Heterologous Expression and Characterization of Plant Wax Ester Producing Enzymes
Abstract
1. Introduction
2. Results
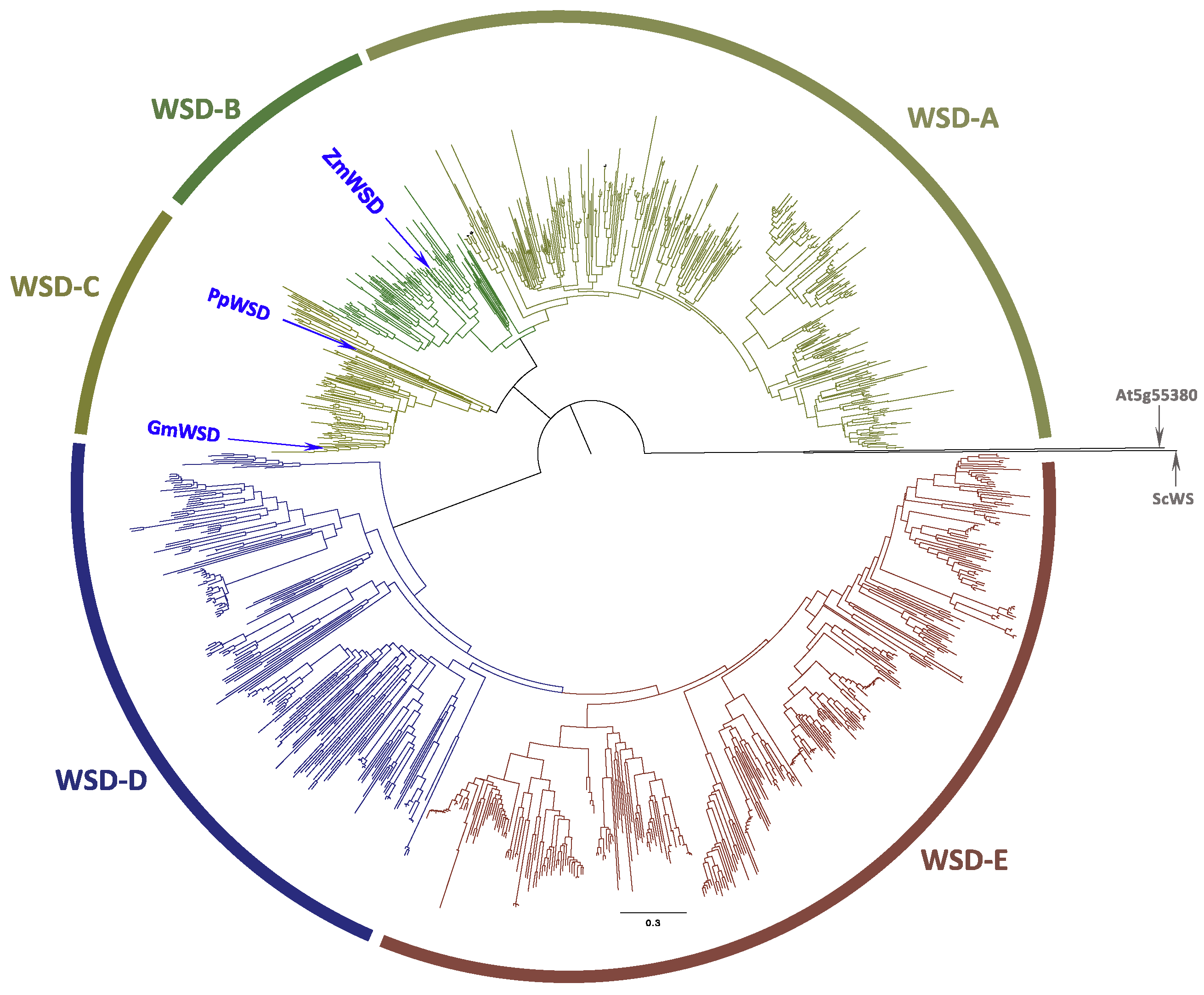
2.1. Sequence Characterization and Phylogenetic Classification of WS and WSD Proteins
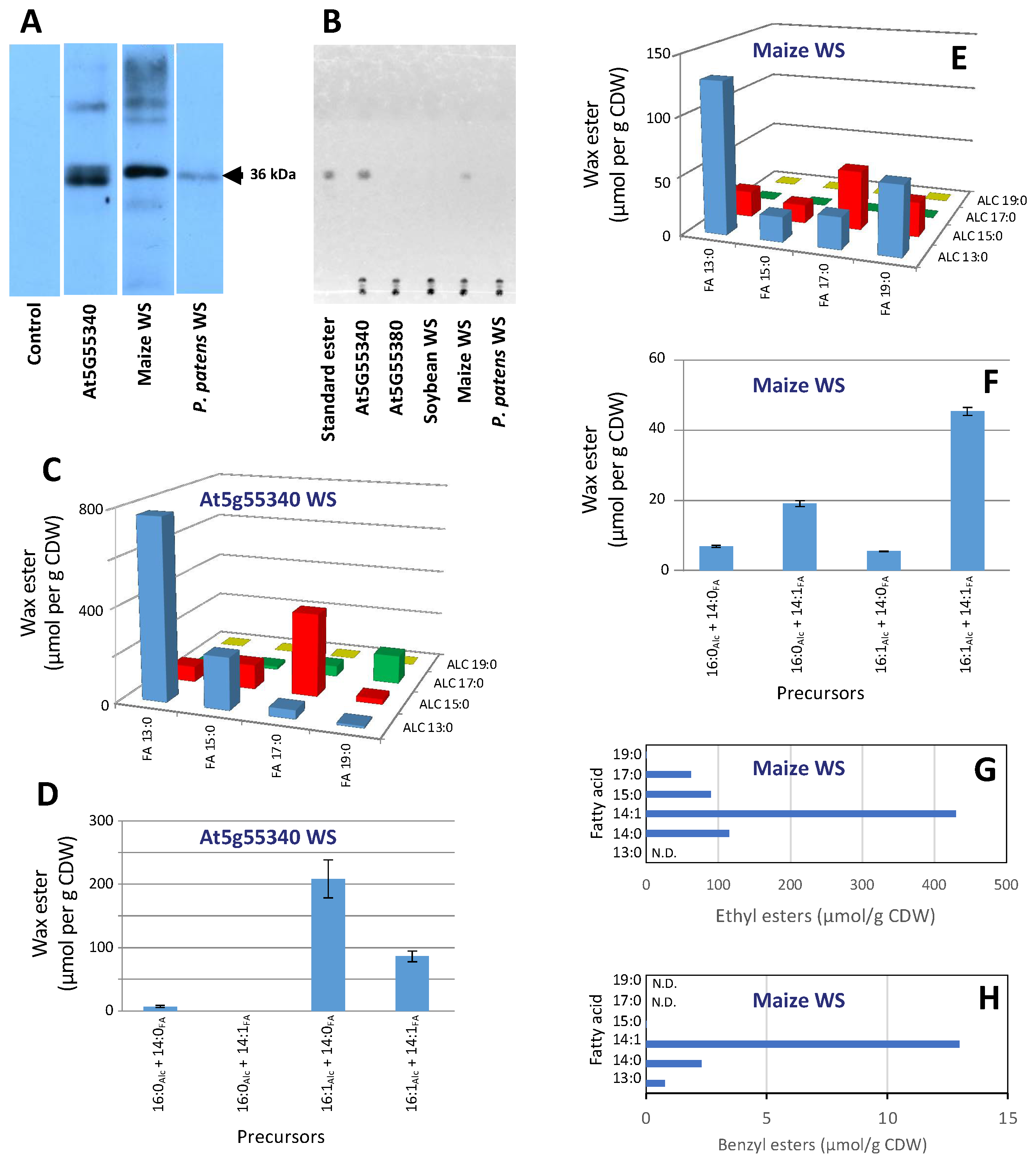
2.2. Functional Characterization of the WS Proteins by Heterologous Expression in Yeast
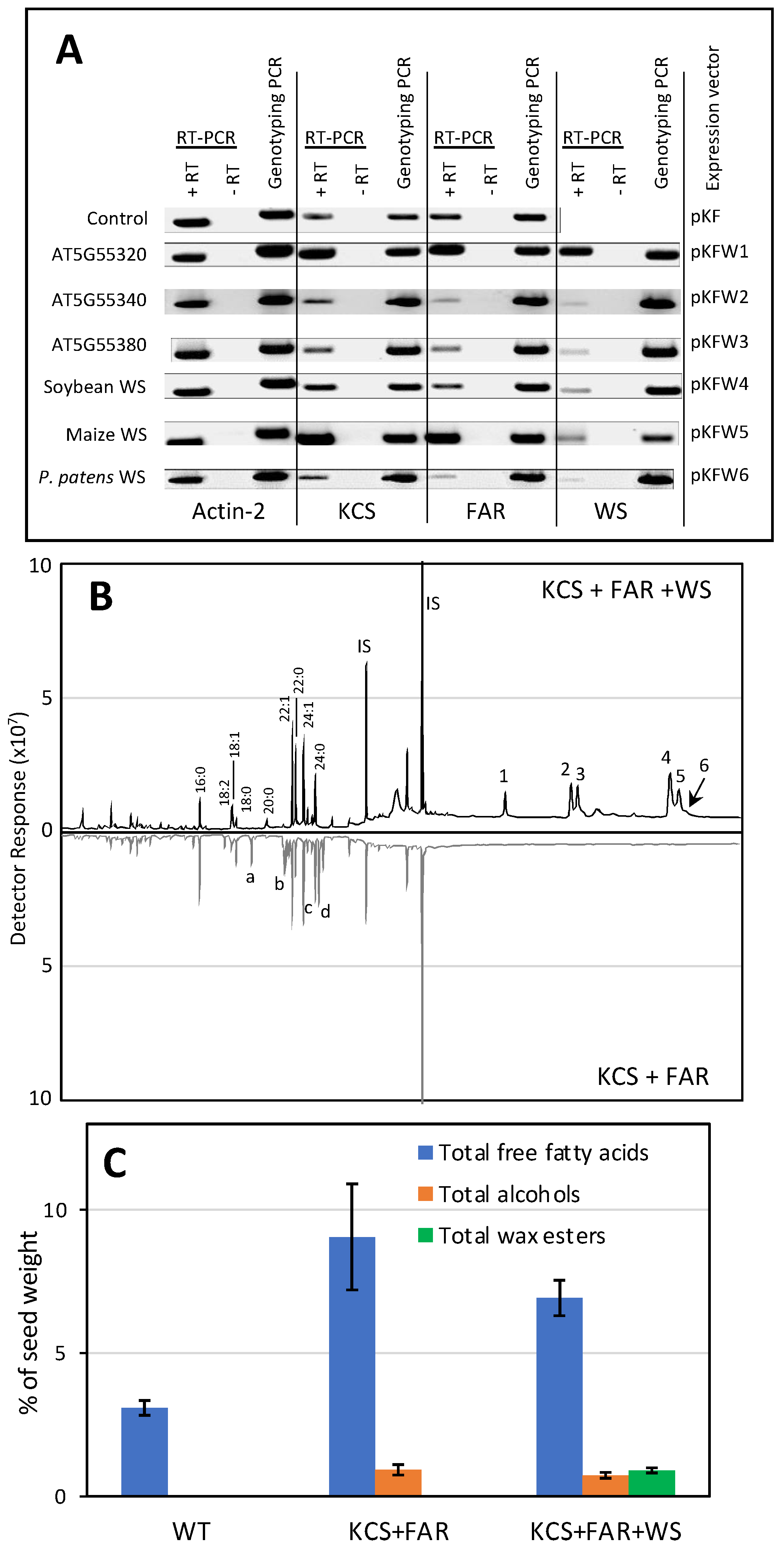
2.3. Functional Characterization of the WS Proteins by Heterologous Expression in Arabidopsis Seeds
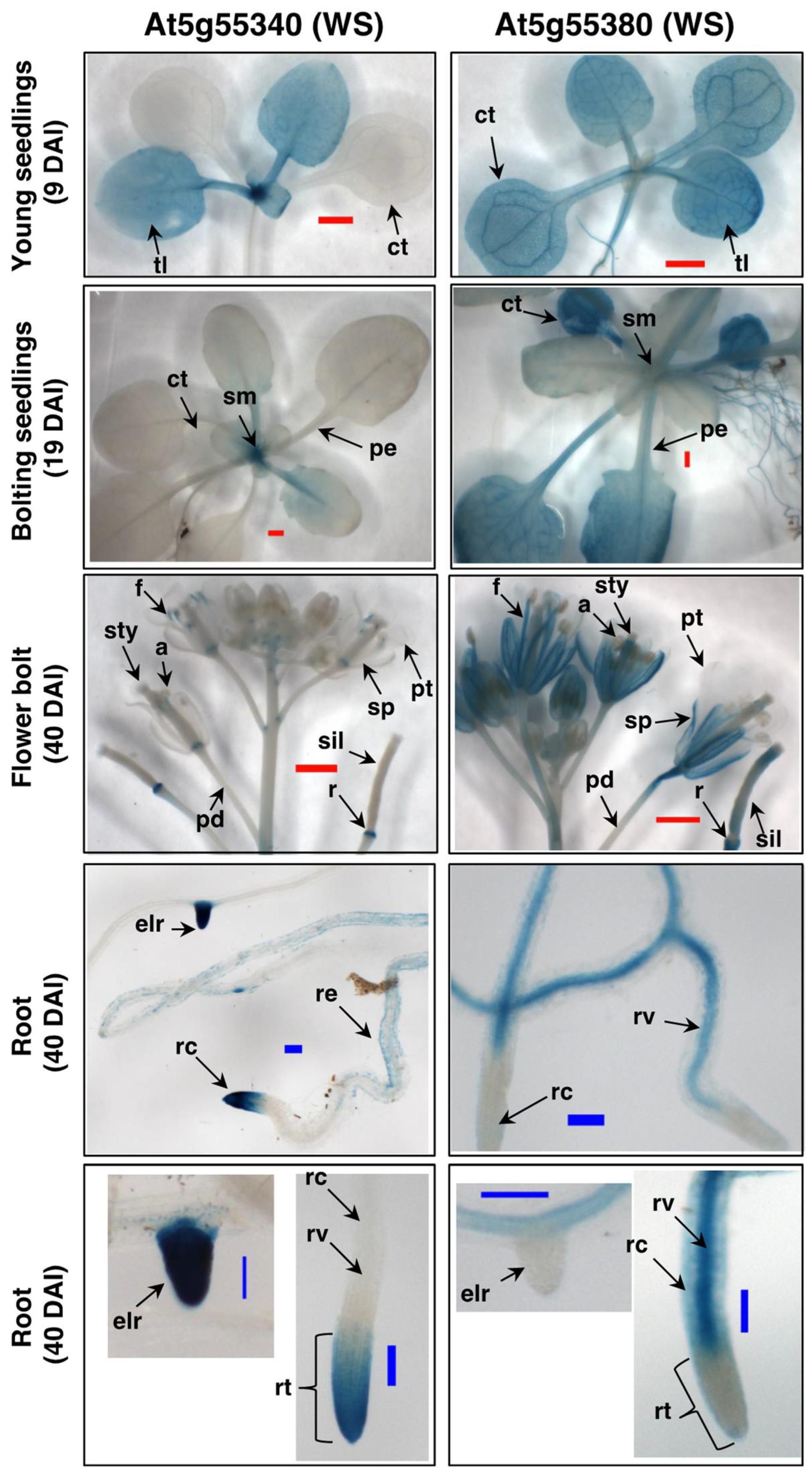
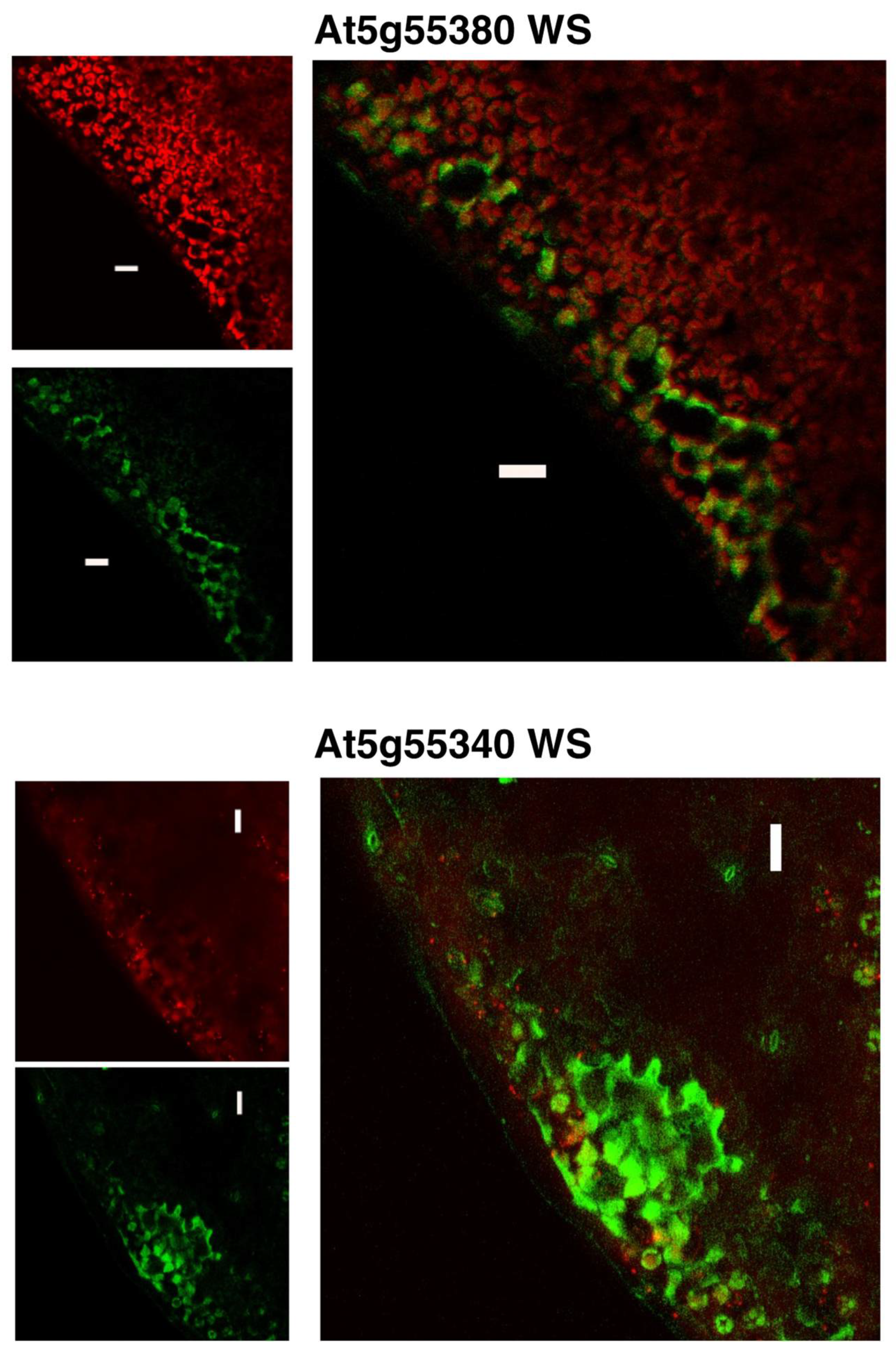
2.4. Expression Patterns of the Arabidopsis WS Genes
2.5. Co-Expression Gene Network Associated with the Arabidopsis MBOAT-Like WS and WSD Coding Genes
3. Discussion
4. Materials and Methods
4.1. Phylogenetic Analysis
4.2. Construction of Plant Expression Cassettes
4.3. Heterologous Expression in Saccharomyces Cerevisiae
4.4. Transgenic Plant Materials
4.5. Reverse Transcription-PCR
4.6. Extraction of Lipids from Seed Samples
4.7. Extraction of Lipids from Yeast Samples
4.8. Thin-Layer Chromatography
4.9. GC-MS Analysis
4.10. Western Blot Analysis
4.11. Histological GUS Activity Analysis
4.12. GFP Fluorescence Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wolfmeier, U.; Schmidt, H.; Heinrichs, F.-L.; Michalczyk, G.; Payer, W.; Dietsche, W.; Boehlke, K.; Hohner, G.; Wildgruber, J. Waxes. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley-VCH Verlag GmbH & Co. KGaA: New York, NY, USA, 2000. [Google Scholar]
- Kolattukudy, P.E. Chemistry and Biochemistry of Natural Waxes; Elsevier: Amsterdam, The Netherlands, 1976. [Google Scholar]
- Crisp, S.; Eaton, R.F.; Tinsley, H.M. Scheme for the identification of sperm whale oil and its products in commercial formulations and in leather articles. Analyst 1984, 109, 1497–1502. [Google Scholar] [CrossRef]
- Dieterichs, E.E.F. A Practical Treatise on Friction, Lubrication, Fats and Oils, 2nd ed.; H.C. Baird & Co.: Philadelphia, PA, USA, 1916; Chapter xvi; 137p. [Google Scholar]
- Phillips, S.J.; Comus, P.W.; Museum, A.-S.D. A Natural History of the Sonoran Desert; Arizona-Sonora Desert Museum Press: Tucson, AZ, USA, 2000. [Google Scholar]
- Gentry, H.S. The natural history of Jojoba (Simmondsia chinensis) and its cultural aspects. Econ. Bot. 1958, 12, 261–295. [Google Scholar] [CrossRef]
- Hepburn, H.R.; Pirk, C.W.W.; Duangphakdee, O. The chemistry of beeswax. In Honeybee Nests: Composition, Structure, Function; Springer: Berlin/Heidelberg, Germany, 2014; pp. 319–339. [Google Scholar]
- Crane, E. The World History of Beekeeping and Honey Hunting; Routledge: New York, NY, USA, 1999; Chapter xxii; 682p. [Google Scholar]
- Benson, G.G.; Hemingway, S.R.; Leach, F.N. Composition of the wrappings of an ancient Egyptian mummy. J. Pharm. Pharmacol. 1978, 30, 78. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.T.; Juniper, B.E. The Cuticle of Plants; Edward Arnold Ltd.: Edinburgh, UK, 1970. [Google Scholar]
- Blomquist, G.J.; Soliday, C.L.; Byers, B.A.; Brakke, J.W.; Jackson, L.L. Cuticular lipids of insects: V. Cuticular wax esters of secondary alcohols from the grasshoppersMelanoplus packardii andMelanoplus sanguinipes. Lipids 1972, 7, 356–362. [Google Scholar] [CrossRef]
- Jackson, L.L.; Blomquist, G.J. Insect waxes. In Chemistry and Biochemistry of Natural Waxes; Kolattukudy, P.E., Ed.; Elsevier: Amsterdam, The Netherlands, 1976; pp. 201–233. [Google Scholar]
- Stránský, K.; Valterová, I.; Kofroňová, E.; Urbanová, K.; Zarevúcka, M.; Wimmer, Z. Non-polar lipid components of human cerumen. Lipids 2011, 46, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Riederer, M.; Schreiber, L. Protecting against water loss: Analysis of the barrier properties of plant cuticles. J. Exp. Bot. 2001, 52, 2023–2032. [Google Scholar] [CrossRef]
- Lolle, S.J.; Hsu, W.; Pruitt, R.E. Genetic analysis of organ fusion in arabidopsis thaliana. Genetics 1998, 149, 607–619. [Google Scholar] [CrossRef]
- Sieber, P.; Schorderet, M.; Ryser, U.; Buchala, A.; Kolattukudy, P.; Métraux, J.-P.; Nawrath, C. Transgenic arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 2000, 12, 721. [Google Scholar] [CrossRef]
- Edlund, A.F.; Swanson, R.; Preuss, D. Pollen and stigma structure and function: The role of diversity in pollination. Plant Cell 2004, 16, S84–S97. [Google Scholar] [CrossRef]
- Reisberg, E.E.; Hildebrandt, U.; Riederer, M.; Hentschel, U. Distinct phyllosphere bacterial communities on arabidopsis wax mutant leaves. PLoS ONE 2013, 8, e78613. [Google Scholar] [CrossRef]
- Ritpitakphong, U.; Falquet, L.; Vimoltust, A.; Berger, A.; Métraux, J.; L’Haridon, F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016, 210, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Jenks, M.A.; Tuttle, H.A.; Eigenbrode, S.; Feldmann, K.A. Leaf epicuticular waxes of the eceriferum mutants in arabidopsis. Plant Physiol. 1995, 108, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Kolattukudy, P.E.; Croteau, R.; Buckner, J.S. Biochemistry of plant waxes. In Chemistry and Biochemistry of Natural Waxes; Kolattukudy, P.E., Ed.; Elsevier: Amsterdam, The Netherlands, 1976; pp. 289–347. [Google Scholar]
- Lardizabal, K.D.; Metz, J.G.; Sakamoto, T.; Hutton, W.C.; Pollard, M.R.; Lassner, M.W. Purification of a jojoba embryo wax synthase, cloning of its cDNA, and production of high levels of wax in seeds of transgenic arabidopsis. Plant Physiol. 2000, 122, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K. A superfamily of membrane-bound O-acyltransferases with implications for Wnt signaling. Trends Biochem. Sci. 2000, 25, 111–112. [Google Scholar] [CrossRef]
- Kalscheuer, R.; Steinbuchel, A. A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J. Biol. Chem. 2003, 278, 8075–8082. [Google Scholar] [CrossRef] [PubMed]
- Stoveken, T.; Kalscheuer, R.; Malkus, U.; Reichelt, R.; Steinbuchel, A. The wax ester synthase/acyl coenzyme A: Diacylglycerol acyltransferase from Acinetobacter sp. strain ADP1: Characterization of a novel type of acyltransferase. J. Bacteriol. 2005, 187, 1369–1376. [Google Scholar] [CrossRef]
- Li, F.; Wu, X.; Lam, P.; Bird, D.; Zheng, H.; Samuels, L.; Jetter, R.; Kunst, L. Identification of the wax ester synthase/acyl-coenzyme A: Diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in arabidopsis. Plant Physiol. 2008, 148, 97–107. [Google Scholar] [CrossRef]
- Patwari, P.; Salewski, V.; Gutbrod, K.; Kreszies, T.; Dresen-Scholz, B.; Peisker, H.; Steiner, U.; Meyer, A.J.; Schreiber, L.; Dörmann, P. Surface wax esters contribute to drought tolerance in Arabidopsis. Plant J. 2019, 98, 727–744. [Google Scholar] [CrossRef]
- Cheng, J.B.; Russell, D.W. Mammalian wax biosynthesis. Expression cloning of wax synthase cDNAs encoding a member of the acyltransferase enzyme family. J. Biol. Chem. 2004, 279, 37798–37807. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- King, A.; Nam, J.-W.; Han, J.; Hilliard, J.; Jaworski, J.G. Cuticular wax biosynthesis in petunia petals: Cloning and characterization of an alcohol-acyltransferase that synthesizes wax-esters. Planta 2007, 226, 381–394. [Google Scholar] [CrossRef]
- Uthoff, S.; Stöveken, T.; Weber, N.; Vosmann, K.; Klein, E.; Kalscheuer, R.; Steinbüchel, A. Thio wax ester biosynthesis utilizing the unspecific bifunctional wax ester synthase/acyl coenzyme A: Diacylglycerol acyltransferase of Acinetobacter sp. strain ADP1. Appl. Environ. Microbiol. 2005, 71, 790–796. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, H.; Rodriguez, J.; Salacup, J.; Castañeda, I.; Schnell, D.; Pareek, A.; Dhankher, O. Increased cuticle waxes by overexpression of WSD1 improves osmotic stress tolerance in Arabidopsis thaliana and Camelina sativa. Int. J. Mol. Sci. 2021, 22, 5173. [Google Scholar] [CrossRef] [PubMed]
- Klypina, N.; Hanson, S.F. Arabidopsis thaliana wax synthase gene homologues show diverse expression patterns that suggest a specialized role for these genes in reproductive organs. Plant Sci. 2008, 175, 312–320. [Google Scholar] [CrossRef]
- Sandager, L.; Gustavsson, M.H.; Ståhl, U.; Dahlqvist, A.; Wiberg, E.; Banas, A.; Lenman, M.; Ronne, H.; Stymne, S. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 2002, 277, 6478–6482. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wang, J.; Zhang, G.; Liu, J.; Manan, S.; Hu, H.; Zhao, J. Two types of soybean diacylglycerol acyltransferases are differentially involved in triacylglycerol biosynthesis and response to environmental stresses and hormones. Sci. Rep. 2016, 6, 28541. [Google Scholar] [CrossRef]
- Kalscheuer, R.; Luftmann, H.; Steinbüchel, A. Synthesis of novel lipids in Saccharomyces cerevisiae by heterologous expression of an unspecific bacterial acyltransferase. Appl. Environ. Microbiol. 2004, 70, 7119–7125. [Google Scholar] [CrossRef]
- Zienkiewicz, K.; Benning, U.; Siegler, H.; Feussner, I. The type 2 acyl-CoA:diacylglycerol acyltransferase family of the oleaginous microalga Lobosphaera incisa. BMC Plant Biol. 2018, 18, 298. [Google Scholar] [CrossRef]
- Trotter, P.J. The genetics of fatty acid metabolism in saccharomyces cerevisiae. Annu. Rev. Nutr. 2001, 21, 97–119. [Google Scholar] [CrossRef]
- Metz, J.G.; Pollard, M.R.; Anderson, L.; Hayes, T.R.; Lassner, M.W. Purification of a jojoba embryo fatty acyl-coenzyme A reductase and expression of its cDNA in high erucic acid rapeseed. Plant Physiol. 2000, 122, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Sims, T.L.; Goldberg, R.B. The glycininGy1gene from soybean. Nucleic Acids Res. 1989, 17, 4386. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ding, S.-H.; Huang, L.-Y.; Wang, Y.-D.; Sun, H.-C.; Xiang, Z.-H. High-level expression of basic fibroblast growth factor in transgenic soybean seeds and characterization of its biological activity. Biotechnol. Lett. 2006, 28, 869–875. [Google Scholar] [CrossRef]
- Lassner, M.; Lardizabal, K.; Metz, J.G. A jojoba beta-Ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell 1996, 8, 281–292. [Google Scholar] [CrossRef]
- Nakai, K.; Horton, P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999, 24, 34–35. [Google Scholar] [CrossRef]
- Hooper, C.M.; Castleden, I.R.; Tanz, S.; Aryamanesh, N.; Millar, A.H. SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 2016, 45, D1064–D1074. [Google Scholar] [CrossRef]
- Singh, U.; Hur, M.; Dorman, K.; Wurtele, E.S. MetaOmGraph: A workbench for interactive exploratory data analysis of large expression datasets. Nucleic Acids Res. 2020, 48, e23. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Dimopoulos, N.; Tindjau, R.; Wong, D.C.J.; Matzat, T.; Haslam, T.; Song, C.; Gambetta, G.A.; Kunst, L.; Castellarin, S.D. Drought stress modulates cuticular wax composition of the grape berry (Vitis vinifera L.). J. Exp. Bot. 2020, 71, 3126–3141. [Google Scholar] [CrossRef]
- Huang, L.; Xiao, Q.; Zhao, X.; Wang, D.; Wei, L.; Li, X.; Liu, Y.; He, Z.; Kang, L.; Guo, Y. Responses of cuticular waxes of faba bean to light wavelengths and selection of candidate genes for cuticular wax biosynthesis. Plant Genome 2020, 13, e20058. [Google Scholar] [CrossRef]
- Zhang, F.; Rodriguez, S.; Keasling, J.D. Metabolic engineering of microbial pathways for advanced biofuels production. Curr. Opin. Biotechnol. 2011, 22, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Röttig, A.; Wenning, L.; Bröker, D.; Steinbüchel, A. Fatty acid alkyl esters: Perspectives for production of alternative biofuels. Appl. Microbiol. Biotechnol. 2009, 85, 1713–1733. [Google Scholar] [CrossRef]
- Wang, T.; Xing, J.; Liu, X.; Yao, Y.; Hu, Z.; Peng, H.; Xin, M.; Zhou, D.-X.; Zhang, Y.; Ni, Z. GCN5 contributes to stem cuticular wax biosynthesis by histone acetylation of CER3 in Arabidopsis. J. Exp. Bot. 2018, 69, 2911–2922. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Zhu, L.-H.; Krens, F.; Smith, M.A.; Li, X.; Qi, W.; Van Loo, E.N.; Iven, T.; Feussner, I.; Nazarenus, T.J.; Huai, D.; et al. Dedicated industrial oilseed crops as metabolic engineering platforms for sustainable industrial feedstock production. Sci. Rep. 2016, 6, 22181. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Landy, A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu. Rev. Biochem. 1989, 58, 913–941. [Google Scholar] [CrossRef]
- Oelkers, P.; Cromley, D.; Padamsee, M.; Billheimer, J.T.; Sturley, S.L. The DGA1 Gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 2002, 277, 8877–8881. [Google Scholar] [CrossRef] [PubMed]
- Neumann, E.; Kakorin, S.; Tsoneva, I.; Nikolova, B.; Tomov, T. Calcium-mediated DNA adsorption to yeast cells and kinetics of cell transformation by electroporation. Biophys. J. 1996, 71, 868–877. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Shockey, J.; Mason, C.; Gilbert, M.; Cao, H.; Li, X.; Cahoon, E.; Dyer, J. Development and analysis of a highly flexible multi-gene expression system for metabolic engineering in Arabidopsis seeds and other plant tissues. Plant Mol. Biol. 2015, 89, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef]
- Heilmann, M.; Iven, T.; Ahmann, K.; Hornung, E.; Stymne, S.; Feussner, I. Production of wax esters in plant seed oils by oleosomal co-targeting of biosynthetic enzymes. J. Lipid Res. 2012, 53, 2153–2161. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Taki, T.; Kasama, T.; Handa, S.; Ishikawa, D. A simple and quantitative purification of glycosphingolipids and phospholipids by thin-layer chromatography blotting. Anal. Biochem. 1994, 223, 232–238. [Google Scholar] [CrossRef]
- Martin, A.; Synge, R. A new form of chromatogram employing two liquid phases: A theory of chromatography. 2. Application to the micro-determination of the higher monoamino-acids in proteins. Biochem. J. 1941, 35, 1358. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]


| Enzyme Type | Gene ID | Protein Length (aa) | Subcellular Location a | Maximum Expression b | |||
|---|---|---|---|---|---|---|---|
| PSORT Prediction | SUBA Prediction | Organ | Expression Level (fpkm) | SRA Accession | |||
| Wax synthase | At1g34490 | 337 | TM or PM | PM | Root cells (isolated) | 22.0 | SRR8206657 |
| At1g34500 | 341 | PM | PM | Leaves (7-d old seedlings) | 19.3 | SRR8742425 | |
| At1g34520 | 336 | TM or PM | PM | Apical Meristem | 233.3 | SRR390310 | |
| At3g51970 | 345 | TM | PM | Stigma (non-pollinated) | 44.3 | ERR2278241 | |
| At5g51420 | 435 | TM or PM | PM | Inflorescence meristem | 66.7 | SRR5681054 | |
| At5g55320 | 339 | PM or TM | PM | Developing seeds (3-d after pollination) | 149.7 | SRR1232482 | |
| At5g55330 | 346 | PM or TM | PM | Siliques (1-d after pollination) | 246.4 | SRR3347475 | |
| At5g55340 | 333 | PM | PM | Developing embryos (globular stage) | 68.4 | SRR8249028 | |
| At5g55350 | 345 | mitoIM or PM | PM | Stem | 578.8 | SRR2037335 | |
| At5g55360 | 342 | PM or TM | PM | Seedlings (10-d after germination) | 25.0 | SRR3707607 | |
| At5g55370 | 343 | PM or TM | PM | Seedlings (7-d after germination) | 27.7 | SRR4734675 | |
| At5g55380 | 341 | TM or PM | PM | Inflorescence meristem | 41.3 | ERR1698199 | |
| WS/ DGAT | At1g72110 | 479 | ER | Perox | Stem | 73.2 | SRR4007446 |
| At2g38995 | 487 | ER | PM | Siliques | 88.7 | SRR3347480 | |
| At3g49190 | 522 | PM | Cysk | Leaves (7-d old seedlings) | 11.9 | SRR8742425 | |
| At3g49200 | 507 | PM | Cysk | Root cells (isolated) | 430.2 | SRR8206660 | |
| At3g49210 | 518 | ER | Cysk | Leaves (7-d old seedlings) | 14.2 | SRR8742425 | |
| At5g12420 | 480 | PM | Cysk | Roots (6-d old seedlings) | 367.6 | SRR5195558 | |
| At5g16350 | 488 | Cytpl | Perox | Hypocotyl | 76.4 | SRR6312333 | |
| At5g22490 | 482 | ER | Cysk | Stem | 619.0 | SRR2037351 | |
| At5g37300 | 481 | Perox | Cysk or Perox | Flowers (6-week old plants) | 184.2 | SRR6179906 | |
| At5g53380 | 483 | Perox | Cysk | Root cells (isolated) | 54.8 | SRR8206659 | |
| At5g53390 | 485 | PM | Cysk | Seedling | 70.4 | ERR1876169 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, D.; Li, L.; Rizhsky, L.; Bhandary, P.; Nikolau, B.J. Heterologous Expression and Characterization of Plant Wax Ester Producing Enzymes. Metabolites 2022, 12, 577. https://doi.org/10.3390/metabo12070577
Cheng D, Li L, Rizhsky L, Bhandary P, Nikolau BJ. Heterologous Expression and Characterization of Plant Wax Ester Producing Enzymes. Metabolites. 2022; 12(7):577. https://doi.org/10.3390/metabo12070577
Chicago/Turabian StyleCheng, Daolin, Ling Li, Ludmila Rizhsky, Priyanka Bhandary, and Basil J. Nikolau. 2022. "Heterologous Expression and Characterization of Plant Wax Ester Producing Enzymes" Metabolites 12, no. 7: 577. https://doi.org/10.3390/metabo12070577
APA StyleCheng, D., Li, L., Rizhsky, L., Bhandary, P., & Nikolau, B. J. (2022). Heterologous Expression and Characterization of Plant Wax Ester Producing Enzymes. Metabolites, 12(7), 577. https://doi.org/10.3390/metabo12070577








