Stability of Metabolomic Content during Sample Preparation: Blood and Brain Tissues
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Sample Collection
2.3. Extract and Homogenate Preparation
2.4. Sample Incubation and Preparation for NMR Analysis
2.5. NMR Measurements
2.6. Statistical Analysis
3. Results
3.1. Incubation of Serum at 4 °C
3.2. Whole Blood Extraction: Cryogenic Lysis versus Methanol Lysis
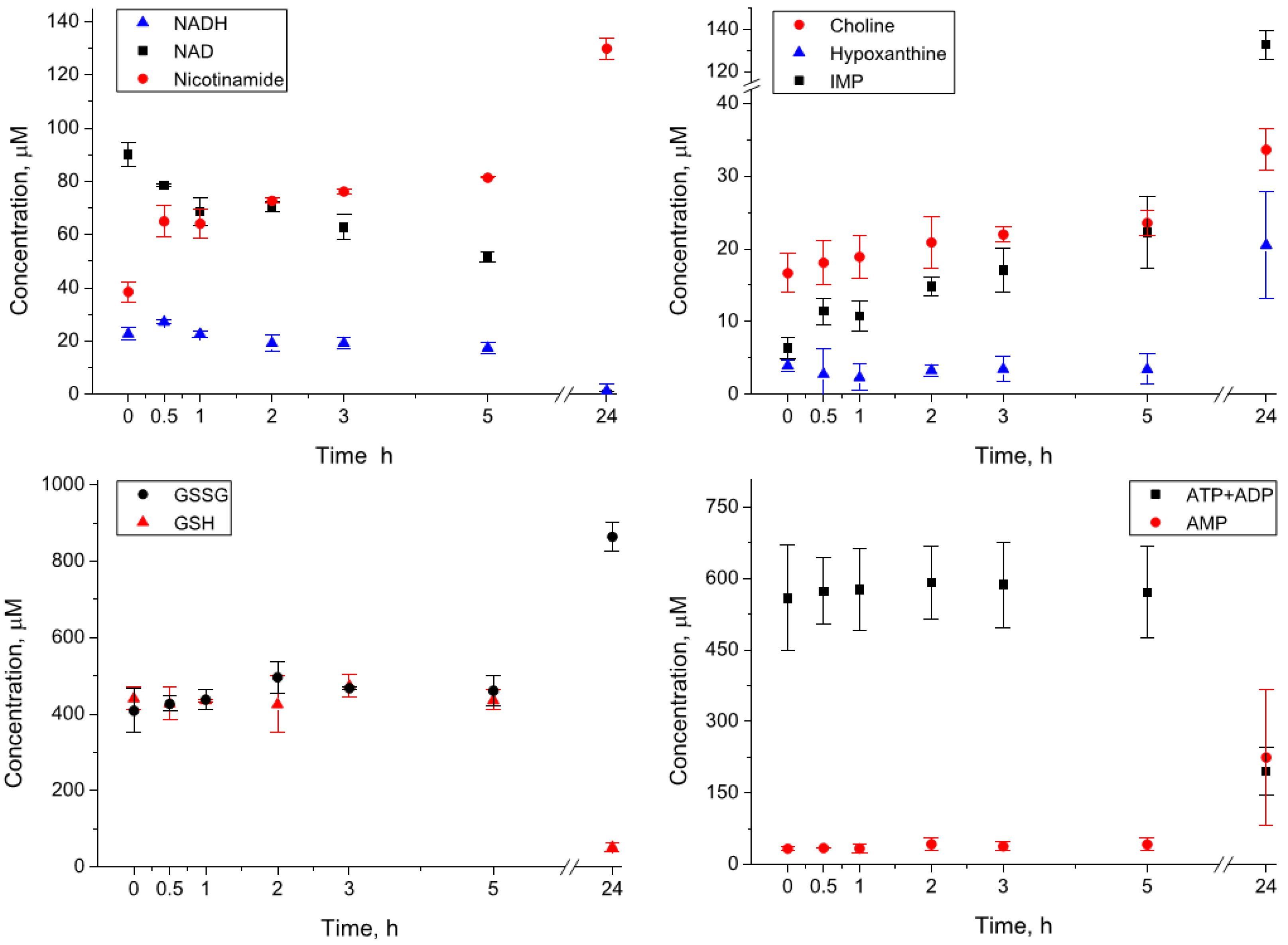
3.3. Incubation of the Whole Blood Extracts at 4 °C
3.4. Incubation of the Whole Blood Homogenates at 4 °C
3.5. Brain Extraction: Homogenization Followed by Enzyme Quenching versus Enzyme Quenching Followed by Homogenization
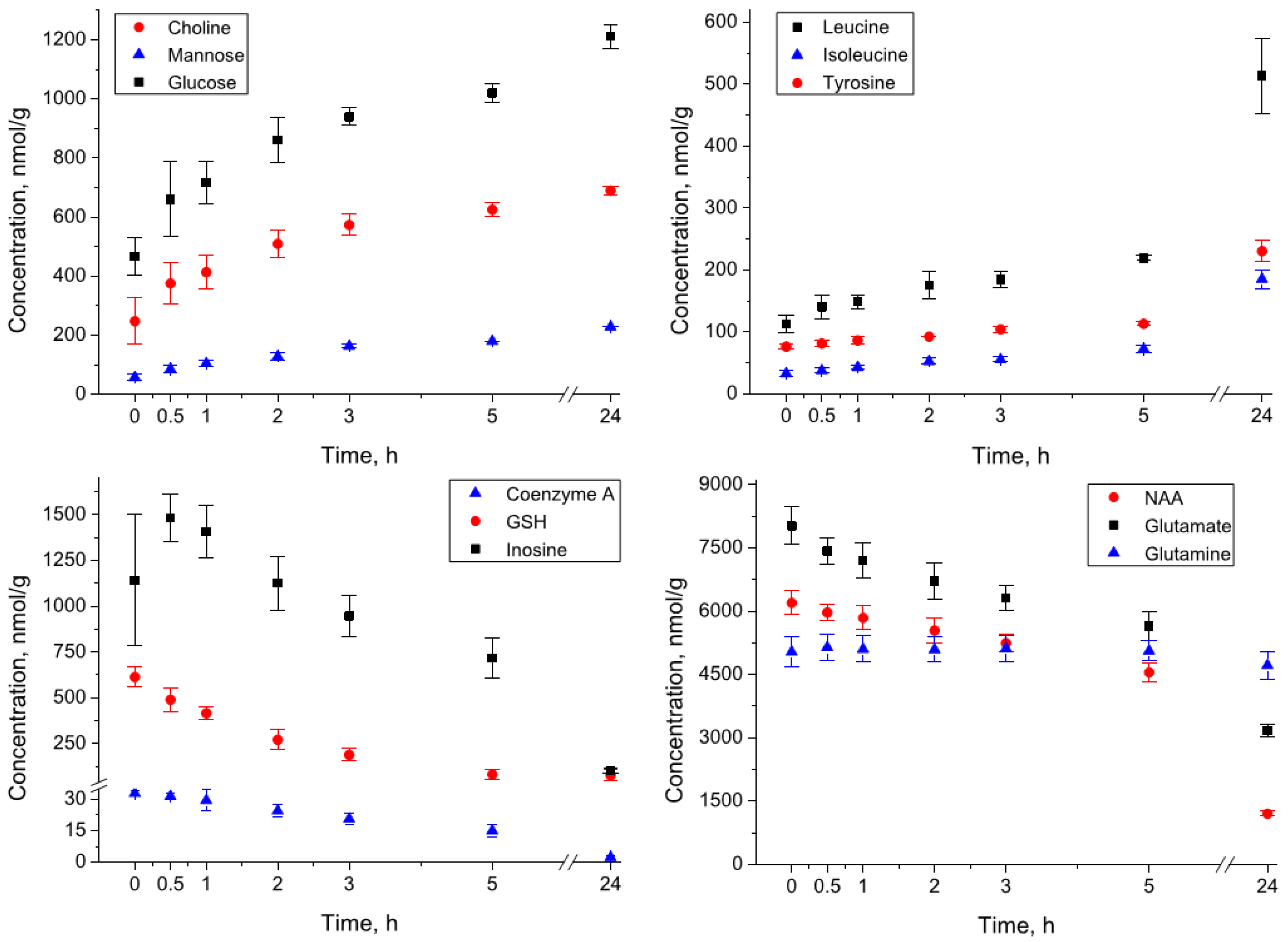
3.6. Incubation of the Brain Extracts at 4 °C
3.7. Incubation of the Brain Homogenates at 4 °C
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Müller, A.; Düchting, P.; Weiler, E.W. A Multiplex GC-MS/MS Technique for the Sensitive and Quantitative Single-Run Analysis of Acidic Phytohormones and Related Compounds, and Its Application to Arabidopsis Thaliana. Planta 2002, 216, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Annesley, T.M. Ion Suppression in Mass Spectrometry. Clin. Chem. 2003, 49, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Nagana Gowda, G.A.; Raftery, D. Overview of NMR Spectroscopy-Based Metabolomics: Opportunities and Challenges. In NMR-Based Metabolomics: Methods and Protocols; Gowda, G.A.N., Raftery, D., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 3–14. ISBN 978-1-4939-9690-2. [Google Scholar]
- Yin, P.; Peter, A.; Franken, H.; Zhao, X.; Neukamm, S.S.; Rosenbaum, L.; Lucio, M.; Zell, A.; Häring, H.-U.; Xu, G.; et al. Preanalytical Aspects and Sample Quality Assessment in Metabolomics Studies of Human Blood. Clin. Chem. 2013, 59, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Sánchez, B.; Priego-Capote, F.; Castro, M.D.L. de Metabolomics Analysis II. Preparation of Biological Samples Prior to Detection. TrAC Trends Anal. Chem. 2010, 29, 120–127. [Google Scholar] [CrossRef]
- Vuckovic, D. Current Trends and Challenges in Sample Preparation for Global Metabolomics Using Liquid Chromatography–Mass Spectrometry. Anal. Bioanal Chem. 2012, 403, 1523–1548. [Google Scholar] [CrossRef] [PubMed]
- Gil, A.; Siegel, D.; Permentier, H.; Reijngoud, D.-J.; Dekker, F.; Bischoff, R. Stability of Energy Metabolites-An Often Overlooked Issue in Metabolomics Studies: A Review: General. Electrophoresis 2015, 36, 2156–2169. [Google Scholar] [CrossRef]
- Zhou, J.; Yin, Y. Strategies for Large-Scale Targeted Metabolomics Quantification by Liquid Chromatography-Mass Spectrometry. Analyst 2016, 141, 6362–6373. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.-G.; Hu, J.; Wu, X.; Xu, Y.-J. The Recent Developments in Sample Preparation for Mass Spectrometry-Based Metabolomics. Crit. Rev. Anal. Chem. 2017, 47, 325–331. [Google Scholar] [CrossRef]
- Stevens, V.L.; Hoover, E.; Wang, Y.; Zanetti, K.A. Pre-Analytical Factors That Affect Metabolite Stability in Human Urine, Plasma, and Serum: A Review. Metabolites 2019, 9, 156. [Google Scholar] [CrossRef]
- González-Domínguez, R.; González-Domínguez, Á.; Sayago, A.; Fernández-Recamales, Á. Recommendations and Best Practices for Standardizing the Pre-Analytical Processing of Blood and Urine Samples in Metabolomics. Metabolites 2020, 10, 229. [Google Scholar] [CrossRef]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, C.J.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; et al. Mass Spectrometry-Based Metabolomics: A Guide for Annotation, Quantification and Best Reporting Practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Chaker, J.; Kristensen, D.M.; Halldorsson, T.I.; Olsen, S.F.; Monfort, C.; Chevrier, C.; Jégou, B.; David, A. Comprehensive Evaluation of Blood Plasma and Serum Sample Preparations for HRMS-Based Chemical Exposomics: Overlaps and Specificities. Anal. Chem. 2022, 94, 866–874. [Google Scholar] [CrossRef] [PubMed]
- Dona, A.C.; Jiménez, B.; Schäfer, H.; Humpfer, E.; Spraul, M.; Lewis, M.R.; Pearce, J.T.M.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Precision High-Throughput Proton NMR Spectroscopy of Human Urine, Serum, and Plasma for Large-Scale Metabolic Phenotyping. Anal. Chem. 2014, 86, 9887–9894. [Google Scholar] [CrossRef] [PubMed]
- Martias, C.; Baroukh, N.; Mavel, S.; Blasco, H.; Lefèvre, A.; Roch, L.; Montigny, F.; Gatien, J.; Schibler, L.; Dufour-Rainfray, D.; et al. Optimization of Sample Preparation for Metabolomics Exploration of Urine, Feces, Blood and Saliva in Humans Using Combined NMR and UHPLC-HRMS Platforms. Molecules 2021, 26, 4111. [Google Scholar] [CrossRef] [PubMed]
- Herrling, T. UV-Induced Free Radicals in the Skin Detected by ESR Spectroscopy and Imaging Using Nitroxides. Free Radic. Biol. Med. 2003, 35, 59–67. [Google Scholar] [CrossRef]
- Satake, M.; Dmochowska, B.; Nishikawa, Y.; Madaj, J.; Xue, J.; Guo, Z.; Reddy, D.V.; Rinaldi, P.L.; Monnier, V.M. Vitamin C Metabolomic Mapping in the Lens with 6-Deoxy-6-Fluoro-Ascorbic Acid and High-Resolution 19 F-NMR Spectroscopy. Invest. Ophthalmol. Vis. Sci. 2003, 44, 2047. [Google Scholar] [CrossRef] [PubMed]
- Bando, N.; Hayashi, H.; Wakamatsu, S.; Inakuma, T.; Miyoshi, M.; Nagao, A.; Yamauchi, R.; Terao, J. Participation of Singlet Oxygen in Ultraviolet-a-Induced Lipid Peroxidation in Mouse Skin and Its Inhibition by Dietary β-Carotene: An Ex Vivo Study. Free Radic. Biol. Med. 2004, 37, 1854–1863. [Google Scholar] [CrossRef]
- Son, H.-S.; Kim, K.M.; van den Berg, F.; Hwang, G.-S.; Park, W.-M.; Lee, C.-H.; Hong, Y.-S. 1H Nuclear Magnetic Resonance-Based Metabolomic Characterization of Wines by Grape Varieties and Production Areas. J. Agric. Food Chem. 2008, 56, 8007–8016. [Google Scholar] [CrossRef]
- Shehadul Islam, M.; Aryasomayajula, A.; Selvaganapathy, P. A Review on Macroscale and Microscale Cell Lysis Methods. Micromachines 2017, 8, 83. [Google Scholar] [CrossRef]
- Hirayama, A.; Sugimoto, M.; Suzuki, A.; Hatakeyama, Y.; Enomoto, A.; Harada, S.; Soga, T.; Tomita, M.; Takebayashi, T. Effects of Processing and Storage Conditions on Charged Metabolomic Profiles in Blood: CE and CEC. Electrophoresis 2015, 36, 2148–2155. [Google Scholar] [CrossRef]
- Miele, M.M.; Irving, B.A.; Wenrich, B.R.; Martin, P.L.; Rovnyak, D. Reproducibility and Stability of Aqueous Metabolite Levels in Extracted Serum by NMR Spectroscopy. Curr. Metab. 2017, 5, 45–54. [Google Scholar] [CrossRef]
- Dienel, G.A. Stop the Rot. Enzyme Inactivation at Brain Harvest Prevents Artifacts. J. Neurochem. 2021, 158, 1007–1031. [Google Scholar] [CrossRef] [PubMed]
- Tsentalovich, Y.P.; Zelentsova, E.A.; Yanshole, L.V.; Yanshole, V.V.; Odud, I.M. Most Abundant Metabolites in Tissues of Freshwater Fish Pike-Perch (Sander Lucioperca). Sci. Rep. 2020, 10, 17128. [Google Scholar] [CrossRef] [PubMed]
- Snytnikova, O.A.; Khlichkina, A.A.; Yanshole, L.V.; Yanshole, V.V.; Iskakov, I.A.; Egorova, E.V.; Stepakov, D.A.; Novoselov, V.P.; Tsentalovich, Y.P. Metabolomics of the Human Aqueous Humor. Metabolomics 2017, 13, 5. [Google Scholar] [CrossRef]
- Glinskikh, A.; Snytnikova, O.; Zelentsova, E.; Borisova, M.; Tsentalovich, Y.; Akulov, A. The Effect of Blood Contained in the Samples on the Metabolomic Profile of Mouse Brain Tissue: A Study by NMR Spectroscopy. Molecules 2021, 26, 3096. [Google Scholar] [CrossRef] [PubMed]
- Nagana Gowda, G.A.; Raftery, D. Whole Blood Metabolomics by 1 H NMR Spectroscopy Provides a New Opportunity To Evaluate Coenzymes and Antioxidants. Anal. Chem. 2017, 89, 4620–4627. [Google Scholar] [CrossRef]
- Paskevich, S.I.; Molchanov, M.V.; Timchenko, M.A.; Kutyshenko, V.P. Sample Pretreatment of Brain Tissues and Cerebrospinal Fluid for NMR Investigations. J. Anal. Chem. 2013, 68, 862–870. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. Can NMR Solve Some Significant Challenges in Metabolomics? J. Magn. Reson. 2015, 260, 144–160. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Djukovic, D.; Bettcher, L.F.; Gu, H.; Raftery, D. NMR-Guided Mass Spectrometry for Absolute Quantitation of Human Blood Metabolites. Anal. Chem. 2018, 90, 2001–2009. [Google Scholar] [CrossRef]
- Snytnikova, O.A.; Yanshole, L.V.; Iskakov, I.A.; Yanshole, V.V.; Chernykh, V.V.; Stepakov, D.A.; Novoselov, V.P.; Tsentalovich, Y.P. Quantitative Metabolomic Analysis of the Human Cornea and Aqueous Humor. Metabolomics 2017, 13, 152. [Google Scholar] [CrossRef]
- Yanshole, V.V.; Yanshole, L.V.; Snytnikova, O.A.; Tsentalovich, Y.P. Quantitative Metabolomic Analysis of Changes in the Lens and Aqueous Humor under Development of Age-Related Nuclear Cataract. Metabolomics 2019, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, M.; Alvarez, B.; Radi, R. One- and Two-Electron Oxidation of Thiols: Mechanisms, Kinetics and Biological Fates. Free Radic. Res. 2016, 50, 150–171. [Google Scholar] [CrossRef] [PubMed]
- Moingt, M.; Bressac, M.; Bélanger, D.; Amyot, M. Role of Ultra-Violet Radiation, Mercury and Copper on the Stability of Dissolved Glutathione in Natural and Artificial Freshwater and Saltwater. Chemosphere 2010, 80, 1314–1320. [Google Scholar] [CrossRef] [PubMed]
- Boyanton, B.L.; Blick, K.E. Stability Studies of Twenty-Four Analytes in Human Plasma and Serum. Clin. Chem. 2002, 48, 2242–2247. [Google Scholar] [CrossRef] [PubMed]
- Breier, M.; Wahl, S.; Prehn, C.; Fugmann, M.; Ferrari, U.; Weise, M.; Banning, F.; Seissler, J.; Grallert, H.; Adamski, J.; et al. Targeted Metabolomics Identifies Reliable and Stable Metabolites in Human Serum and Plasma Samples. PLoS ONE 2014, 9, e89728. [Google Scholar] [CrossRef]
- Haid, M.; Muschet, C.; Wahl, S.; Römisch-Margl, W.; Prehn, C.; Möller, G.; Adamski, J. Long-Term Stability of Human Plasma Metabolites during Storage at −80 °C. J. Proteome Res. 2018, 17, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Suzuki, M.; Kobayashi, T.; Yoshida, M. Differences in Metabolite Profiles Caused by Pre-Analytical Blood Processing Procedures. J. Biosci. Bioeng. 2018, 125, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Ledvina, A.R.; Ewles, M.; Pang, Y.; Cape, S. Whole Blood Stability in Quantitative Bioanalysis. Bioanalysis 2019, 11, 1885–1897. [Google Scholar] [CrossRef]
- Bernini, P.; Bertini, I.; Luchinat, C.; Nincheri, P.; Staderini, S.; Turano, P. Standard Operating Procedures for Pre-Analytical Handling of Blood and Urine for Metabolomic Studies and Biobanks. J. Biomol. NMR 2011, 49, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Loo, R.L.; Lodge, S.; Kimhofer, T.; Bong, S.-H.; Begum, S.; Whiley, L.; Gray, N.; Lindon, J.C.; Nitschke, P.; Lawler, N.G.; et al. Quantitative In-Vitro Diagnostic NMR Spectroscopy for Lipoprotein and Metabolite Measurements in Plasma and Serum: Recommendations for Analytical Artifact Minimization with Special Reference to COVID-19/SARS-CoV-2 Samples. J. Proteome Res. 2020, 19, 4428–4441. [Google Scholar] [CrossRef] [PubMed]
- Dienel, G.A. Metabolomic Assays of Postmortem Brain Extracts: Pitfalls in Extrapolation of Concentrations of Glucose and Amino Acids to Metabolic Dysregulation In Vivo in Neurological Diseases. Neurochem. Res. 2019, 44, 2239–2260. [Google Scholar] [CrossRef] [PubMed]
- Dienel, G.A. Metabolomic and Imaging Mass Spectrometric Assays of Labile Brain Metabolites: Critical Importance of Brain Harvest Procedures. Neurochem. Res. 2020, 45, 2586–2606. [Google Scholar] [CrossRef] [PubMed]
- Ogushi, S.; Lawson, J.W.; Dobson, G.P.; Veech, R.L.; Uyeda, K. A New Transient Activator of Phosphofructokinase during Initiation of Rapid Glycolysis in Brain. J. Biol. Chem. 1990, 265, 10943–10949. [Google Scholar] [CrossRef]
- Gruetter, R.; Ugurbil, K.; Seaquist, E.R. Steady-State Cerebral Glucose Concentrations and Transport in the Human Brain. J. Neurochem. 2002, 70, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Reinstrup, P.; Mellergård, P.; Uski, T. Intracerebral Microdialysis in Clinical Practice: Baseline Values for Chemical Markers during Wakefulness, Anesthesia, and Neurosurgery. Neurosurgery 2000, 47, 10. [Google Scholar]
- Boumezbeur, F.; Petersen, K.F.; Cline, G.W.; Mason, G.F.; Behar, K.L.; Shulman, G.I.; Rothman, D.L. The Contribution of Blood Lactate to Brain Energy Metabolism in Humans Measured by Dynamic 13C Nuclear Magnetic Resonance Spectroscopy. J. Neurosci. 2010, 30, 13983–13991. [Google Scholar] [CrossRef]
- Stavinoha, W.B.; Weintraub, S.T.; Modak, A.T. The use of microwave heating to inactivate cholinesterase in the rat brain prior to analysis for acetylcholine. J. Neurochem. 1973, 20, 361–371. [Google Scholar] [CrossRef]
- Pontén, U.; Ratcheson, R.A.; Siesjö, B.K. Metabolic changes in the brains of mice frozen in liquid nitrogen. J. Neurochem. 1973, 21, 1121–1126. [Google Scholar] [CrossRef]
- Veech, R.L.; Harris, R.L.; Veloso, D.; Veech, E.H. Freeze-blowing: A new technique for the study of brain In Vivo. J. Neurochem. 1973, 20, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Levine, J.; Panchalingam, K.; McClure, R.J.; Gershon, S.; Pettegrew, J.W. Stability of CSF Metabolites Measured by Proton NMR: Short Communication. J. Neural. Transm. 2000, 107, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Rosenling, T.; Slim, C.L.; Christin, C.; Coulier, L.; Shi, S.; Stoop, M.P.; Bosman, J.; Suits, F.; Horvatovich, P.L.; Stockhofe-Zurwieden, N.; et al. The Effect of Preanalytical Factors on Stability of the Proteome and Selected Metabolites in Cerebrospinal Fluid (CSF). J. Proteome Res. 2009, 8, 5511–5522. [Google Scholar] [CrossRef] [PubMed]
- Rosenling, T.; Stoop, M.P.; Smolinska, A.; Muilwijk, B.; Coulier, L.; Shi, S.; Dane, A.; Christin, C.; Suits, F.; Horvatovich, P.L.; et al. The Impact of Delayed Storage on the Measured Proteome and Metabolome of Human Cerebrospinal Fluid. Clin. Chem. 2011, 57, 1703–1711. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, B.; Voronina, E.; Schipke, C.; Peters, O.; Parr, M.K.; Díaz-Hernández, M.D.; Schlörer, N.E. Pursuing Experimental Reproducibility: An Efficient Protocol for the Preparation of Cerebrospinal Fluid Samples for NMR-Based Metabolomics and Analysis of Sample Degradation. Metabolites 2020, 10, 251. [Google Scholar] [CrossRef] [PubMed]

| Sample Type | Serum | Extract | Homogenate | ||
|---|---|---|---|---|---|
| Time, h | 0 | 0 | 24 | 0 | 24 |
| Amino acids and their derivatives, peptides | |||||
| Alanine | 471 ± 22 | 284 ± 19 | 275 ± 15 | 330 ± 60 | 400 ± 50 |
| Asparagine | 120 ± 50 | 105 ± 9 | 94 ± 5 | 105 ± 9 | 94 ± 6 |
| Aspartate | 260 ± 40 | 109 ± 17 | 90 ± 7 | 95 ± 15 | 102 ± 12 |
| Ac-Carnitine | 10 ± 3 | 19 ± 1 | 16.5 ± 0.4 | 14.9 ± 2.5 | 15.3 ± 2.2 |
| Betaine | 45.4 ± 2.9 | 51.9 ± 2.8 | 61.2 ± 2.9 | 50.9 ± 1.1 | 55 ± 6 |
| Creatine | 43 ± 8 | 160 ± 7 | 172 ± 12 | 138 ± 16 | 144 ± 28 |
| Creatinine | 97 ± 11 | 90 ± 4 | 87 ± 6 | 86 ± 5 | 92 ± 9 |
| Ergothioneine | <LOD 1 | 154 ± 25 | 98 ± 4 | 70 ± 50 | 70 ± 60 |
| Glutamate | 160 ± 60 | 183 ± 11 | 213 ± 12 | 230 ± 23 | 259 ± 16 |
| Glutamine | 680 ± 70 | 520 ± 30 | 530 ± 40 | 500 ± 80 | 460 ± 90 |
| Glycine | 210 ± 4 | 241 ± 21 | 233 ± 13 | 250 ± 40 | 353 ± 29 * |
| GSH | <LOD | 383 ± 19 | 175 ± 29 * | 430 ± 50 | 46 ± 11 * |
| GSSG | 671 ± 9 | 575 ± 17 | 420 ± 50 * | 554 ± 25 | 810 ± 50 * |
| Histidine | 114 ± 28 | 73.3 ± 1.2 | 77 ± 4 | 75 ± 8 | 78.4 ± 2.1 |
| Isoleucine | 65 ± 3 | 61 ± 7 | 51 ± 5 | 50 ± 9 | 63.3 ± 1.9 * |
| Leucine | 122 ± 7 | 107 ± 7 | 105 ± 6 | 104 ± 9 | 118.8 ± 2.8 * |
| Lysine | 189 ± 25 | 134 ± 8 | 131 ± 12 | 135 ± 7 | 142 ± 18 |
| Ornithine | 91 ± 6 | 87 ± 8 | 78 ± 4 | 126 ± 17 | 153 ± 6 |
| Phenylalanine | 61 ± 17 | 49.6 ± 1.9 | 49 ± 3 | 50.4 ± 0.8 | 62 ± 4 * |
| Proline | 356 ± 17 | 192 ± 17 | 161 ± 5 | 214 ± 27 | 221 ± 23 |
| Serine | 110 ± 50 | 75 ± 11 | 62 ± 6 | 76 ± 10 | 117 ± 7 * |
| Threonine | 178 ± 7 | 136 ± 14 | 135 ± 8 | 153 ± 7 | 165 ± 11 |
| Tryptophan | 70 ± 9 | 38.2 ± 2.6 | 38.5 ± 0.9 | 40 ± 5 | 50 ± 5 |
| Tyrosine | 66 ± 14 | 48.1 ± 2.1 | 48 ± 3 | 54 ± 3 | 62 ± 4 |
| Valine | 186.0 ± 1.2 | 186 ± 17 | 182 ± 13 | 151 ± 8 | 186.3 ± 2.9 * |
| Organic acids | |||||
| Acetate | 580 ± 40 | 66 ± 11 | 104 ± 6 * | 67 ± 20 | 92 ± 9 * |
| α-Aminobutyrate | 26 ± 11 | 13.7 ± 1 | 15.97 ± 0.15 | 12.4 ± 1.4 | 15.1 ± 1.3 |
| β-Hydroxybutyrate | 33 ± 11 | 27.2 ± 2.6 | 22.4 ± 1.1 | 17 ± 6 | 16.3 ± 1.5 |
| Formate | 69 ± 29 | 46 ± 20 | 67 ± 4 | 51 ± 6 | 44 ± 11 |
| Fumarate | 4.3 ± 2.0 | 1.6 ± 0.3 | 1.8 ± 0.3 | 2.4 ± 0.4 | 4 ± 1 |
| Isobutyrate | 6.6 ± 2.5 | 7.4 ± 1.2 | 9.1 ± 0.9 | 6.1 ± 1.9 | 8.8 ± 1.1 |
| α-Ketoisovalerate | 13 ± 5 | 9.2 ± 1.2 | 7.6 ± 1.2 | 6 ± 0.21 | 6.6 ± 1.6 |
| Lactate | 2770 ± 60 | 1230 ± 30 | 1270 ± 90 | 1210 ± 50 | 2120 ± 70 * |
| Pyroglutamate | 110 ± 70 | 13.1 ± 2.4 | 22 ± 4 | 20 ± 4 | 77 ± 6 * |
| Pyruvate | 11 ± 6 | 17 ± 4 | 29.8 ± 1.8 * | 12 ± 7 | 86 ± 15 * |
| Succinate | 3.2 ± 2.7 | 3.91 ± 0.13 | 4.6 ± 0.6 | 3.9 ± 0.3 | 9.4 ± 1.6 |
| Alcohols, amines, amides, sugars | |||||
| Acetone | 11.4 ± 1.2 | 4.9 ± 0.5 | 4.3 ± 0.5 | 3.7 ± 1.2 | 3.7 ± 1.5 |
| Choline | 13.8 ± 2.9 | 10.2 ± 2.0 | 11.0 ± 2.0 | 16.7 ± 2.7 | 33.7 ± 2.8 * |
| Dimethylamine | 18.3 ± 1.3 | 8.1 ± 0.4 | 7.3 ± 0.4 | 7.4 ± 0.4 | 7.7 ± 0.9 |
| Glucose | 3740 ± 60 | 3700 ± 400 | 4400 ± 900 | 3390 ± 110 | 5190 ± 70 * |
| Gl-PhCholine | 22.1 ± 1.0 | 21.8 ± 0.6 | 19.3 ± 1.7 | 18.3 ± 0.5 | 17.1 ± 2.0 |
| Mannose | 33 ± 15 | 42.0 ± 2.0 | 31.2 ± 2.3 * | 41 ± 3 | <LOD * |
| Nicotinamide | <LOD | 3.5 ± 2.5 | 5.1 ± 0.9 | 39 ± 4 3 | 130 ± 4 * |
| PhCholine | 4.08 ± 0.23 | 5.7 ± 0.7 | 5.2 ± 1 | 7.1 ± 1.3 | 8.7 ± 1.1 |
| Nitrogenous bases, nucleotides, nucleosides | |||||
| ADP + ATP | <LOD | 664 ± 14 | 460 ± 50 * | 560 ± 110 | 200 ± 50 * |
| ADP ribose | <LOD | 7 ± 6 | 11 ± 4 | 41 ± 3 3 | 78 ± 5 * |
| AMP | <LOD | 8.9 ± 0.7 | 15.6 ± 1.3 * | 33 ± 5 | 220 ± 140 * |
| Hypoxanthine | <LOD | 2.1 ± 2.1 | 3.2 ± 0.6 | 3.9 ± 0.8 | 21 ± 7 * |
| IMP | <LOD | 3.2 ± 1.9 | 2.8 ± 1.6 | 6.4 ± 1.5 | 133 ± 7 * |
| NAD | <LOD | 22 ± 6 | 28.1 ± 0.3 | 90 ± 4 3 | <LOD * |
| NADH | <LOD | 4.6 ± 2.0 | 0.8 ± 1.0 * | 22.8 ± 2.2 2 | 1.3 ± 2.8 * |
| NADP | <LOD | 14.9 ± 1.8 | 12.8 ± 1.1 | 6.5 ± 1.6 | 8 ± 0.4 |
| Sample type | Extract | Homogenate | ||
|---|---|---|---|---|
| Time, h | 0 | 24 | 0 | 24 |
| Amino acids and their derivatives, peptides | ||||
| Alanine | 384 ± 15 | 388 ± 10 | 530 ± 40 | 1043 ± 29 * |
| Asparagine | 480 ± 100 | 360 ± 150 | 143 ± 11 | 224 ± 5 * |
| Aspartate | 2270 ± 80 | 2260 ± 70 | 3200 ± 150 | 8030 ± 240 * |
| Ac-Carnitine | 25 ± 5 | 23.7 ± 2.4 | 22 ± 6 | 8.4 ± 1.3 * |
| Carnosine | 74.4 ± 2.2 | 69.9 ± 1.8 | 85 ± 8 | 89 ± 6 |
| Creatine | 7490 ± 190 | 7540 ± 250 | 8570 ± 130 | 7600 ± 280 * |
| Glutamate | 7430 ± 160 | 7670 ± 230 | 8000 ± 400 | 3170 ± 150 * |
| Glutamine | 4110 ± 80 | 4150 ± 150 | 5020 ± 110 | 4700 ± 150 |
| Glycine | 830 ± 120 | 820 ± 110 | 940 ± 50 | 2550 ± 190 * |
| GSH | 740 ± 70 | 690 ± 80 | 610 ± 50 | 80 ± 30 * |
| GSSG | 170 ± 30 | 168 ± 25 | <LOD | <LOD |
| Histidine | 43.9 ± 2.5 | 49 ± 5 | 69 ± 8 | 370 ± 40 * |
| Isoleucine | 17.5 ± 2.4 | 20.9 ± 2.7 | 33 ± 4 | 185 ± 15 * |
| Leucine | 65 ± 7 | 68 ± 5 | 113 ± 14 | 510 ± 60 * |
| N-acetylaspartate | 5630 ± 90 | 5650 ± 100 | 6200 ± 270 | 1210 ± 50 * |
| Pantothenate | 15.3 ± 0.8 | 17.1 ± 0.9 | 23.7 ± 2.8 | 42 ± 5 * |
| Phenylalanine | 81.9 ± 1.6 | 80 ± 12 | 94 ± 11 | 700 ± 60 * |
| Serine | 510 ± 40 | 493 ± 16 | 550 ± 30 | 1248 ± 16 * |
| Threonine | 420 ± 70 | 430 ± 50 | 340 ± 50 | 610 ± 80 * |
| Tryptophan | 16 ± 5 | 20 ± 4 | 27.5 ± 2.7 | 68 ± 7 * |
| Tyrosine | 54 ± 4 | 51.2 ± 1.9 | 76 ± 4 | 231 ± 17 * |
| Valine | 42 ± 5 | 44 ± 4 | 58 ± 8 | 197 ± 8 * |
| Organic acids | ||||
| Acetate | 158.7 ± 1.4 | 168 ± 10 | 390 ± 90 | 4050 ± 100 * |
| α-Aminobutyrate | 5.4 ± 1.6 | 8.6 ± 2.3 | 5.6 ± 1.0 | 5.7 ± 1.2 |
| γ-Aminobutyrate | 1710 ± 180 | 1670 ± 150 | 2800 ± 400 | 7890 ± 230 * |
| Ascorbate | 640 ± 40 | 644 ± 28 | 844 ± 10 | 517 ± 28 * |
| Formate | 130 ± 11 | 132 ± 9 | 135 ± 6 | 132 ± 15 |
| Fumarate | 12.7 ± 2.1 | 15.4 ± 0.8 | 76.1 ± 2.5 | 48.8 ± 0.7 * |
| Isobutyrate | 8.4 ± 1.6 | 6.6 ± 1.1 | 5.2 ± 1.3 | 5.6 ± 1.9 |
| Lactate | 6500 ± 300 | 6500 ± 300 | 8000 ± 210 | 7040 ± 280 * |
| Pyroglutamate | <LOD | <LOD | 96 ± 21 | 473 ± 11 * |
| Pyruvate | 5.3 ± 1.6 | 11.3 ± 1.3 * | 5.5 ± 2.3 | 5.7 ± 1.3 |
| Succinate | 416 ± 12 | 416 ± 8 | 16.8 ± 2.0 | 33.6 ± 0.6 * |
| Taurine | 3060 ± 140 | 3060 ± 130 | 3800 ± 50 | 3622 ± 27 * |
| Alcohols, amines, amides, sugars | ||||
| Choline | 52 ± 6 | 52 ± 3 | 250 ± 80 | 691 ± 14 * |
| Glucose | <LOD | <LOD | 470 ± 60 | 1210 ± 40 * |
| Gl-PhCholine | 650 ± 40 | 650 ± 40 | 610 ± 60 | 10 ± 8 * |
| Glycerol | 164 ± 15 | 158 ± 9 | 320 ± 40 | 1060 ± 40 * |
| myo-Inositol | 6190 ± 160 | 6190 ± 160 | 6763 ± 23 | 6763 ± 23 |
| scyllo-Inositol | 96 ± 6 | 90 ± 14 | 84.8 ± 2.6 | 86 ± 4 |
| Mannose | <LOD | <LOD | 58 ± 13 | 229.3 ± 1.1 * |
| Nicotinamide | 80 ± 30 | 81 ± 21 | 159 ± 22 | 110 ± 24 |
| PhCholine | 305 ± 16 | 301 ± 19 | 480 ± 60 | 833 ± 14 * |
| Phosphoethanolamine | 800 ± 30 | 830 ± 40 | 800 ± 17 | 600 ± 30 * |
| Nitrogenous bases, nucleotides, nucleosides | ||||
| Adenosine | 193 ± 6 | 201 ± 16 | 500 ± 400 | 7 ± 6 * |
| ADP | 236 ± 17 | 230 ± 40 | 53 ± 11 | 10.9 ± 1.9 * |
| ADP ribose | 110 ± 30 | 120 ± 30 | 120 ± 90 | 4 ± 3 * |
| AMP | 1430 ± 70 | 1430 ± 60 | 81 ± 27 | 32 ± 6 * |
| ATP | 161 ± 28 | 144 ± 15 | 60 ± 50 | 12 ± 3 * |
| Coenzyme A | 26.3 ± 0.6 | 25.3 ± 2.8 | 32.9 ± 1.1 | 2.4 ± 0.8 * |
| Cytidine | 19 ± 8 | 24 ± 6 | 46 ± 6 | 67 ± 6 * |
| GMP | 196 ± 27 | 191 ± 18 | <LOD | <LOD |
| GTP | 136 ± 7 | 127 ± 2.9 | 69 ± 16 | 21 ± 5 * |
| Guanosine | 8.5 ± 1.2 | 5 ± 3 | 35 ± 23 | 1 ± 0.6 * |
| Hypoxanthine | <LOD | <LOD | 290 ± 120 | 1230 ± 70 * |
| IMP | 140 ± 40 | 140 ± 30 | <LOD | <LOD |
| Inosine | 120 ± 16 | 110 ± 18 | 1100 ± 400 | 100 ± 9 * |
| NAD | 160 ± 40 | 167 ± 30 | <LOD | <LOD |
| NADH | 19.5 ± 2.8 | 15 ± 3 | <LOD | <LOD |
| NADPH | 10.2 ± 1.1 | 4.3 ± 1.3 * | <LOD | <LOD |
| UMP | 90 ± 6 | 95 ± 9 | <LOD | <LOD |
| Uracyl | <LOD | <LOD | 44 ± 12 | 261 ± 6 * |
| Uridine | 48 ± 4 | 49 ± 3 | 128 ± 20 | 12 ± 10 * |
| Xanthine | <LOD | <LOD | 117 ± 30 | 280 ± 13 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fomenko, M.V.; Yanshole, L.V.; Tsentalovich, Y.P. Stability of Metabolomic Content during Sample Preparation: Blood and Brain Tissues. Metabolites 2022, 12, 811. https://doi.org/10.3390/metabo12090811
Fomenko MV, Yanshole LV, Tsentalovich YP. Stability of Metabolomic Content during Sample Preparation: Blood and Brain Tissues. Metabolites. 2022; 12(9):811. https://doi.org/10.3390/metabo12090811
Chicago/Turabian StyleFomenko, Maxim V., Lyudmila V. Yanshole, and Yuri P. Tsentalovich. 2022. "Stability of Metabolomic Content during Sample Preparation: Blood and Brain Tissues" Metabolites 12, no. 9: 811. https://doi.org/10.3390/metabo12090811
APA StyleFomenko, M. V., Yanshole, L. V., & Tsentalovich, Y. P. (2022). Stability of Metabolomic Content during Sample Preparation: Blood and Brain Tissues. Metabolites, 12(9), 811. https://doi.org/10.3390/metabo12090811







