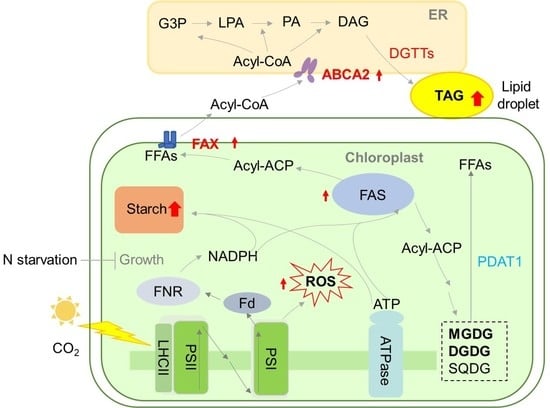
Co-Expression of Lipid Transporters Simultaneously Enhances Oil and Starch Accumulation in the Green Microalga Chlamydomonas reinhardtii under Nitrogen Starvation
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Cultivation Conditions
2.2. RNA Isolation and Quantification
2.3. Lipid Extraction and Quantification
2.4. Starch Content Quantification
2.5. AGPase Activity Assays
2.6. Determination of ROS Levels
2.7. Biomass Determination
2.8. Analysis of Photosynthetic Parameters
2.9. Statistical Analysis
3. Results
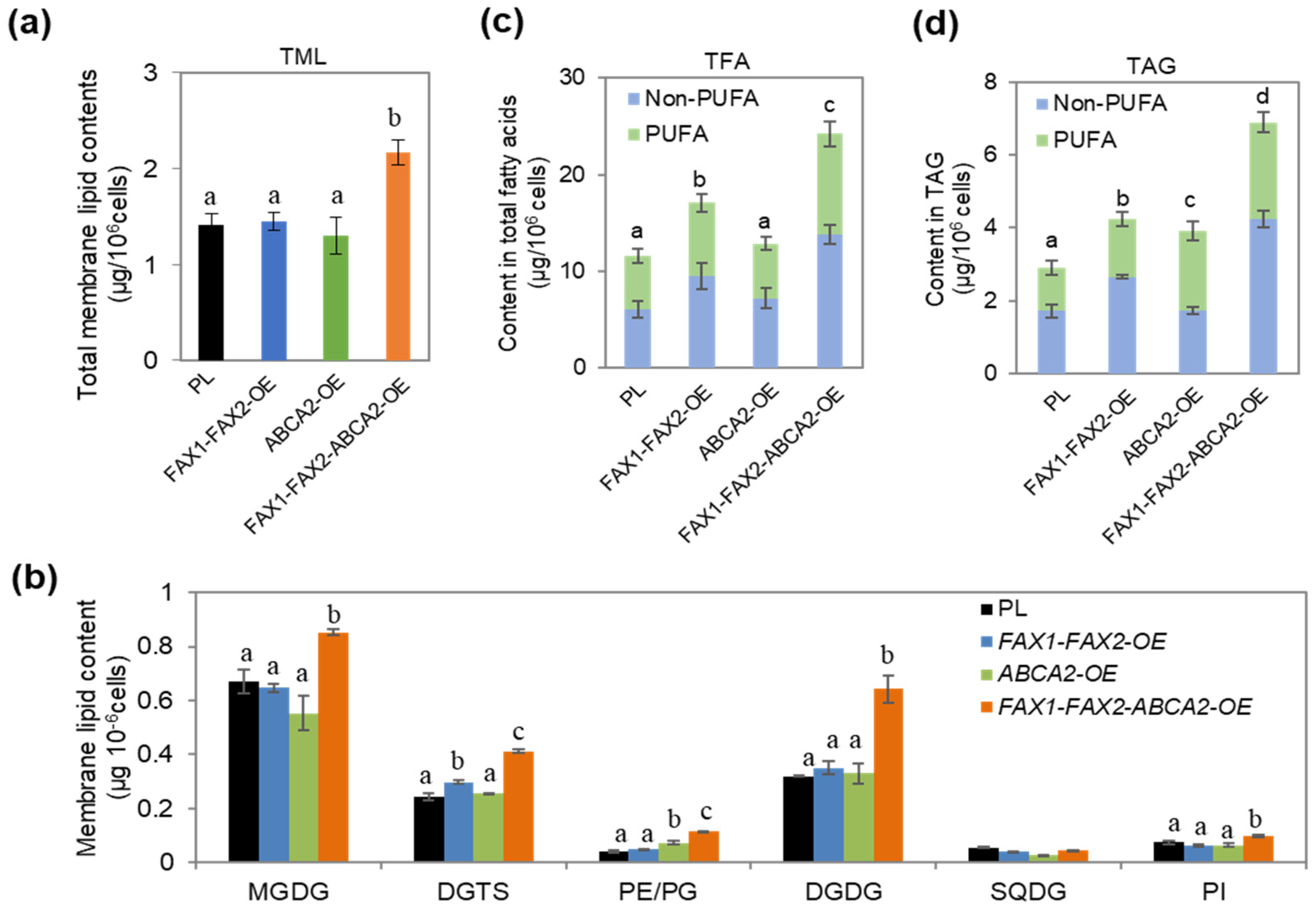
3.1. Oil Content Increased in FAX1-FAX2-ABCA2 OE under N Starvation
3.2. Major Membrane Lipid Levels Were Enhanced in FAX1-FAX2-ABCA2-OE
3.3. Expression Levels of Key Genes in Lipid Metabolism in FAX1-FAX2-ABCA2-OE
3.4. Starch Content Increases in FAX1-FAX2-ABCA2 OE under N Starvation
3.5. Physiological Characteristics of FAX1-FAX2-ABCA2 OE under N Starvation
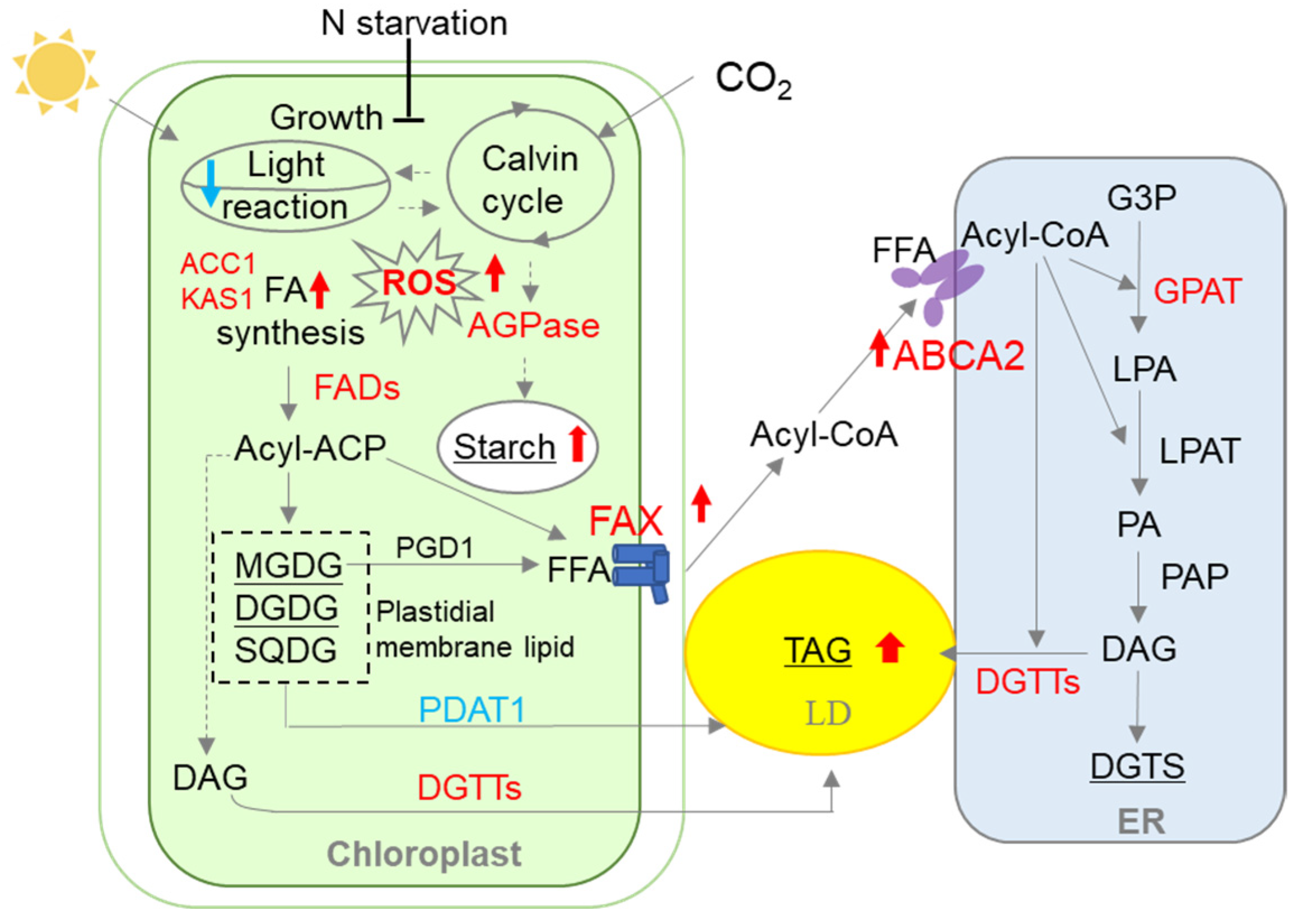
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muñoz, C.F.; Südfeld, C.; Naduthodi, M.I.S.; Weusthuis, R.A.; Barbosa, M.J.; Wijffels, R.H.; D’Adamo, S. Genetic Engineering of Microalgae for Enhanced Lipid Production. Biotechnol. Adv. 2021, 52, 107836. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Danquah, M.K.; Chen, X.D.; Lu, Y. Microalgae Bioengineering: From CO2 Fixation to Biofuel Production. Renew. Sustain. Energy Rev. 2011, 15, 3252–3260. [Google Scholar] [CrossRef]
- Saroussi, S.; Sanz-Luque, E.; Kim, R.G.; Grossman, A.R. Nutrient Scavenging and Energy Management: Acclimation Responses in Nitrogen and Sulfur Deprived Chlamydomonas. Curr. Opin. Plant Biol. 2017, 39, 114–122. [Google Scholar] [CrossRef]
- Niyogi, K.K. Safety Valves for Photosynthesis. Curr. Opin. Plant Biol. 2000, 3, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.-Y.; Benning, C. Triacylglycerol Accumulation in Photosynthetic Cells in Plants and Algae. Subcell. Biochem. 2016, 86, 179–205. [Google Scholar] [CrossRef] [PubMed]
- Vonlanthen, S.; Dauvillée, D.; Purton, S. Evaluation of Novel Starch-Deficient Mutants of Chlorella Sorokiniana for Hyper-Accumulation of Lipids. Algal. Res. 2015, 12, 109–118. [Google Scholar] [CrossRef]
- Chisti, Y. Biodiesel from Microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef]
- Kong, F.; Romero, I.T.; Warakanont, J.; Li-Beisson, Y. Lipid Catabolism in Microalgae. New Phytol. 2018, 218, 1340–1348. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Kong, F.; Wang, P.; Lee, Y.; Kang, B.-H. The Disassembly of Lipid Droplets in Chlamydomonas. New Phytol. 2021, 231, 1359–1364. [Google Scholar] [CrossRef]
- Kang, N.K.; Baek, K.; Koh, H.G.; Atkinson, C.A.; Ort, D.R.; Jin, Y.-S. Microalgal Metabolic Engineering Strategies for the Production of Fuels and Chemicals. Bioresour. Technol. 2022, 345, 126529. [Google Scholar] [CrossRef]
- Kumar, G.; Shekh, A.; Jakhu, S.; Sharma, Y.; Kapoor, R.; Sharma, T.R. Bioengineering of Microalgae: Recent Advances, Perspectives, and Regulatory Challenges for Industrial Application. Front. Bioeng. Biotechnol. 2020, 8, 914. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Yamaoka, Y.; Ohama, T.; Lee, Y.; Li-Beisson, Y. Molecular Genetic Tools and Emerging Synthetic Biology Strategies to Increase Cellular Oil Content in Chlamydomonas Reinhardtii. Plant Cell Physiol. 2019, 60, 1184–1196. [Google Scholar] [CrossRef] [PubMed]
- Benning, C. Mechanisms of Lipid Transport Involved in Organelle Biogenesis in Plant Cells. Annu. Rev. Cell Dev. Biol. 2009, 25, 71–91. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, C.; Li-Beisson, Y.; Philippar, K. Fatty Acid and Lipid Transport in Plant Cells. Trends Plant Sci. 2016, 21, 145–158. [Google Scholar] [CrossRef]
- Jang, S.; Kong, F.; Lee, J.; Choi, B.Y.; Wang, P.; Gao, P.; Yamano, T.; Fukuzawa, H.; Kang, B.-H.; Lee, Y. CrABCA2 Facilitates Triacylglycerol Accumulation in Chlamydomonas Reinhardtii under Nitrogen Starvation. Mol. Cells 2020, 43, 48–57. [Google Scholar] [CrossRef]
- Li, N.; Zhang, Y.; Meng, H.; Li, S.; Wang, S.; Xiao, Z.; Chang, P.; Zhang, X.; Li, Q.; Guo, L.; et al. Characterization of Fatty Acid Exporters Involved in Fatty Acid Transport for Oil Accumulation in the Green Alga Chlamydomonas Reinhardtii. Biotechnol. Biofuels 2019, 12, 14. [Google Scholar] [CrossRef]
- Peter, J.; Huleux, M.; Spaniol, B.; Sommer, F.; Neunzig, J.; Schroda, M.; Li-Beisson, Y.; Philippar, K. Fatty Acid Export (FAX) Proteins Contribute to Oil Production in the Green Microalga Chlamydomonas reinhardtii. Front. Mol. Biosci. 2022, 9, 939834. [Google Scholar] [CrossRef]
- Scranton, M.A.; Ostrand, J.T.; Fields, F.J.; Mayfield, S.P. Chlamydomonas as a Model for Biofuels and Bio-Products Production. Plant J. 2015, 82, 523–531. [Google Scholar] [CrossRef]
- Takeuchi, T.; Benning, C. Nitrogen-Dependent Coordination of Cell Cycle, Quiescence and TAG Accumulation in Chlamydomonas. Biotechnol. Biofuels 2019, 12, 292. [Google Scholar] [CrossRef]
- Chen, R.; Yang, M.; Li, M.; Zhang, H.; Lu, H.; Dou, X.; Feng, S.; Xue, S.; Zhu, C.; Chi, Z.; et al. Enhanced Accumulation of Oil through Co-Expression of Fatty Acid and ABC Transporters in Chlamydomonas under Standard Growth Conditions. Biotechnol. Biofuels Bioprod. 2022, 15, 54. [Google Scholar] [CrossRef]
- Neupert, J.; Karcher, D.; Bock, R. Generation of Chlamydomonas Strains That Efficiently Express Nuclear Transgenes. Plant J. 2009, 57, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- Hoober, J.K. The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Use. Science 1989, 246, 1503–1504. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kong, F.; Liang, Y.; Légeret, B.; Beyly-Adriano, A.; Blangy, S.; Haslam, R.P.; Napier, J.A.; Beisson, F.; Peltier, G.; Li-Beisson, Y. Chlamydomonas Carries out Fatty Acid β-Oxidation in Ancestral Peroxisomes Using a Bona Fide Acyl-CoA Oxidase. Plant J. 2017, 90, 358–371. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Burlacot, A.; Liang, Y.; Légeret, B.; Alseekh, S.; Brotman, Y.; Fernie, A.R.; Krieger-Liszkay, A.; Beisson, F.; Peltier, G.; et al. Interorganelle Communication: Peroxisomal MALATE DEHYDROGENASE2 Connects Lipid Catabolism to Photosynthesis through Redox Coupling in Chlamydomonas. Plant Cell 2018, 30, 1824–1847. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Han, D.; Yoon, K.; Hu, Q.; Li, Y. Characterization of Type 2 Diacylglycerol Acyltransferases in Chlamydomonas reinhardtii Reveals Their Distinct Substrate Specificities and Functions in Triacylglycerol Biosynthesis. Plant J. 2016, 86, 3–19. [Google Scholar] [CrossRef]
- Klein, U.; Betz, A. Fermentative Metabolism of Hydrogen-Evolving Chlamydomonas Moewusii. Plant Physiol. 1978, 61, 953–956. [Google Scholar] [CrossRef]
- Yang, M.; Kong, F.; Xie, X.; Wu, P.; Chu, Y.; Cao, X.; Xue, S. Galactolipid DGDG and Betaine Lipid DGTS Direct De Novo Synthesized Linolenate into Triacylglycerol in a Stress-Induced Starchless Mutant of Chlamydomonas reinhardtii. Plant Cell Physiol. 2020, 61, 851–862. [Google Scholar] [CrossRef]
- Kong, F.; Yamasaki, T.; Kurniasih, S.D.; Hou, L.; Li, X.; Ivanova, N.; Okada, S.; Ohama, T. Robust Expression of Heterologous Genes by Selection Marker Fusion System in Improved Chlamydomonas Strains. J. Biosci. Bioeng. 2015, 120, 239–245. [Google Scholar] [CrossRef]
- Xi, Y.; Kong, F.; Chi, Z. ROS Induce β-Carotene Biosynthesis Caused by Changes of Photosynthesis Efficiency and Energy Metabolism in Dunaliella Salina Under Stress Conditions. Front. Bioeng. Biotechnol. 2021, 8, 1447. [Google Scholar] [CrossRef]
- Ran, W.; Wang, H.; Liu, Y.; Qi, M.; Xiang, Q.; Yao, C.; Zhang, Y.; Lan, X. Storage of Starch and Lipids in Microalgae: Biosynthesis and Manipulation by Nutrients. Bioresour. Technol. 2019, 291, 121894. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Wang, G.; Kim, S.-C.; Li, J.; Zhou, Y.; Wang, X. Increased Expression of Fatty Acid and ABC Transporters Enhances Seed Oil Production in Camelina. Biotechnol. Biofuels 2021, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Gügel, I.L.; Giavalisco, P.; Zeisler, V.; Schreiber, L.; Soll, J.; Philippar, K. FAX1, a novel membrane protein mediating plastid fatty acid export. PLoS Biol. 2015, 13, e1002053. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yamaoka, Y.; Ono, H.; Kim, H.; Shim, D.; Maeshima, M.; Martinoia, E.; Cahoon, E.B.; Nishida, I.; Lee, Y. AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2013, 110, 773–778. [Google Scholar] [CrossRef]
- Takemura, T.; Imamura, S.; Tanaka, K. Identification of a chloroplast fatty acid exporter protein, CmFAX1, and triacylglycerol accumulation by its overexpression in the unicellular red alga Cyanidioschyzon merolae. Algal. Res. 2019, 38, 101396. [Google Scholar] [CrossRef]
- Li, X.; Moellering, E.R.; Liu, B.; Johnny, C.; Fedewa, M.; Sears, B.B.; Kuo, M.-H.; Benning, C. A Galactoglycerolipid Lipase Is Required for Triacylglycerol Accumulation and Survival Following Nitrogen Deprivation in Chlamydomonas reinhardtii. Plant Cell 2012, 24, 4670–4686. [Google Scholar] [CrossRef] [PubMed]
- Takatani, N.; Use, K.; Kato, A.; Ikeda, K.; Kojima, K.; Aichi, M.; Maeda, S.-I.; Omata, T. Essential Role of Acyl-ACP Synthetase in Acclimation of the Cyanobacterium Synechococcus Elongatus Strain PCC 7942 to High-Light Conditions. Plant Cell Physiol. 2015, 56, 1608–1615. [Google Scholar] [CrossRef]
- Liu, J.; Yao, C.; Meng, Y.; Cao, X.; Wu, P.; Xue, S. The ΔF/Fm’-Guided Supply of Nitrogen in Culture Medium Facilitates Sustainable Production of TAG in Nannochloropsis Oceanica IMET1. Biotechnol. Biofuels 2018, 11, 168. [Google Scholar] [CrossRef]
- Yao, C.; Ai, J.; Cao, X.; Xue, S.; Zhang, W. Enhancing Starch Production of a Marine Green Microalga Tetraselmis Subcordiformis through Nutrient Limitation. Bioresour. Technol. 2012, 118, 438–444. [Google Scholar] [CrossRef]
- Moellering, E.R.; Benning, C. Galactoglycerolipid Metabolism under Stress: A Time for Remodeling. Trends Plant Sci. 2011, 16, 98–107. [Google Scholar] [CrossRef]
- Hu, Q.; Sommerfeld, M.; Jarvis, E.; Ghirardi, M.; Posewitz, M.; Seibert, M.; Darzins, A. Microalgal Triacylglycerols as Feedstocks for Biofuel Production: Perspectives and Advances. Plant J. 2008, 54, 621–639. [Google Scholar] [CrossRef]
- Yang, W.; Wittkopp, T.M.; Li, X.; Warakanont, J.; Dubini, A.; Catalanotti, C.; Kim, R.G.; Nowack, E.C.M.; Mackinder, L.C.M.; Aksoy, M.; et al. Critical Role of Chlamydomonas reinhardtii Ferredoxin-5 in Maintaining Membrane Structure and Dark Metabolism. Proc. Natl. Acad. Sci. USA 2015, 112, 14978–14983. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.-Y.; Lucker, B.F.; Zienkiewicz, K.; Miller, T.E.; Zienkiewicz, A.; Sears, B.B.; Kramer, D.M.; Benning, C. Galactoglycerolipid Lipase PGD1 Is Involved in Thylakoid Membrane Remodeling in Response to Adverse Environmental Conditions in Chlamydomonas. Plant Cell 2018, 30, 447–465. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Fal, S.; Aasfar, A.; Rabie, R.; Smouni, A.; Arroussi, H.E. Salt Induced Oxidative Stress Alters Physiological, Biochemical and Metabolomic Responses of Green Microalga Chlamydomonas reinhardtii. Heliyon 2022, 8, e08811. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Han, D.; Hu, G.; Dauvillee, D.; Sommerfeld, M.; Ball, S.; Hu, Q. Chlamydomonas Starchless Mutant Defective in ADP-Glucose Pyrophosphorylase Hyper-Accumulates Triacylglycerol. Metab. Eng. 2010, 12, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Ballicora, M.A.; Iglesias, A.A.; Preiss, J. ADP-Glucose Pyrophosphorylase: A Regulatory Enzyme for Plant Starch Synthesis. Photosynth. Res. 2004, 79, 1–24. [Google Scholar] [CrossRef]
- Zabawinski, C.; Van Den Koornhuyse, N.; D’Hulst, C.; Schlichting, R.; Giersch, C.; Delrue, B.; Lacroix, J.M.; Preiss, J.; Ball, S. Starchless Mutants of Chlamydomonas reinhardtii Lack the Small Subunit of a Heterotetrameric ADP-Glucose Pyrophosphorylase. J. Bacteriol. 2001, 183, 1069–1077. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, R.; Yamaoka, Y.; Feng, Y.; Chi, Z.; Xue, S.; Kong, F. Co-Expression of Lipid Transporters Simultaneously Enhances Oil and Starch Accumulation in the Green Microalga Chlamydomonas reinhardtii under Nitrogen Starvation. Metabolites 2023, 13, 115. https://doi.org/10.3390/metabo13010115
Chen R, Yamaoka Y, Feng Y, Chi Z, Xue S, Kong F. Co-Expression of Lipid Transporters Simultaneously Enhances Oil and Starch Accumulation in the Green Microalga Chlamydomonas reinhardtii under Nitrogen Starvation. Metabolites. 2023; 13(1):115. https://doi.org/10.3390/metabo13010115
Chicago/Turabian StyleChen, Ru, Yasuyo Yamaoka, Yanbin Feng, Zhanyou Chi, Song Xue, and Fantao Kong. 2023. "Co-Expression of Lipid Transporters Simultaneously Enhances Oil and Starch Accumulation in the Green Microalga Chlamydomonas reinhardtii under Nitrogen Starvation" Metabolites 13, no. 1: 115. https://doi.org/10.3390/metabo13010115
APA StyleChen, R., Yamaoka, Y., Feng, Y., Chi, Z., Xue, S., & Kong, F. (2023). Co-Expression of Lipid Transporters Simultaneously Enhances Oil and Starch Accumulation in the Green Microalga Chlamydomonas reinhardtii under Nitrogen Starvation. Metabolites, 13(1), 115. https://doi.org/10.3390/metabo13010115