk fat globule; SCD = stearoyl-CoA desaturase; TAG = triglyceride; VLDL = very-low-density lipoprotein.
Figure 1.
Milk fat synthesis in the ruminant mammary epithelial cell [15]. Abbreviations: ACC = acetyl-CoA carboxylase; AGPAT = acyl glycerol phosphate acyl transferase; CD36 = fatty acid transferase (CD36); CLD = cytoplasmic lipid droplet; CoA = coenzyme A; CM = chylomicron; DGAT = diacylglycerol acyltransferase; ER = endoplasmic reticulum; FA = fatty acid; FABP = fatty acid binding protein; FAS = fatty acid synthase; Glut1 = glucose transporter1; GPAT = glycerol-3 phosphate acyltransferase; LPL = lipoprotein lipase; MFG = milk fat globule; SCD = stearoyl-CoA desaturase; TAG = triglyceride; VLDL = very-low-density lipoprotein.
3. PUFA Dietary Supplementation Modifies a Complex Gene Network in Ruminants’ Mammary Glands
PUFA inclusion in ruminants’ diets appears to be an effective strategy for improving dairy products’ nutritional quality since beneficial milk FAs, namely cis-9-18:1, vaccenic, conjugated linoleic acid (CLA), and α-linolenic acid (ALA), can be increased [24], and they are associated with the prevention or decrease of the risk of chronic diseases in humans [25,26]. There is a noteworthy suggestion that UFA supplementation in ruminants’ diets downregulates mRNA abundance of ACC, FAS, and SCD1, as well as SREBF1 and PPARγ [27,28]. In this section, a summary of studies concerning the effect of PUFA supplementation in ruminants’ diet in key genes’ expression in the mammary gland, which participate in de novo lipogenesis (FAS, ACC, ACSS, and ACSL), FA uptake (LPL, CD36), FA desaturation (SCD1, FADS), long-chain FA elongation (ELOVL5 and ELOVL6) and transport (FABP3 and FABP4), TAG synthesis (GPAT, AGPAT, DGAT1, LPIN1, and GPAM), as well as transcriptional regulation (SREBF1, PPARγ, and INSIG1) and lipid droplet formation (BTN1A1 and XDH), are presented (see Table 2). The PUFA dietary treatments cited subsequently focus on the inclusion of fish oil, rich in EPA (eicosapentaenoic acid, 20:5 n-3) and DHA (docosahexaenoic acid, 22:6 n-3), plant-derived oils, and seeds such as soybean, sunflower, safflower (Carthamus tinctorius L.), rapeseed, canola, and linseed, rich in linoleic acid or α-linolenic acid, respectively, as well as the infusion of trans-10, cis-12 CLA in cows, ewes, and goats (Table 2).
Beginning with fish oil, its supplementation in dairy cows’ diets downregulated the expression of SCD1, FAS, ACC, and LPL [29]. On the contrary, in another study, the inclusion of fish oil or DHA-rich microalgae in dairy cows’ diet reduced SREBF1 expression in the mammary tissue, but this reduction was inadequate to substantially alter the gene expression of enzymes implicated in milk fat synthesis [30]. Vargas-Bello-Pérez et al. [31] investigated the inclusion of fish and soybean oil in dairy cows’ diets and found that fish oil reduced the expression of ACC, PPARγC1, LPIN1, and FABP3 in the mammary gland, while soybean oil downregulated ACC, INSIG1, DGAT1, and LPIN1. Other authors found that the supplementation with fish and soybean oil [32], as well as with marine DHA-rich algae, along with sunflower and linseed oil [33], downregulated the expression of SREBF1, SCD1, FAS, and LPL in the bovine mammary gland. In dairy ewes, fish oil supplementation tended (p < 0.10) to reduce the mRNA abundance of ACSS2, FAS, LPIN1, FADS2, and INSIG1, while a downregulation observed in ACC and ACSS1 genes’ expression was not significant [34]. Moreover, the expression of ACC, ACSL1, ACSS1, ACSS2, ELOVL6, FADS2, AGPAT2, and LPIN1 genes was reduced in the ovine mammary gland under fish oil-induced milk fat depression (MFD) [35]. ACC, ACSS1, and AGPAT6, as well as SREBF1 gene expression, was also decreased by fish oil inclusion in ewes’ diet, whereas FABP3, LPL, SCD1, and INSIG1 expression tended to be decreased [36]. Furthermore, in dairy goats, the mRNA abundance tended to decrease for SCD1 (p < 0.10) in the mammary gland when they were fed with fish and linseed oil simultaneously, while SREBF1 and PPARγ were upregulated [37]. On the contrary, fish and linseed oil supplementation had no effect on FAS and G6PDH expression. A study examining the effect of fish oil inclusion in different goats’ tissues demonstrated a decline in the expression of FAS, SCD1, and FADS2 only in the liver, but not in the mammary or omental adipose tissue [38]. Another study also showed that the inclusion of fish and soybean oil in dairy cows’ diets had no significant difference in the expression of either SREBF1 or PPRAγ [39].
Summarizing the above results, it seems that, even though the inclusion of fish oils in ruminants’ diets is an effective strategy for increasing PUFA content in milk and therefore offering a functional product for consumers, a danger to the animals’ productivity is lurking as fish oils are associated with MFD syndrome. The development of MFD seems to be related to the downregulation of key genes involved in mammary glands’ lipogenesis, notably in de novo synthesis; a reduced expression of these genes was observed in most studies. In addition, the animal species per se may constitute a key factor affecting gene expression in the mammary gland, as goats, compared to cows and ewes, were less affected by fish oil supplementation in their diet.
Apart from fish oil, other studies exclusively examined the effect of plant-derived oils and seeds in key genes’ expression in the mammary gland, and they mostly found a tendency for reduction in the mRNA abundance of those genes (Table 2). Specifically, the inclusion of sunflower oil in dairy cow diets reduced ACC, FAS, GPAT, and AGPAT mRNA abundance, while SCD1 and LPL tended to be decreased (p < 0.08) [40]. Regarding dairy goats, supplementation with sunflower oil or linseed oil following hay-based diets caused an inhibition in mammary SCD1 and LPL activity [41]. Additionally, inclusion of linseed or safflower oil downregulated the expression of SREBF1, FAS, and ACSS1 significantly in the bovine mammary gland, while the decrease observed in ACC and SCD1 was not statistically significant [42]. Lastly, the expression of SCD1 was significantly decreased by the oil supplementation in the dairy cows’ diet, especially by soybean oil compared with rapeseed or linseed oil [43], and this result was associated with lower desaturase indexes, used for mammary SCD1 activity, observed in milk. On the contrary, rapeseed and sunflower oil supplementation had no effect on the mRNA abundance of either LPL, ACC, FAS, SCD1, FABP3 and FABP4, and XDH [44,45] or GPAM, DGAT1, CD36, INSIG1 [45], and BTN1A1 [44] in bovine mammary tissue. The same pattern was observed in goats’ mammary tissue as the inclusion of sunflower oil and linseed oil in maize silage-based diets had no effect on the abundance of mRNA encoding for LPL, ACC, FAS, or SCD1 [46]. Neither were alterations observed in the mRNA abundance of ACC, FAS, LPL, or SCD1 in the mammary tissue of lactating beef cows under the effect of dietary linoleate–safflower seed oil [47]. On the contrary, formaldehyde-treated linseed and oleic sunflower oils in lactating goats’ diets decreased mammary SCD1 mRNA, but no notable change was observed for ACC or FAS [48]. Instead, this research indicated that mammary LPL mRNA increased with oleic sunflower oil supplementation, but the activity of G6PD was not affected by the lipid supplements [48]. Recently, a study showed that dietary inclusion of linseed reduced the SCD1, LPIN3, ELOVL5, and ELOVL6 genes’ regulation, while the mRNA abundance of ACC, FAS, DGAT1, SREBF1, and FADS1 did not change significantly [49]. In contrast, an increase in the transcripts of other genes implicated in TAG synthesis, apart from DGAT1, such as GPAT4, AGPAT1, AGPAT2, and AGPAT3, was observed. The authors suggested that the inclusion of linseed reduced the overall metabolic activity of fat and protein, in contrast to carbohydrate, which increased, and as a result, the pathways of glycolysis, gluconeogenesis, and pentose phosphate were upregulated. On the other hand, inclusion of canola seeds in dairy cows’ diets did not alter the SCD1 or ACC gene expression [50], nor did the addition of dietary long-chain FAs, provided by soybeans, have a significant effect on the mRNA abundance of ACC or FAS [51]. Lastly, supplementing goats’ diets with safflower or linseed oil increased the mRNA expression of SCD1, ACC, and LPL in the mammary tissue, but the mRNA expression of FAS was not affected by the oil supplementation [52].
To sum up, the effect of plant oil inclusion in ruminants’ diets seems to still be ambiguous as, in many studies, no significant effects were noted in the mRNA abundance of key genes in mammary tissue. It could be hypothesized that plant oils in comparison with fish oils are less capable of altering genes’ expression, and as a result, plant oils contribute to MFD to a lesser extent. This phenomenon could be attributed to the DHA and other very-long-chain PUFAs, such as EPA and DPA (docosapentaenoic acid, 22:5 n-3), found in fish and marine oils, which inhibit the rumen biohydrogenation of vaccenic acid (C18:1 trans-11) to C18:0, and consequently plenty of intermediates (such as trans-10, cis-12 CLA and trans-10 18:1) accumulate in the rumen and act as lipogenesis inhibitors in the mammary gland. It could be suggested that a combination of fish oils, for modifying the biohydrogenation process in the rumen, and plant oils, as a substrate for C18:1 trans-11 formation, in ruminants’ diets is a promising strategy for increasing cis-9, trans-11 CLA content in milk fat. However, there is a need for further investigation as the effect of plant oils depends on their composition and origin, as well as on the experiments’ duration (rumen microbiome adaptation).
Three studies examining the infusion of trans-10, cis-12 CLA in dairy cows indicated a reduced expression of SREBF1 and FAS [53,54,55], along with LPL [54], ACC [55], or SCD1 [53]. A similar pattern was also observed in trans-10, cis-12 CLA-induced MFD in lactating ewes, which resulted in FAS, ACC, ACSS2, ACSS3, and FADS2 downregulation [56]. Hussein et al. [57], also experimenting with CLA-induced MFD in lactating ewes, incorporated a lipid-encapsulated CLA supplement containing cis-9, trans-11 and trans-10, cis-12 CLA isomers, and they found that the mRNA abundance of ACC, FAS, LPL, SCD1, AGPAT6, SREBF1, and INSIG1 was reduced. Moreover, a comparative study of the inclusion of fish oil- or trans-10, cis-12 CLA in the mammary mRNA abundance of some lipogenic genes and TFs showed that fish oil downregulated the expression of ACSS2, FAS, and LPIN1, while ACC had no significant difference. Additionally, LPL and LPIN1 were lower with fish oil than with CLA, whereas PPARγ was higher in ewes receiving CLA. Furthermore, treatment with CLA upregulated GPAT4, while the abundance of FABP3, SCD1, GPAM, and SREBF1 did not variate [58]. Tsiplakou et al. [59] evaluated the key genes’ expression in sheep and goats’ mammary glands under two feeding strategies and found that the mRNA abundance of ACC and FAS was lower in sheep compared to goats when animals were fed on either a group or an individual basis. In contrast, LPL mRNA levels in the mammary gland did not vary between sheep and goats fed on a group basis. However, the mRNA level of SCD1 was higher in sheep compared to goats, independent of the way they received their diets (group or individual), linked to the higher concentration of cis-9 trans-11 CLA content in sheep milk fat compared with that of goats.
Overall, fish and plant oil supplementation in ruminants’ diets confirms the initial statement that PUFA supplementation in ruminants’ diet downregulates, in most studies, the expression of key genes involved in the pathways of de novo synthesis (e.g., ACC and FAS) and FA activation (e.g., ACSS1, ACSS2 and ACSL1), FA uptake and transport (e.g., LPL and FABP3), FA desaturation (e.g., SCD1 and FADS2), TAG synthesis (e.g., AGAPT6 and LPIN1), FA elongation (e.g., ELOVL6), as well as TFs (e.g., SREBF1, INSIG1, and PPARγ) involved in their regulation. However, according to the aforementioned literature, it seems that genes involved in de novo synthesis (e.g., FAS and ACC) have more plasticity compared to those implicated in other pathways, such as desaturation (e.g., SCD1). Additionally, the alterations of the former gene expressions in dairy cows’ mammary glands under the impact of such dietary strategies depicted higher consistencies compared with those observed in small ruminants. The TFs were also modified by PUFA supplementation in most cases. Generally, the synchronized downregulation in key gene expression, observed in many studies in dairy ruminants [60,61,62], encourages the idea that there is a central regulator, probably SREBF1 or PPARγ, of milk fat synthesis. SREBF1 downregulation is frequently detected during MFD induced by marine lipid supplementation [32,33,36,39] or CLA [55,57]. The reduction of SREBF1 mRNA abundance has shown an inhibition of the SREBP1 signaling pathway and its target genes [15,63], which partly explains the negative effects of dietary oil supplementation on many genes’ expression, including ACC, FAS, LPL, FABP3, and SCD1. PPARγ could partially regulate the expression of SREBF1, which in turn could affect the activity of PPARγ by increasing the production of its natural agonists [62]. The downregulation of SREBF1 is also correlated with INSIG1 reduction, as INSIG1 is known to be involved in SREBF1 activation to its mature form [64]. The capability of diet to alter the mRNA abundance of SREBF1 and INSIG1 has been previously reported by several researchers [32,36,57,65].
The intense inhibition of de novo synthesis and the resulting milk fat depression (MFD) observed in many cases could be attributed to the reduction of SCFAs as this reduction can reach up to 60% [29]. These effects can be attributed to the synthesis of specific rumen biohydrogenation intermediates associated with the ruminal trans-10 pathway that exerts antilipogenic effects in bovines [60]. The downregulation of the genes, implicated in de novo synthesis, has been suggested as decreasing the milk fat percentage. However, in the study of Ahnadi et al. [29] the milk fat content was significantly associated only with ACC, FAS, and SCD1 gene expression, but not with that of LPL. On the contrary, provided data in ruminants suggest that the feeding strategies which increase LCFA in the produced milk, e.g., the inclusion of plant oils [15,66], do not reduce the mRNA abundance of LPL. Besides, the downregulation of LPL has been related to a reduction in milk fat synthesis [32,33,36,57]. Concerning SCD1 gene expression, its nutritional regulation involves complex interactions between dietary factors and regulatory events which have not been intensively described [67]. Few studies have shown that SCD1 is downregulated when induced-MFD diets are provided to ruminants, such as those containing fish and plant oils [29,33,40], as well as CLA [53,55,57]. A possible explanation for that reduction might be the increases observed in EPA and trans-10, cis-12 CLA [67,68] and the reduction of cis-9 18:1 in milk, which is related to milk fat fluidity and to the negative effect of marine lipids on mammary lipogenesis [60,69,70]. However, in other studies, the inclusion of fish and soybean oils causing MFD is associated with a lack of alteration or even an increase in SCD1 mRNA levels [32,39,58,71]. Thus, there is a suggestion that these variations in SCD1 expression could be due to the nutritional regulation of its expression [67], depending on the origin and doses of the supplemented oils, as well as the period of adaptation to the treatments. Apart from SCD1 regulation, Conte et al. [49] showed that the inclusion of linseed resulted in downregulation of FADS1 and FADS2 expression related to the increase in milk PUFAs due to diet or ruminal activity (CLA) and the reduction in very-long-chain PUFAs (such as EPA and DHA). According to Mele et al. [72], these two genes (FADS1 and FADS2) engage in the synthesis of EPA and DHA by adding double bonds at the Δ5 and Δ6 positions.
Finally, among the genes involved in the last steps of milk fat synthesis, FA esterification to glycerol and the secretion of milk fat globules, DGAT1 abundance does not vary significantly during MFD [36,39,57,71], perhaps due to its known post-transcriptional regulation [15]. On the contrary, the downregulation of AGPAT6 observed in many studies shows that there is a kind of inhibition of de novo lipogenesis, probably due to its greater affinity for short- and medium-chain SFAs [15].

Table 2.
Selected studies presenting the impact of PUFA-rich diets and supplements on mammary gland gene network that regulate fat synthesis.
Table 2.
Selected studies presenting the impact of PUFA-rich diets and supplements on mammary gland gene network that regulate fat synthesis.
| Method | Diet Supplement | Level/Detail | Effect | Reference |
|---|---|---|---|---|
| Dairy cows | ||||
| In vivo | Fish oil | 3.7% of DM unprotected fish oil, or 1.5% or 3.0% of DM glutaraldehyde-protected microcapsules of fish oil |
| [29] |
| In vivo | Fish oil or DHA-rich microalgae | 200 g/d |
| [30] |
| In vivo | Soybean oil or fish oil | 2.9% of DM unrefined soybean oil or 2.9% DM of fish oil manufactured from salmon oil |
| [31] |
| In vivo | Short-term administration of trans-10, cis-12 CLA, and a low forage/high oil (LF/HO) diet | 10 g/d infusion of trans-10, cis-12 CLA and LF/HO diet, including 3.0% soybean oil and 1.5% fish oil |
| [32] |
| In vivo | Silage-based diets supplemented with palm fat, linseed oil plus DHA-rich algae, or sunflower oil plus DHA-rich algae | 3.1% of the basal diet DM of rumen-stable fractionated palm fat, a mixture of linseed oil (2.7% of the basal diet DM) plus DHA-rich algae (0.4% of the basal diet DM), or a mixture of sunflower oil (2.7% of the basal diet DM) plus DHA-rich algae (0.4% of the basal diet DM) |
| [33] |
| In vivo | Fish and soybean oil | 3.5% of DM (1% fish oil and 2.5% soybean oil) |
| [39] |
| In vivo | Sunflower oil | 1% of DM sunflower oil |
| [40] |
| In vivo | Linseed or safflower oil | 5% linseed or safflower oil on DM |
| [42] |
| In vivo | Soybean, rapeseed, or linseed oil | 2.7% soybean, rapeseed, or linseed oil on DM basis, or 2.7% of a 1:1:1 mixture of the 3 oils |
| [43] |
| In vivo | Rapeseed or sunflower oil | 130 g/d of oil from whole, intact rapeseeds in a HF diet (F:C = 64:36), or 130 g/d of sunflower oil in a LF diet (F:C = 43:57) |
| [44] |
| In vivo | Rapeseed or sunflower oil | 13.9% of DM whole, intact rapeseeds in a HF diet (F:C = 65:35) or 4% of DM sunflower oil in a LF diet (F:C = 46:54) |
| [45] |
| In vivo | Linoleate–safflower seed | low-fat control supplement (64.2% cracked corn, 32.1% safflower seed meal, and 3.7% liquid molasses) fed at 0.35% of BW daily (DM basis) or a cracked, high-linoleate safflower seed supplement (94.0% cracked, high-linoleate safflower seeds and 6% liquid molasses) at 0.23% of BW daily (DM basis) |
| [47] |
| In vivo | Canola seeds | 4.8% of DM canola meal, 3.3% of DM unprotected canola seeds plus 1.5% canola meal, or 4.8% of DM formaldehyde-protected canola seeds |
| [50] |
| In vivo | Infusion of trans-10, cis-12 CLA | 13.6 g/d |
| [53] |
| In vivo | Infusion of trans-10, cis-12 CLA | 7.5 g/d |
| [54] |
| In vivo | Infusion of trans-10, cis-12 CLA | 10 g/d |
| [55] |
| Dairy ewes | ||||
| In vivo | Fish oil | 20 g of fish oil/kg of DM |
| [34] |
| In vivo | Fish oil | Transcriptome analysis in milk somatic cell (MECs) of ewes suffering from fish oil-induced MFD |
| [35] |
| In vivo | Fish oil | 17 g of fish oil/kg of diet DM |
| [36] |
| In vivo | Linseed | 20% of DM linseed panel |
| [49] |
| In vivo | Trans-10, cis-12 CLA-induced MFD | 10 g of a rumen-protected CLA product/kg of diet DM |
| [56] |
| In vivo | Lipid-encapsulated CLA supplement containing cis-9, trans-11 and trans-10, cis-12 CLA isomers | 15 g/d |
| [57] |
| In vivo | Comparative study between the inclusion of fish oil- or trans-10, cis-12 CLA | 2.4% of DM fish oil or 1% of DM a commercial product rich in trans-10,cis-12 CLA |
| [58] |
| Dairy goats | ||||
| In vivo | Fish and linseed oil | 530 g/day of extruded linseeds (EL) or 340 g/day of extruded linseeds plus 39 g/day of fish oil (ELFO) | ELFO:
| [37] |
| In vivo | Fish oil | 90 g/day of sunflower oil and fish oil (2:1) plus additional starch |
| [38] |
| In vivo | Sunflower oil or linseed oil following hay-based diets | 55 g/kg diet DM sunflower oil or linseed oil |
| [41] |
| In vivo | Sunflower oil and linseed oil in maize silage-based diets | 6.1% of diet DM sunflower oil or 6.2% of diet DM linseed oil |
| [46] |
| In vivo | Formaldehyde-treated linseed or oleic sunflower oil | 11.2% of DM intake formaldehyde-treated linseed or 3.5% of DM intake oleic sunflower oil |
| [48] |
| In vivo | Soybeans | 22% of DM soybeans |
| [51] |
| In vivo | Safflower or linseed oil | 50 g/kg of TMR DM safflower oil or 50 g/kg of TMR DM linseed oil |
| [52] |
| Dairy ewes and goats | ||||
| In vivo | Two feeding strategies: group or individual basis | Basic diets |
| [59] |
↓ = decrease, ↑ = increase.
4. Bioactive Nutrients: An Ally of Mammary Gland Homeostasis
The inclusion of dietary FAs in the diet of ruminants, apart from the beneficial role in the development of dairy products enriched with bioactive molecules, such as omega-3 FAs (functional foods), can cause some negative effects, such as oxidative stress and consequently homeostatic imbalances on the mammary gland. Therefore, it is acknowledged as being of utmost importance to both examine the potential of various feed antioxidant components to counteract these effects and to comprehend the mechanisms underlying these features, will be valuable as a contribution to the alleviation of such phenomena.
Cells produce both ROS (reactive oxygen species) and RNS (reactive nitrogen species), as part of their physiological metabolism in order to regulate their homeostasis, as signaling molecules [73]. However, ROS overproduction can induce severe oxidative imbalance by impairing proteins, lipids, or even DNA, causing a situation broadly known as oxidative stress [73], which has a highly negative relation with the performance, health, and welfare of ruminants [74,75]. PUFAs are generally prone to non-enzymatic (autoxidation and photooxidation) and enzymatic oxidation [9]. During PUFA oxidation, various metabolites are generated which induce oxidative stress mainly by increasing the concentration of lipid peroxidation aldehydes, such as malondialdehyde (MDA), and activating superoxide anion generators, such as NADPH oxidases [9], affecting the cellular homeostasis of the mammary gland. The source of PUFAs, as well as their dietary inclusion levels, can influence the oxidative status of target tissues. Therefore, dietary supplementation of antioxidant compounds along with PUFA-enriched rations may have the potential to lessen the appearance of oxidative stress incidence and consequently mammary gland homeostasis. Indeed, the supply of antioxidant compounds, namely whole sesame seeds or sesame meal combined with vitamin E and selenium [76,77] delivered some promising results for minimizing oxidative stress along with inflammation in ruminants [78,79]. Apart from the contribution of antioxidant compounds in ruminants’ nutrition on the animal level, there is a greater need for fully understanding the changes and influence occurring on the molecular level as part of the expression of specific genes in the mammary gland.
4.1. NFE2L2 Factor and Its Significance in Antioxidant Cellular Defense
In whatever manner, the redox balance can be manipulated through nutrition by stimulating the expression of specific genes for antioxidant properties. A crucial mechanism for preventing the hazardous aftermath of oxidative stress is the NFE2L2 pathway, activated under stressful conditions, supplying oxidant compounds, or under inflammation [80]. The antioxidant defense and cellular response appear to be controlled by a set of TFs, including nuclear factor erythroid 2-like 2 (NFE2L2), nuclear factor-kappa B (NFKB), PPARs, and PPARγC1A. The Kelch-like epichlorohydrin-associated protein 1 (KEAP1)–NFE2L2–antioxidant response element (ARE) signaling pathway significantly preserves cellular homeostasis through the regulation of genes related to cellular redox balance [81]. The activation of NFE2L2 reflects on the complex formed by KEAP1 and CUL3 [82]. Concisely, under homeostatic conditions, KEAP1 attaches with NFE2L2, contributing to NFE2L2 degradation, and thus overcoming its activation [83]. Conversely, the overproduction of ROS results in the unbinding of KEAP1 and NFE2L2, subsequently leading to NFE2L2 activation and an increase in glutathione (GSH) production [84]. NFE2L2, as a TF, belongs to the leucine zipper family and can be activated by antioxidant elements located in the regulatory regions of many antioxidant genes [85]. After, NFE2L2 is released from KEAP1 and transferred to the nucleus, where it stimulates the transcription of genes associated with the antioxidant response [86,87]. More specifically, it appears to have a crucial role in the mammary gland antioxidant status, through regulating its mRNA abundance [88]. Additionally, in vitro studies of cultured MECs showed that, after the activation of the NFE2L2 pathway by several antioxidant compounds, the expression of antioxidant genes was stimulated [89]. However, not only nutrients [90] but also the existence of ROS can activate the NFE2L2 pathway [91]. Nonetheless, the target genes of NFE2L2, although well-established in monogastric animals, have not been fully confirmed in ruminants. Nevertheless, there is a need for more precise experimental approaches and new techniques to further understand the mechanism of this TF and its dynamic in the lessening of oxidative stress [92].
4.2. Nutrients Affect Homeostatic Mammary Gland Gene Networks
According to the accessed literature, plant compounds; AAs, such as methionine (Met) [93]; trace minerals and vitamins [79,94]; and plant bioactive compounds [95] seem to efficiently contribute to lessening the burden of oxidative stress (Table 3). Aside from that, the impaired antioxidant status can cause severe damage to the immune system; thus, nutrition research also targets the mechanisms regulating gene expression on the immune system, as has been shown by several studies that will be presented in the following section. To begin with, AAs, apart from contributing to protein synthesis, may effectively regulate various critical metabolic processes in the mammary gland [93]. So, the concept behind supplying animal diets with AAs with known antioxidant protection is to alleviate oxidative cellular damage [96,97], especially during the peripartum period. In addition, AAs can be metabolized by ruminants for creating sub-products, for instance, Met, to synthesize sulfur-containing antioxidants such as GSH [93]. Moreover, Arg has a crucial role in both innate and acquired immunity by modifying the activity, proliferation, and apoptosis of immune cells via the mTOR pathway [98]. In detail, Met enhanced antioxidant protection against heat stress in bovine MECs in vitro [99,100]. In another study by Han et al. [88], methionine upregulated the mRNA abundance of genes that control antioxidant mechanisms. More specifically, significant upregulations of NFE2L2 target genes such as GCLC, GCLM, GPX1, GSR, ME1, FECH, FTH1, and NQO1 were found. In contrast, the supply of Met had no effect on HMOX1, TXN, GSTM1, TALDO1, BCL2, or PIR. Additionally, the expression of NFE2L2, CUL3, NFKB1, and MAPK14 was upregulated. As a result, the research group indicated that Met has a significant contribution to alleviate oxidative stress by regulating GSH metabolism in MECs.
In addition to the well-known importance of trace elements and vitamins as essential nutrients, they have also immunoregulatory and antioxidant properties. Deficiencies in these kinds of micronutrients are strictly associated with various disorders such as mastitis [101]. The mechanism that underscores the contribution of micronutrients to health is usually caused by their antioxidant features [102]. Selenium (Se) is a naturally derived antioxidant, easily supplied through feed. Since it is an essential component of 25 animals’ selenoproteins, it operates as a fundamental regulator for their activity and expression [103]. One of them is glutathione peroxidase (GPX), crucial for the antioxidant defense of cells and tissues. In a study by Sun et al. [104], the organic form of Se (hydroxy-selenοmethionine, HMSeBA, and selenomethionine, SeMet) resulted not only in higher GPX activity, but also in the enhanced mRNA abundance of GPX3, as compared to the sodium selenite (SS) form in bovine MECs. However, no effect was noted for CAT and SOD gene expression. Moreover, Han et al. [105], in an experiment with nano-selenium (nano-Se) and SS supplementation in dairy cows, found that dietary nano-Se upregulated the mRNA expression of GPX1, GPX2 and GPX4, as well as that of thioredoxin reductase 2 and 3, in the mammary gland, compared with the SS group. The common stage for both was the significance of the proper selection of Se source.
Bioactive compounds derived from plants are widely known as potential contributors to decreasing oxidative stress in ruminants [106]. In several experiments with the use of polyphenols, some promising results have been published. For instance, the treatment of bovine MECs with tea polyphenols (100 μg/mL) enhanced the protein abundance of NFE2L2 and HMOX1 [95]. Thoroughly, the co-treatment of cells with tea polyphenols and H2O2 increased the mRNA abundance of NFE2L2, HMOX1, and NQO1 compared to H2O2 treatment. In the same manner, the co-treatment of cells with tea-polyphenols and H2O2 caused an upsurge in BCL2, but it significantly diminished BAX, CASPASE3, and CASPASE9 mRNA abundance compared with the H2O2 treatment [95]. In the same experiment, the mRNA expression of TNFA, IL6, and IL1B, was decreased in the tea-polyphenols plus H2O2-treated group compared with the H2O2-treated group, along with a reduced apoptosis rate [95]. Therefore, Ma et al. [95] concluded that tea-polyphenols may effectively set up a favorable antioxidant by inhibiting the activation of CASPASE3 and the expression of BAX and by strengthening the mRNA expression of BCL2. In addition, tea-polyphenols can lessen the inflammatory burden caused by ROS by inhibiting the expression of inflammatory cytokines. Moreover, resveratrol (50 µM) induced similar effects in bovine MECs challenged with H2O2 [89]. However, it is worth mentioning that the use of polyphenols extracted from green tea in vivo had no effect on oxidative stress in peripartal cows [107]. Considering the in vivo trials, it has been reported that the post-ruminal bioavailability of tea-polyphenols may play a critical role in their antioxidant impact [108].
Among plant compounds with known antioxidant properties are the plant lignans due to their polyphenolic content, such as those contained in flax. Flax hulls have been assessed for their influence upon some target genes in the mammary gland specific to the expression of antioxidant features [109]. In this experiment, the dietary inclusion of flax hull in cows increased the mRNA abundance of CAT, GPX1, and SOD1, but lessened that of GPX3, SOD2, and SOD3 in the mammary gland [110]. Moreover, when flax hulls were combined with 500 g/d flax oil in cows’ diets, the mRNA levels of CAT, GPX1, GPX3, SOD2, and SOD3 were decreased, unveiling the prooxidant effect of PUFAs (flax oil) [109]. In another experiment, the inclusion of flax meal in dairy cows’ diets gave some prominent results on the regulation of specific target genes and TFs in the mammary gland [110]. The NFE2L2 mRNA abundance was linearly increased among the inclusion levels, while the CAT, GPX1, GPX3, SOD1, SOD2, SOD3, and NFKB genes remained unaffected [110]. Palin et al. [111] also performed an experiment on dairy cows, supplementing flax hulls, with or without flax oil, for the effect of this nutritional treatment upon the expression of lipogenic genes. The inclusion of flax hulls alone upregulated the mRNA abundance of SREBPF1, FAS, LPL, PPARγ1, SCD, and ACC [111]. The results of this study demonstrated that the lignans present in flax hulls not only have significant antioxidant potential, but they can also rebalance key lipogenic gene networks that are generally downregulated by dietary PUFA overload.
Lastly, the inclusion of piper meal, which is rich in bioactive polyphenols, increased the mRNA abundance of SOD1, SOD2, and SOD3, and for NFE2L2, while it did not affect that of CAT, GPX1, GPX2, or GPX3 in dairy goats [112]. Furthermore, dietary supplementation with piper meal affected the immune-transcriptional profile of goats’ mammary glands by downregulating the expression of NFKB1 [112]. Thus, it could be concluded that polyphenolic compounds have both immune-oxidative properties.

Table 3.
Selected studies presenting the impact of bioactive components and feed additives on MECs and the mammary gland gene network.
Table 3.
Selected studies presenting the impact of bioactive components and feed additives on MECs and the mammary gland gene network.
| Animal Species | Experimental Method | Nutrition Tested | Level | Effect | Reference |
|---|---|---|---|---|---|
| Bovine MECs | In vitro | Tea-polyphenols (GTP), H2O2, combination | 500 µM H2O2 100 µg/mL GTP 100 µg/mL GTP plus 500 µM H2O2 | GTP:
| [95] |
| Bovine MECs | In vitro | -Organic Se source hydroxy-selenomethionine (HMSeBA) -Selenomethionine (SeMet) -Sodium selenite (SS) | 0 20 nM HMSeBA 50 nM HMSeBA 100 nM HMSeBA 150 nM HMSeBA 100 nM SeMet 100 nM SS | HMSeBA:
| [104] |
| Dairy cows | In vivo | Rumen-protected methionine | 0.09% and 0.10% of DMI |
| [88] |
| Dairy cows | In vivo | Nano-selenium (nano-Se) Sodium selenite (SS) | 0.30 mg Se/kg DM SS 0.30 mg Se/kg DM Nano-Se |
| [105] |
| Dairy cows | In vivo | Flax hulls | no flax hulls (CONT) 9·88% flax hulls (HULL) 500 g flax oil/d (COFO) 9·88% flax hulls and 500 g flax oil/d (HUFO) | HULL vs. CONT:
| [109] |
| Dairy cows | In vivo | Flax hulls | no flax hulls (CONT) 9·88% flax hulls (HULL) 500 g flax oil/d (COFO) 9·88% flax hulls and 500 g flax oil/d (HUFO) | HULL:
| [111] |
| Dairy cows | In vivo | Flax meal (FM) | 5% (5FM) 10% (10FM) 15% (15FM) |
| [110] |
| Dairy goats | In vivo | Piper meal | CPM diet (1.3% piper meal per kg dry matter) |
| [112] |
↓ = decrease, ↑ = increase.
5. Further Prospects
Considering the aforementioned research evidence, on the path towards promoting functional dairy products enriched with biomolecules beneficial for human health, such as omega-3 FAs, unfavorable cellular conditions may occur. However, it could be concluded that there are certain gene biomarkers that are more firmly linked to lipogenesis in the mammary gland, while these genes are also variously affected by different dietary fatty acids (PUFA source). Additionally, it was also unveiled that PUFA levels, sources, and animal species appear to also have crucial importance considering the regulation of the mammary gland lipogenic network. On the other hand, ample evidence suggests that dietary antioxidant compounds, and especially polyphenols and flavonoids, can strengthen the immune-oxidative status of the mammary gland, and their simultaneous supplementation may alleviate any side effect caused by dietary PUFA overload. Future studies should evaluate the effect of PUFA-rich diets, and especially the combination of different PUFA sources, achieving a sustainable transfer efficiency as a strategy to develop functional dairy products. Simultaneously, bioactive compounds with prominent antioxidant potential capable of neutralizing PUFAs’ adverse effects on mammary gland oxidative status should be also assessed in vivo.
Author Contributions
Conceptualization, A.M., E.T. and A.C.P.; writing—original draft preparation, P.K. and F.Z.; writing—review and editing, A.M., E.T. and A.C.P. visualization, P.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Abbreviation | Word or phrase in full |
| AAs | amino acids |
| ACC | acetyl-CoA carboxylase |
| ACL | ATP-citrate lyase |
| ACSL1 | acyl-CoA synthetase long chain family member 1 |
| ACSS2 | acyl-CoA synthetase short-chain family member 2 |
| AGPAT | acyl glycerol phosphate acyl transferase; lysophosphatidic acid acyltransferase |
| ALA | α-linolenic acid |
| ARE | antioxidant response element |
| Arg | arginine |
| BAX | Bcl-2 associated X-protein |
| BCL2 | B-cell lymphoma-2 |
| BHBA | β-hydroxybutyrate |
| BTN1A1 | butyrophilin subfamily 1 member A1 |
| CAT | catalase |
| CD36 | CD36 molecule; fatty acid transferase |
| CLA | conjugated linoleic acid |
| CLD | cytoplasmic lipid droplet |
| CM | chylomicrons |
| CoA | coenzyme A |
| CUL3 | cullin-3 |
| DAG | diacylglycerol |
| DGAT | diacylglycerol acyltransferase |
| DHA | docosahexaenoic acid |
| DNA | deoxyribonucleic acid |
| DPA | docosapentaenoic acid |
| ECM | extra-cellular matrix proteins |
| ELOVL | fatty acid elongase |
| EPA | eicosapentaenoic acid |
| ER | endoplasmic reticulum |
| FABP | fatty acid binding protein |
| FADS | fatty acid desaturase |
| FAs | fatty acids |
| FAS | fatty acid synthase |
| FATP | fatty acid transport protein |
| FECH | ferrochelatase |
| FFARs | free fatty acid receptors |
| FFAs | free fatty acids |
| FTH1 | ferritin heavy chain 1 |
| G6PDH | glucose-6-phosphate dehydrogenase |
| GCLC | glutamate-cysteine ligase catalytic subunit |
| GCLM | glutamate-cysteine ligase modifier subunit |
| Glut1 | glucose transporter1 |
| glycerol-3-P | glycerol-3-phosphate |
| GPAM | glycerol-3-phosphate acyltransferase, mitochondrial |
| GPAT | glycerol-3-phosphate acyltransferase |
| GPX | glutathione peroxidase |
| GSH | glutathione |
| GSR | glutathione-disulfide reductase |
| GSTM1 | glutathione s-transferase mu 1 |
| HMOX1 | heme oxygenase 1 |
| HMSeBA | hydroxy-selenomethionine |
| IL | interleukin |
| INSIG | insulin-induced gene |
| KEAP1 | Kelch-like epichlorohydrin-associated protein 1 |
| LCFAs | long-chain fatty acids |
| LPA | lysophosphatidic acid |
| LPIN | lipin |
| LPL | lipoprotein lipase |
| MAPK14 | mitogen-activated protein kinase 14 |
| MCFAs | medium chain fatty acids |
| MDA | malondialdehyde |
| ME1 | malic enzyme 1 |
| MECs | mammary epithelial cells |
| Met | methionine |
| MFD | milk fat depression |
| MFG | milk fat globule |
| mRNA | messenger ribonucleic acid |
| mTOR | mechanistic target of rapamycin |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| NEFA | non-esterified FA |
| NFE2L2 | nuclear factor erythroid 2-like 2 |
| NFKB | nuclear factor-kappa B |
| NQO1 | NAD(P)H dehydrogenase [quinone] 1 |
| O2 | oxygen |
| PA | phosphatidate |
| PIR | pirin |
| PLIN2 | perilipin 2 |
| PPARs | proliferator-activated receptors |
| PPARγ | proliferator-activated receptor γ |
| PPARγC1A | PPARγ coactivator 1 alpha |
| PUFAs | polyunsaturated fatty acids |
| RNS | reactive nitrogen species |
| ROS | reactive oxygen species |
| SCD | stearoyl-CoA desaturase |
| SCFAs | short chain fatty acids |
| Se | selenium |
| SeMet | selenomethionine |
| SFAs | saturated fatty acids |
| SLC27A6 | solute carrier family 27-member 6 |
| SOD | superoxide dismutase |
| SREBF1 | sterol regulatory element binding transcription factor 1 |
| SS | sodium selenite |
| TAGs | triglycerides |
| TALDO1 | transaldolase 1 |
| TFs | transcription factors |
| TNFA | tumor necrosis factor alpha |
| TXN | thioredoxin |
| UFAs | unsaturated fatty acids |
| VLDL | very-low-density lipoprotein |
| XDH | xanthine dehydrogenase |
References
- Srour, B.; Fezeu, L.K.; Kesse-Guyot, E.; Allès, B.; Méjean, C.; Andrianasolo, R.M.; Chazelas, E.; Deschasaux, M.; Hercberg, S.; Galan, P.; et al. Ultra-Processed Food Intake and Risk of Cardiovascular Disease: Prospective Cohort Study (NutriNet-Santé). BMJ 2019, 365, l1451. [Google Scholar] [CrossRef] [PubMed]
- Schwingshackl, L.; Hoffmann, G.; Lampousi, A.-M.; Knüppel, S.; Iqbal, K.; Schwedhelm, C.; Bechthold, A.; Schlesinger, S.; Boeing, H. Food Groups and Risk of Type 2 Diabetes Mellitus: A Systematic Review and Meta-Analysis of Prospective Studies. Eur. J. Epidemiol. 2017, 32, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Brondel, L.; Quilliot, D.; Mouillot, T.; Khan, N.A.; Bastable, P.; Boggio, V.; Leloup, C.; Pénicaud, L. Taste of Fat and Obesity: Different Hypotheses and Our Point of View. Nutrients 2022, 14, 555. [Google Scholar] [CrossRef] [PubMed]
- King, H. Hippocrates Now: The “Father of Medicine” in the Internet Age; Bloomsbury Studies in Classical Reception; Bloomsbury Academic: London, UK, 2019; ISBN 9781350005891. [Google Scholar]
- Kholif, A.; Olafadehan, O. Dietary strategies to enrich milk with healthy fatty acids—A review. Ann. Anim. Sci. 2022, 22, 523–536. [Google Scholar] [CrossRef]
- Plata-Pérez, G.; Angeles-Hernandez, J.C.; Morales-Almaráz, E.; Del Razo-Rodríguez, O.E.; López-González, F.; Peláez-Acero, A.; Campos-Montiel, R.G.; Vargas-Bello-Pérez, E.; Vieyra-Alberto, R. Oilseed Supplementation Improves Milk Composition and Fatty Acid Profile of Cow Milk: A Meta-Analysis and Meta-Regression. Animals 2022, 12, 1642. [Google Scholar] [CrossRef] [PubMed]
- Toral, P.G.; Monahan, F.J.; Hervás, G.; Frutos, P.; Moloney, A.P. Review: Modulating ruminal lipid metabolism to improve the fatty acid composition of meat and milk. Challenges and opportunities. Animal 2018, 12, s272–s281. [Google Scholar] [CrossRef]
- Urrutia, O.; Mendizabal, J.A.; Alfonso, L.; Soret, B.; Insausti, K.; Arana, A. Adipose Tissue Modification through Feeding Strategies and Their Implication on Adipogenesis and Adipose Tissue Metabolism in Ruminants. Int. J. Mol. Sci. 2020, 21, 3183. [Google Scholar] [CrossRef]
- Mavrommatis, A.; Chronopoulou, E.G.; Sotirakoglou, K.; Labrou, N.E.; Zervas, G.; Tsiplakou, E. The Impact of the Dietary Supplementation Level with Schizochytrium sp., on the Oxidative Capacity of Both Goats’ Organism and Milk. Livest. Sci. 2018, 218, 37–43. [Google Scholar] [CrossRef]
- Barrea, L.; Annunziata, G.; Bordoni, L.; Muscogiuri, G.; Colao, A.; Savastano, S. Nutrigenetics-Personalized Nutrition in Obesity and Cardiovascular Diseases. Int. J. Obes. Suppl. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Ordovas, J.M.; Ferguson, L.R.; Tai, E.S.; Mathers, J.C. Personalised Nutrition and Health. BMJ 2018, 361, bmj.k2173. [Google Scholar] [CrossRef]
- Kizilaslan, M.; Arzik, Y.; Cinar, M.U.; Konca, Y. Genome-Wise Engineering of Ruminant Nutrition—Nutrigenomics: Applications, Challenges, and Future Perspectives—A Review. Ann. Anim. Sci. 2022, 22, 511–521. [Google Scholar] [CrossRef]
- Nayeri, S.; Stothard, P. Tissues, Metabolic Pathways and Genes of Key Importance in Lactating Dairy Cattle. Springer Sci. Rev. 2016, 4, 49–77. [Google Scholar] [CrossRef]
- Jensen, R.G. The Composition of Bovine Milk Lipids: January 1995 to December 2000. J. Dairy Sci. 2002, 85, 295–350. [Google Scholar] [CrossRef]
- Bernard, L.; Leroux, C.; Chilliard, Y. Expression and Nutritional Regulation of Lipogenic Genes in the Ruminant Lactating Mammary Gland. Adv. Exp. Med. Biol. 2008, 606, 67–108. [Google Scholar] [CrossRef]
- Guo, W.; Choi, J.-K.; Kirkland, J.L.; Corkey Barbara, E.; Hamilton, J.A. Esterification of Free Fatty Acids in Adipocytes: A Comparison between Octanoate and Oleate. Biochem. J. 2000, 349, 463. [Google Scholar] [CrossRef]
- Mu, T.; Hu, H.; Ma, Y.; Feng, X.; Zhang, J.; Gu, Y. Regulation of Key Genes for Milk Fat Synthesis in Ruminants. Front. Nutr. 2021, 8, 765147. [Google Scholar] [CrossRef]
- Bernard, L.; Leroux, C.; Chilliard, Y. Nutritional Regulation of Mammary Lipogenesis and Milk Fat in Ruminant: Contribution to Sustainable Milk Production. Rev. Colomb. Cienc. Pecu. 2013, 26, 292–302. [Google Scholar]
- Loften, J.R.; Linn, J.G.; Drackley, J.K.; Jenkins, T.C.; Soderholm, C.G.; Kertz, A.F. Invited Review: Palmitic and Stearic Acid Metabolism in Lactating Dairy Cows. J. Dairy Sci. 2014, 97, 4661–4674. [Google Scholar] [CrossRef]
- Burns, T. Fatty Acids and Lipogenesis in Ruminant Adipocytes. Ph.D. Thesis, Clemson University, Clemson, SC, USA, 2011. [Google Scholar]
- Osorio, J.S.; Lohakare, J.; Bionaz, M. Biosynthesis of Milk Fat, Protein, and Lactose: Roles of Transcriptional and Posttranscriptional Regulation. Physiol. Genom. 2016, 48, 231–256. [Google Scholar] [CrossRef]
- Bionaz, M.; Vargas-Bello-Pérez, E.; Busato, S. Advances in Fatty Acids Nutrition in Dairy Cows: From Gut to Cells and Effects on Performance. J. Anim. Sci. Biotechnol. 2020, 11, 110. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. Gene Networks Driving Bovine Milk Fat Synthesis during the Lactation Cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bello-Pérez, E.; Geldsetzer-Mendoza, C.; Morales, M.S.; Toro-Mujica, P.; Fellenberg, M.A.; Ibáñez, R.A.; Gómez-Cortés, P.; Garnsworthy, P.C. Effect of Olive Oil in Dairy Cow Diets on the Fatty Acid Profile and Sensory Characteristics of Cheese. Int. Dairy J. 2018, 85, 8–15. [Google Scholar] [CrossRef]
- Nguyen, D.v.; Malau-Aduli, B.S.; Cavalieri, J.; Nichols, P.D.; Malau-Aduli, A.E.O. Supplementation with Plant-Derived Oils Rich in Omega-3 Polyunsaturated Fatty Acids for Lamb Production. Vet. Anim. Sci. 2018, 6, 29–40. [Google Scholar] [CrossRef] [PubMed]
- González-Becerra, K.; Ramos-Lopez, O.; Barrón-Cabrera, E.; Riezu-Boj, J.I.; Milagro, F.I.; Martínez-López, E.; Martínez, J.A. Fatty Acids, Epigenetic Mechanisms and Chronic Diseases: A Systematic Review. Lipids Health Dis. 2019, 18, 178. [Google Scholar] [CrossRef] [PubMed]
- Bauman, D.E.; Mather, I.H.; Wall, R.J.; Lock, A.L. Major Advances Associated with the Biosynthesis of Milk. J. Dairy Sci. 2006, 89, 1235–1243. [Google Scholar] [CrossRef]
- Thering, B.J.; Graugnard, D.E.; Piantoni, P.; Loor, J.J. Adipose Tissue Lipogenic Gene Networks Due to Lipid Feeding and Milk Fat Depression in Lactating Cows. J. Dairy Sci. 2009, 92, 4290–4300. [Google Scholar] [CrossRef]
- Ahnadi, C.E.; Beswick, N.; Delbecchi, L.; Kennelly, J.J.; Lacasse, P. Addition of Fish Oil to Diets for Dairy Cows. II. Effects on Milk Fat and Gene Expression of Mammary Lipogenic Enzymes. J. Dairy Res. 2002, 69, 521–531. [Google Scholar] [CrossRef]
- Vahmani, P.; Glover, K.E.; Fredeen, A.H. Effects of Pasture versus Confinement and Marine Oil Supplementation on the Expression of Genes Involved in Lipid Metabolism in Mammary, Liver, and Adipose Tissues of Lactating Dairy Cows. J. Dairy Sci. 2014, 97, 4174–4183. [Google Scholar] [CrossRef]
- Vargas-Bello-Pérez, E.; Cancino-Padilla, N.; Geldsetzer-Mendoza, C.; Morales, M.S.; Leskinen, H.; Garnsworthy, P.C.; Loor, J.J.; Romero, J. Effects of Dietary Polyunsaturated Fatty Acid Sources on Expression of Lipid-Related Genes in Bovine Milk Somatic Cells. Sci. Rep. 2020, 10, 14850. [Google Scholar] [CrossRef]
- Harvatine, K.J.; Bauman, D.E. SREBP1 and Thyroid Hormone Responsive Spot 14 (S14) Are Involved in the Regulation of Bovine Mammary Lipid Synthesis during Diet-Induced Milk Fat Depression and Treatment with CLA. J. Nutr. 2006, 136, 2468–2474. [Google Scholar] [CrossRef]
- Angulo, J.; Mahecha, L.; Nuernberg, K.; Nuernberg, G.; Dannenberger, D.; Olivera, M.; Boutinaud, M.; Leroux, C.; Albrecht, E.; Bernard, L. Effects of Polyunsaturated Fatty Acids from Plant Oils and Algae on Milk Fat Yield and Composition Are Associated with Mammary Lipogenic and SREBF1 Gene Expression. Animal 2012, 6, 1961–1972. [Google Scholar] [CrossRef]
- Frutos, P.; Toral, P.G.; Hervás, G. Individual Variation of the Extent of Milk Fat Depression in Dairy Ewes Fed Fish Oil: Milk Fatty Acid Profile and MRNA Abundance of Candidate Genes Involved in Mammary Lipogenesis. J. Dairy Sci. 2017, 100, 9611–9622. [Google Scholar] [CrossRef]
- Suárez-Vega, A.; Toral, P.G.; Gutiérrez-Gil, B.; Hervás, G.; Arranz, J.J.; Frutos, P. Elucidating Fish Oil-Induced Milk Fat Depression in Dairy Sheep: Milk Somatic Cell Transcriptome Analysis. Sci. Rep. 2017, 7, 45905. [Google Scholar] [CrossRef]
- Carreño, D.; Hervás, G.; Toral, P.G.; Castro-Carrera, T.; Frutos, P. Fish Oil-Induced Milk Fat Depression and Associated Downregulation of Mammary Lipogenic Genes in Dairy Ewes. J. Dairy Sci. 2016, 99, 7971–7981. [Google Scholar] [CrossRef]
- Faulconnier, Y.; Bernard, L.; Boby, C.; Domagalski, J.; Chilliard, Y.; Leroux, C. Extruded Linseed Alone or in Combination with Fish Oil Modifies Mammary Gene Expression Profiles in Lactating Goats. Animal 2018, 12, 1564–1575. [Google Scholar] [CrossRef]
- Toral, P.G.; Bernard, L.; Delavaud, C.; Gruffat, D.; Leroux, C.; Chilliard, Y. Effects of Fish Oil and Additional Starch on Tissue Fatty Acid Profile and Lipogenic Gene MRNA Abundance in Lactating Goats Fed a Diet Containing Sunflower-Seed Oil. Animal 2013, 7, 948–956. [Google Scholar] [CrossRef]
- Invernizzi, G.; Thering, B.J.; McGuire, M.A.; Savoini, G.; Loor, J.J. Sustained Upregulation of Stearoyl-CoA Desaturase in Bovine Mammary Tissue with Contrasting Changes in Milk Fat Synthesis and Lipogenic Gene Networks Caused by Lipid Supplements. Funct. Integr. Genom. 2010, 10, 561–575. [Google Scholar] [CrossRef]
- Peterson, D.G.; Matitashvili, E.A.; Bauman, D.E. Diet-Induced Milk Fat Depression in Dairy Cows Results in Increased Trans-10, Cis-12 CLA in Milk Fat and Coordinate Suppression of MRNA Abundance for Mammary Enzymes Involved in Milk Fat Synthesis. J. Nutr. 2003, 133, 3098–3102. [Google Scholar] [CrossRef]
- Bernard, L.; Leroux, C.; Faulconnier, Y.; Durand, D.; Shingfield, K.J.; Chilliard, Y. Effect of Sunflower-Seed Oil or Linseed Oil on Milk Fatty Acid Secretion and Lipogenic Gene Expression in Goats Fed Hay-Based Diets. J. Dairy Res. 2009, 76, 241–248. [Google Scholar] [CrossRef]
- Ibeagha-Awemu, E.M.; Li, R.; Ammah, A.A.; Dudemaine, P.-L.; Bissonnette, N.; Benchaar, C.; Zhao, X. Transcriptome Adaptation of the Bovine Mammary Gland to Diets Rich in Unsaturated Fatty Acids Shows Greater Impact of Linseed Oil over Safflower Oil on Gene Expression and Metabolic Pathways. BMC Genom. 2016, 17, 104. [Google Scholar] [CrossRef]
- Jacobs, A.A.A.; van Baal, J.; Smits, M.A.; Taweel, H.Z.H.; Hendriks, W.H.; van Vuuren, A.M.; Dijkstra, J. Effects of Feeding Rapeseed Oil, Soybean Oil, or Linseed Oil on Stearoyl-CoA Desaturase Expression in the Mammary Gland of Dairy Cows. J. Dairy Sci. 2011, 94, 874–887. [Google Scholar] [CrossRef] [PubMed]
- Ollier, S.; Leroux, C.; de la Foye, A.; Bernard, L.; Rouel, J.; Chilliard, Y. Whole Intact Rapeseeds or Sunflower Oil in High-Forage or High-Concentrate Diets Affects Milk Yield, Milk Composition, and Mammary Gene Expression Profile in Goats. J. Dairy Sci. 2009, 92, 5544–5560. [Google Scholar] [CrossRef] [PubMed]
- Leroux, C.; Bernard, L.; Faulconnier, Y.; Rouel, J.; de la Foye, A.; Domagalski, J.; Chilliard, Y. Bovine Mammary Nutrigenomics and Changes in the Milk Composition Due to Rapeseed or Sunflower Oil Supplementation of High-Forage or High-Concentrate Diets. Lifestyle Genom. 2016, 9, 65–82. [Google Scholar] [CrossRef] [PubMed]
- Bernard, L.; Bonnet, M.; Leroux, C.; Shingfield, K.J.; Chilliard, Y. Effect of Sunflower-Seed Oil and Linseed Oil on Tissue Lipid Metabolism, Gene Expression, and Milk Fatty Acid Secretion in Alpine Goats Fed Maize Silage–Based Diets. J. Dairy Sci. 2009, 92, 6083–6094. [Google Scholar] [CrossRef] [PubMed]
- Murrieta, C.M.; Hess, B.W.; Scholljegerdes, E.J.; Engle, T.E.; Hossner, K.L.; Moss, G.E.; Rule, D.C. Evaluation of Milk Somatic Cells as a Source of MRNA for Study of Lipogenesis in the Mammary Gland of Lactating Beef Cows Supplemented with Dietary High-Linoleate Safflower Seeds1. J. Anim. Sci. 2006, 84, 2399–2405. [Google Scholar] [CrossRef]
- Bernard, L.; Rouel, J.; Leroux, C.; Ferlay, A.; Faulconnier, Y.; Legrand, P.; Chilliard, Y. Mammary Lipid Metabolism and Milk Fatty Acid Secretion in Alpine Goats Fed Vegetable Lipids. J. Dairy Sci. 2005, 88, 1478–1489. [Google Scholar] [CrossRef]
- Conte, G.; Giordani, T.; Vangelisti, A.; Serra, A.; Pauselli, M.; Cavallini, A.; Mele, M. Transcriptome Adaptation of the Ovine Mammary Gland to Dietary Supplementation of Extruded Linseed. Animals 2021, 11, 2707. [Google Scholar] [CrossRef]
- Delbecchi, L.; Ahnadi, C.E.; Kennelly, J.J.; Lacasse, P. Milk Fatty Acid Composition and Mammary Lipid Metabolism in Holstein Cows Fed Protected or Unprotected Canola Seeds. J. Dairy Sci. 2001, 84, 1375–1381. [Google Scholar] [CrossRef]
- Bernard, L.; Leroux, C.; Bonnet, M.; Rouel, J.; Martin, P.; Chilliard, Y. Expression and Nutritional Regulation of Lipogenic Genes in Mammary Gland and Adipose Tissues of Lactating Goats. J. Dairy Res. 2005, 72, 250–255. [Google Scholar] [CrossRef]
- Li, X.Z.; Yan, C.G.; Lee, H.G.; Choi, C.W.; Song, M.K. Influence of Dietary Plant Oils on Mammary Lipogenic Enzymes and the Conjugated Linoleic Acid Content of Plasma and Milk Fat of Lactating Goats. Anim. Feed Sci. Technol. 2012, 174, 26–35. [Google Scholar] [CrossRef]
- Baumgard, L.H.; Matitashvili, E.; Corl, B.A.; Dwyer, D.A.; Bauman, D.E. Trans-10, Cis-12 Conjugated Linoleic Acid Decreases Lipogenic Rates and Expression of Genes Involved in Milk Lipid Synthesis in Dairy Cows. J. Dairy Sci. 2002, 85, 2155–2163. [Google Scholar] [CrossRef]
- Harvatine, K.J.; Perfield, J.W.; Bauman, D.E. Expression of Enzymes and Key Regulators of Lipid Synthesis Is Upregulated in Adipose Tissue during CLA-Induced Milk Fat Depression in Dairy Cows. J. Nutr. 2009, 139, 849–854. [Google Scholar] [CrossRef]
- Gervais, R.; McFadden, J.W.; Lengi, A.J.; Corl, B.A.; Chouinard, P.Y. Effects of Intravenous Infusion of Trans-10, Cis-12 18:2 on Mammary Lipid Metabolism in Lactating Dairy Cows. J. Dairy Sci. 2009, 92, 5167–5177. [Google Scholar] [CrossRef]
- Suárez-Vega, A.; Gutiérrez-Gil, B.; Toral, P.G.; Hervás, G.; Arranz, J.J.; Frutos, P. Conjugated Linoleic Acid (CLA)-Induced Milk Fat Depression: Application of RNA-Seq Technology to Elucidate Mammary Gene Regulation in Dairy Ewes. Sci. Rep. 2019, 9, 4473. [Google Scholar] [CrossRef]
- Hussein, M.; Harvatine, K.H.; Weerasinghe, W.M.P.B.; Sinclair, L.A.; Bauman, D.E. Conjugated Linoleic Acid-Induced Milk Fat Depression in Lactating Ewes Is Accompanied by Reduced Expression of Mammary Genes Involved in Lipid Synthesis. J. Dairy Sci. 2013, 96, 3825–3834. [Google Scholar] [CrossRef]
- Toral, P.G.; Hervás, G.; Belenguer, A.; Carreño, D.; Frutos, P. MRNA Abundance of Genes Involved in Mammary Lipogenesis during Fish Oil- or Trans-10, Cis-12 CLA-Induced Milk Fat Depression in Dairy Ewes. J. Dairy Sci. 2017, 100, 3182–3192. [Google Scholar] [CrossRef]
- Tsiplakou, E.; Flemetakis, E.; Kalloniati, C.; Papadomichelakis, G.; Katinakis, P.; Zervas, G. Sheep and Goats Differences in CLA and Fatty Acids Milk Fat Content in Relation with MRNA Stearoyl-CoA Desaturase and Lipogenic Genes Expression in Their Mammary Gland. J. Dairy Res. 2009, 76, 392–401. [Google Scholar] [CrossRef]
- Shingfield, K.J.; Bernard, L.; Leroux, C.; Chilliard, Y. Role of Trans Fatty Acids in the Nutritional Regulation of Mammary Lipogenesis in Ruminants. Animal 2010, 4, 1140–1166. [Google Scholar] [CrossRef]
- Bauman, D.E.; Harvatine, K.J.; Lock, A.L. Nutrigenomics, Rumen-Derived Bioactive Fatty Acids, and the Regulation of Milk Fat Synthesis. Annu. Rev. Nutr. 2011, 31, 299–319. [Google Scholar] [CrossRef]
- Bionaz, M.; Osorio, J.; Loor, J.J. Triennial Lactation Symposium: Nutrigenomics in Dairy Cows: Nutrients, Transcription Factors, and Techniques. J. Anim. Sci. 2015, 93, 5531–5553. [Google Scholar] [CrossRef]
- Ma, L.; Corl, B.A. Transcriptional Regulation of Lipid Synthesis in Bovine Mammary Epithelial Cells by Sterol Regulatory Element Binding Protein-1. J. Dairy Sci. 2012, 95, 3743–3755. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Espenshade, P.J.; Wright, M.E.; Yabe, D.; Gong, Y.; Aebersold, R.; Goldstein, J.L.; Brown, M.S. Crucial Step in Cholesterol Homeostasis. Cell 2002, 110, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Mach, N.; Jacobs, A.A.A.; Kruijt, L.; van Baal, J.; Smits, M.A. Alteration of Gene Expression in Mammary Gland Tissue of Dairy Cows in Response to Dietary Unsaturated Fatty Acids. Animal 2011, 5, 1217–1230. [Google Scholar] [CrossRef] [PubMed]
- Castro-Carrera, T.; Frutos, P.; Leroux, C.; Chilliard, Y.; Hervás, G.; Belenguer, A.; Bernard, L.; Toral, P.G. Dietary Sunflower Oil Modulates Milk Fatty Acid Composition without Major Changes in Adipose and Mammary Tissue Fatty Acid Profile or Related Gene MRNA Abundance in Sheep. Animal 2015, 9, 582–591. [Google Scholar] [CrossRef]
- Bernard, L.; Leroux, C.; Chilliard, Y. Expression and Nutritional Regulation of Stearoyl-CoA Desaturase Genes in the Ruminant Mammary Gland: Relationship with Milk Fatty Acid Composition. In Stearoyl-CoA Desaturase Genes in Lipid Metabolism; Springer: New York, NY, USA, 2013; pp. 161–193. [Google Scholar]
- Kadegowda, A.K.G.; Bionaz, M.; Piperova, L.S.; Erdman, R.A.; Loor, J.J. Peroxisome Proliferator-Activated Receptor-γ Activation and Long-Chain Fatty Acids Alter Lipogenic Gene Networks in Bovine Mammary Epithelial Cells to Various Extents. J. Dairy Sci. 2009, 92, 4276–4289. [Google Scholar] [CrossRef]
- Gama, M.A.S.; Garnsworthy, P.C.; Griinari, J.M.; Leme, P.R.; Rodrigues, P.H.M.; Souza, L.W.O.; Lanna, D.P.D. Diet-Induced Milk Fat Depression: Association with Changes in Milk Fatty Acid Composition and Fluidity of Milk Fat. Livest. Sci. 2008, 115, 319–331. [Google Scholar] [CrossRef]
- Bichi, E.; Hervás, G.; Toral, P.G.; Loor, J.J.; Frutos, P. Milk Fat Depression Induced by Dietary Marine Algae in Dairy Ewes: Persistency of Milk Fatty Acid Composition and Animal Performance Responses. J. Dairy Sci. 2013, 96, 524–532. [Google Scholar] [CrossRef]
- Bichi, E.; Frutos, P.; Toral, P.G.; Keisler, D.; Hervás, G.; Loor, J.J. Dietary Marine Algae and Its Influence on Tissue Gene Network Expression during Milk Fat Depression in Dairy Ewes. Anim. Feed Sci. Technol. 2013, 186, 36–44. [Google Scholar] [CrossRef][Green Version]
- Mele, M.; Serra, A.; Conte, G.; Pollicardo, A.; del Viva, M.; Secchiari, P. Whole Extruded Linseed in the Diet of Dairy Ewes during Early Lactation: Effect on the Fatty Acid Composition of Milk and Cheese. Ital. J. Anim. Sci. 2007, 6, 560–562. [Google Scholar] [CrossRef][Green Version]
- Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef]
- Loor, J.J.; Bionaz, M.; Drackley, J.K. Systems Physiology in Dairy Cattle: Nutritional Genomics and Beyond. Annu. Rev. Anim. Biosci. 2013, 1, 365–392. [Google Scholar] [CrossRef]
- Roche, J.R.; Bell, A.W.; Overton, T.R.; Loor, J.J. Nutritional Management of the Transition Cow in the 21st Century—A Paradigm Shift in Thinking. Anim. Prod. Sci. 2013, 53, 1000. [Google Scholar] [CrossRef]
- Mitsiopoulou, C.; Sotirakoglou, K.; Labrou, N.E.; Tsiplakou, E. The Effect of Whole Sesame Seeds on Milk Chemical Composition, Fatty Acid Profile and Antioxidant Status in Goats. Livest. Sci. 2021, 245, 104452. [Google Scholar] [CrossRef]
- Tsiplakou, E.; Mitsiopoulou, C.; Karaiskou, C.; Simoni, M.; Pappas, A.C.; Righi, F.; Sotirakoglou, K.; Labrou, N.E. Sesame Meal, Vitamin E and Selenium Influence Goats’ Antioxidant Status. Antioxidants 2021, 10, 392. [Google Scholar] [CrossRef]
- Sordillo, L.M. Symposium Review: Oxylipids and the Regulation of Bovine Mammary Inflammatory Responses. J. Dairy Sci. 2018, 101, 5629–5641. [Google Scholar] [CrossRef]
- Surai, P.F.; Kochish, I.I.; Fisinin, V.I.; Juniper, D.T. Revisiting Oxidative Stress and the Use of Organic Selenium in Dairy Cow Nutrition. Animals 2019, 9, 462. [Google Scholar] [CrossRef]
- Taguchi, K.; Yamamoto, M. The KEAP1–NRF2 System as a Molecular Target of Cancer Treatment. Cancers 2020, 13, 46. [Google Scholar] [CrossRef]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell Survival Responses to Environmental Stresses via the Keap1-Nrf2-ARE Pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Canning, P.; Sorrell, F.J.; Bullock, A.N. Structural Basis of Keap1 Interactions with Nrf2. Free Radic. Biol. Med. 2015, 88, 101–107. [Google Scholar] [CrossRef]
- Kansanen, E.; Kuosmanen, S.M.; Leinonen, H.; Levonen, A.-L. The Keap1-Nrf2 Pathway: Mechanisms of Activation and Dysregulation in Cancer. Redox Biol. 2013, 1, 45–49. [Google Scholar] [CrossRef]
- Bataille, A.M.; Manautou, J.E. Nrf2: A Potential Target for New Therapeutics in Liver Disease. Clin. Pharmacol. Ther. 2012, 92, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Yamamoto, M. Molecular Basis of the Keap1–Nrf2 System. Free Radic. Biol. Med. 2015, 88, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; O’Connor, T.; Yamamoto, M. Keap1 Regulates Both Cytoplasmic-Nuclear Shuttling and Degradation of Nrf2 in Response to Electrophiles. Genes Cells 2003, 8, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Manda, G.; Hassan, A.; Alcaraz, M.J.; Barbas, C.; Daiber, A.; Ghezzi, P.; León, R.; López, M.G.; Oliva, B.; et al. Transcription Factor NRF2 as a Therapeutic Target for Chronic Diseases: A Systems Medicine Approach. Pharmacol. Rev. 2018, 70, 348–383. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Batistel, F.; Ma, Y.; Alharthi, A.S.M.; Parys, C.; Loor, J.J. Methionine Supply Alters Mammary Gland Antioxidant Gene Networks via Phosphorylation of Nuclear Factor Erythroid 2-like 2 (NFE2L2) Protein in Dairy Cows during the Periparturient Period. J. Dairy Sci. 2018, 101, 8505–8512. [Google Scholar] [CrossRef]
- Jin, X.; Wang, K.; Liu, H.; Hu, F.; Zhao, F.; Liu, J. Protection of Bovine Mammary Epithelial Cells from Hydrogen Peroxide-Induced Oxidative Cell Damage by Resveratrol. Oxid. Med. Cell. Longev. 2016, 2016, 2572175. [Google Scholar] [CrossRef]
- Inoue, Y.; Shimazawa, M.; Nagano, R.; Kuse, Y.; Takahashi, K.; Tsuruma, K.; Hayashi, M.; Ishibashi, T.; Maoka, T.; Hara, H. Astaxanthin Analogs, Adonixanthin and Lycopene, Activate Nrf2 to Prevent Light-Induced Photoreceptor Degeneration. J. Pharmacol. Sci. 2017, 134, 147–157. [Google Scholar] [CrossRef]
- Nguyen, T.; Nioi, P.; Pickett, C.B. The Nrf2-Antioxidant Response Element Signaling Pathway and Its Activation by Oxidative Stress. J. Biol. Chem. 2009, 284, 13291–13295. [Google Scholar] [CrossRef]
- Ford, H.R.; Busato, S.; Bionaz, M. In Vitro–In Vivo Hybrid Approach for Studying Modulation of NRF2 in Immortalized Bovine Mammary Cells. Front. Anim. Sci. 2021, 2, 674355. [Google Scholar] [CrossRef]
- Coleman, D.N.; Lopreiato, V.; Alharthi, A.; Loor, J.J. Amino Acids and the Regulation of Oxidative Stress and Immune Function in Dairy Cattle. J. Anim. Sci. 2020, 98, S175–S193. [Google Scholar] [CrossRef]
- Yang, F.; Li, X. Role of Antioxidant Vitamins and Trace Elements in Mastitis in Dairy Cows. J. Adv. Vet. Anim. Res 2015, 2, 1. [Google Scholar] [CrossRef]
- Ma, Y.; Ma, X.; An, Y.; Sun, Y.; Dou, W.; Li, M.; Bao, H.; Zhang, C. Green Tea Polyphenols Alleviate Hydrogen Peroxide-Induced Oxidative Stress, Inflammation, and Apoptosis in Bovine Mammary Epithelial Cells by Activating ERK1/2–NFE2L2–HMOX1 Pathways. Front. Vet. Sci. 2022, 8, 804241. [Google Scholar] [CrossRef]
- Osorio, J.S.; Trevisi, E.; Ji, P.; Drackley, J.K.; Luchini, D.; Bertoni, G.; Loor, J.J. Biomarkers of Inflammation, Metabolism, and Oxidative Stress in Blood, Liver, and Milk Reveal a Better Immunometabolic Status in Peripartal Cows Supplemented with Smartamine M or MetaSmart. J. Dairy Sci. 2014, 97, 7437–7450. [Google Scholar] [CrossRef]
- Zhou, Z.; Vailati-Riboni, M.; Trevisi, E.; Drackley, J.K.; Luchini, D.N.; Loor, J.J. Better Postpartal Performance in Dairy Cows Supplemented with Rumen-Protected Methionine Compared with Choline during the Peripartal Period. J. Dairy Sci. 2016, 99, 8716–8732. [Google Scholar] [CrossRef]
- Wu, T.; Wang, C.; Ding, L.; Shen, Y.; Cui, H.; Wang, M.; Wang, H. Arginine Relieves the Inflammatory Response and Enhances the Casein Expression in Bovine Mammary Epithelial Cells Induced by Lipopolysaccharide. Mediat. Inflamm. 2016, 2016, 9618795. [Google Scholar] [CrossRef]
- Mu, T.; Kong, G.-H.; Han, Z.-Y.; Li, H.-X. Cytoprotection of Methionine on Hyperthermia-Induced Damage in Bovine Mammary Epithelial Cells. Cell Biol. Int. 2014, 38, 971–976. [Google Scholar] [CrossRef]
- Han, Z.-Y.; Mu, T.; Yang, Z. Methionine Protects against Hyperthermia-Induced Cell Injury in Cultured Bovine Mammary Epithelial Cells. Cell Stress Chaperones 2015, 20, 109–120. [Google Scholar] [CrossRef]
- Zhao, F.F.; Wu, T.Y.; Wang, H.R.; Ding, L.Y.; Ahmed, G.; Li, H.W.; Tian, W.; Shen, Y.Z. Jugular Arginine Infusion Relieves Lipopolysaccharide-Triggered Inflammatory Stress and Improves Immunity Status of Lactating Dairy Cows. J. Dairy Sci. 2018, 101, 5961–5970. [Google Scholar] [CrossRef]
- Sordillo, L.M. Nutritional Strategies to Optimize Dairy Cattle Immunity. J. Dairy Sci. 2016, 99, 4967–4982. [Google Scholar] [CrossRef]
- Surai, P.F. Selenium in Nutrition and Health; Nottingham University Press: Nottingham, UK, 2006; ISBN 190476116X. [Google Scholar]
- Sun, L.; Wang, F.; Wu, Z.; Ma, L.; Baumrucker, C.; Bu, D. Comparison of Selenium Source in Preventing Oxidative Stress in Bovine Mammary Epithelial Cells. Animals 2020, 10, 842. [Google Scholar] [CrossRef]
- Han, L.; Pang, K.; Fu, T.; Phillips, C.J.C.; Gao, T. Nano-Selenium Supplementation Increases Selenoprotein (Sel) Gene Expression Profiles and Milk Selenium Concentration in Lactating Dairy Cows. Biol. Trace Elem. Res. 2021, 199, 113–119. [Google Scholar] [CrossRef]
- Elolimy, A.A.; Liang, Y.; Lopes, M.G.; Loor, J.J. Antioxidant Networks and the Microbiome as Components of Efficiency in Dairy Cattle. Livest. Sci. 2021, 251, 104656. [Google Scholar] [CrossRef]
- Gessner, D.K.; Brock, C.; Hof, L.M.; Most, E.; Koch, C.; Eder, K. Effects of Supplementation of Green Tea Extract on the Milk Performance of Peripartal Dairy Cows and the Expression of Stress Response Genes in the Liver. J. Anim. Sci. Biotechnol. 2020, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Gessner, D.K.; Ringseis, R.; Eder, K. Potential of Plant Polyphenols to Combat Oxidative Stress and Inflammatory Processes in Farm Animals. J. Anim. Physiol. Anim. Nutr. 2017, 101, 605–628. [Google Scholar] [CrossRef] [PubMed]
- Côrtes, C.; Palin, M.-F.; Gagnon, N.; Benchaar, C.; Lacasse, P.; Petit, H.v. Mammary Gene Expression and Activity of Antioxidant Enzymes and Concentration of the Mammalian Lignan Enterolactone in Milk and Plasma of Dairy Cows Fed Flax Lignans and Infused with Flax Oil in the Abomasum. Br. J. Nutr. 2012, 108, 1390–1398. [Google Scholar] [CrossRef]
- Schogor, A.L.B.; Palin, M.-F.; dos Santos, G.T.; Benchaar, C.; Lacasse, P.; Petit, H.v. Mammary Gene Expression and Activity of Antioxidant Enzymes and Oxidative Indicators in the Blood, Milk, Mammary Tissue and Ruminal Fluid of Dairy Cows Fed Flax Meal. Br. J. Nutr. 2013, 110, 1743–1750. [Google Scholar] [CrossRef]
- Palin, M.-F.; Côrtes, C.; Benchaar, C.; Lacasse, P.; Petit, H.v. MRNA Expression of Lipogenic Enzymes in Mammary Tissue and Fatty Acid Profile in Milk of Dairy Cows Fed Flax Hulls and Infused with Flax Oil in the Abomasum. Br. J. Nutr. 2014, 111, 1011–1020. [Google Scholar] [CrossRef]
- Purba, R.A.P.; Paengkoum, S.; Yuangklang, C.; Paengkoum, P.; Salem, A.Z.M.; Juan Boo, L. Mammary Gene Expressions and Oxidative Indicators in Ruminal Fluid, Blood, Milk, and Mammary Tissue of Dairy Goats Fed a Total Mixed Ration Containing Piper Meal (Piper betle L.). Ital. J. Anim. Sci. 2022, 21, 129–141. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
