Volatilomics-Based Microbiome Evaluation of Fermented Dairy by Prototypic Headspace-Gas Chromatography–High-Temperature Ion Mobility Spectrometry (HS-GC-HTIMS) and Non-Negative Matrix Factorization (NNMF)
Abstract
:1. Introduction
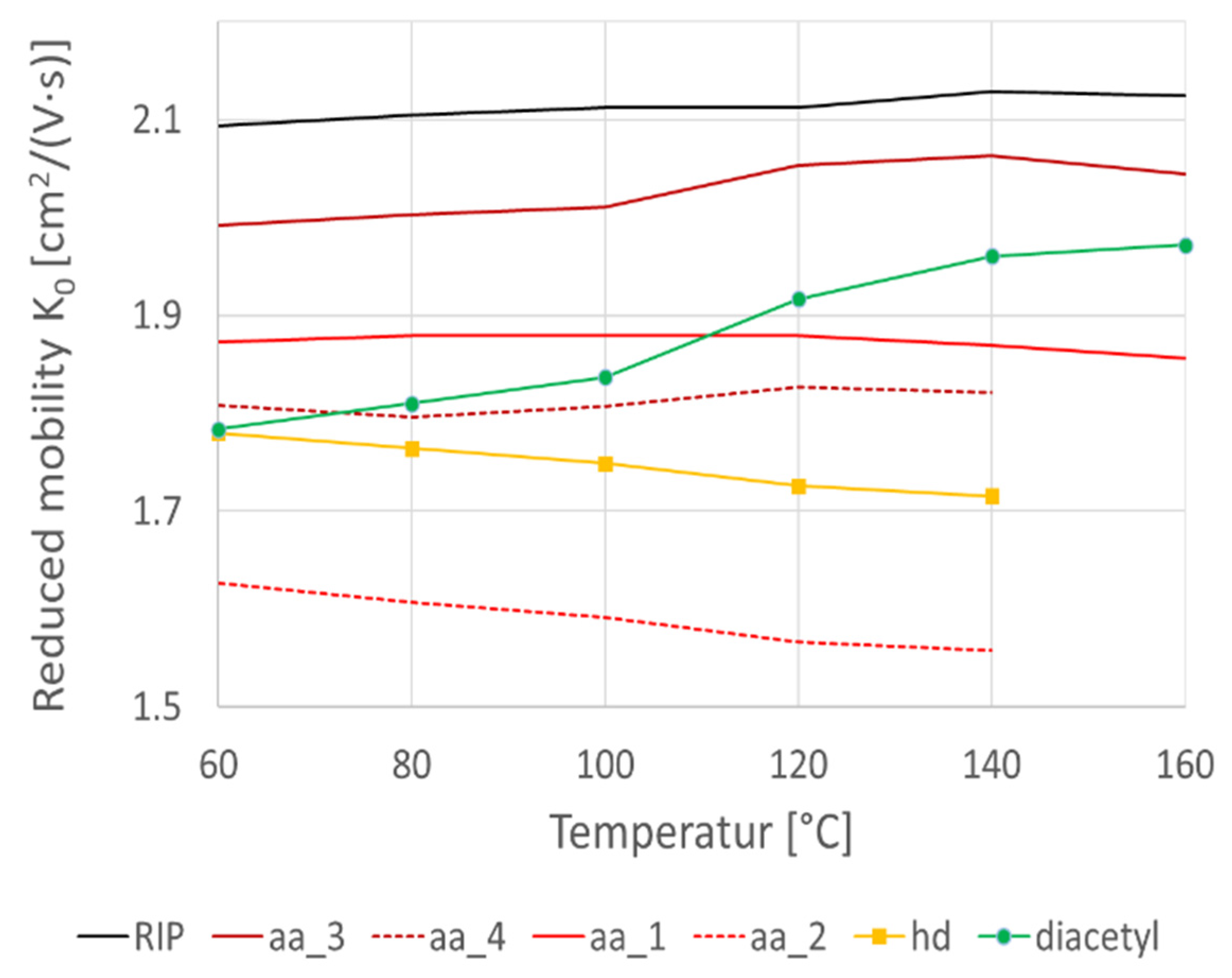
1.1. HS-GC-IMS for VOC Profiling
- K0 = reduced ion mobility in cm2 V−1 s−1
- L = drift length in cm
- E = electric field strength in V cm−1
- tD = drift time in s
- p = pressure of the drift gas in hPa
- p0 = ambient pressure: p0 = 1013.2 hPa
- T = temperature of the drift gas in K
- T0 = ambient temperature: T0 = 273.2 K
1.2. Pattern Recognition and Dimension Reduction Techniques
1.3. Microbial Composition and Flavor Profiles of Fermented Dairy
1.4. Overall Research Objective
2. Results and Discussion
2.1. Optimization of VOC Profiling for Analysis of Fermented Foods
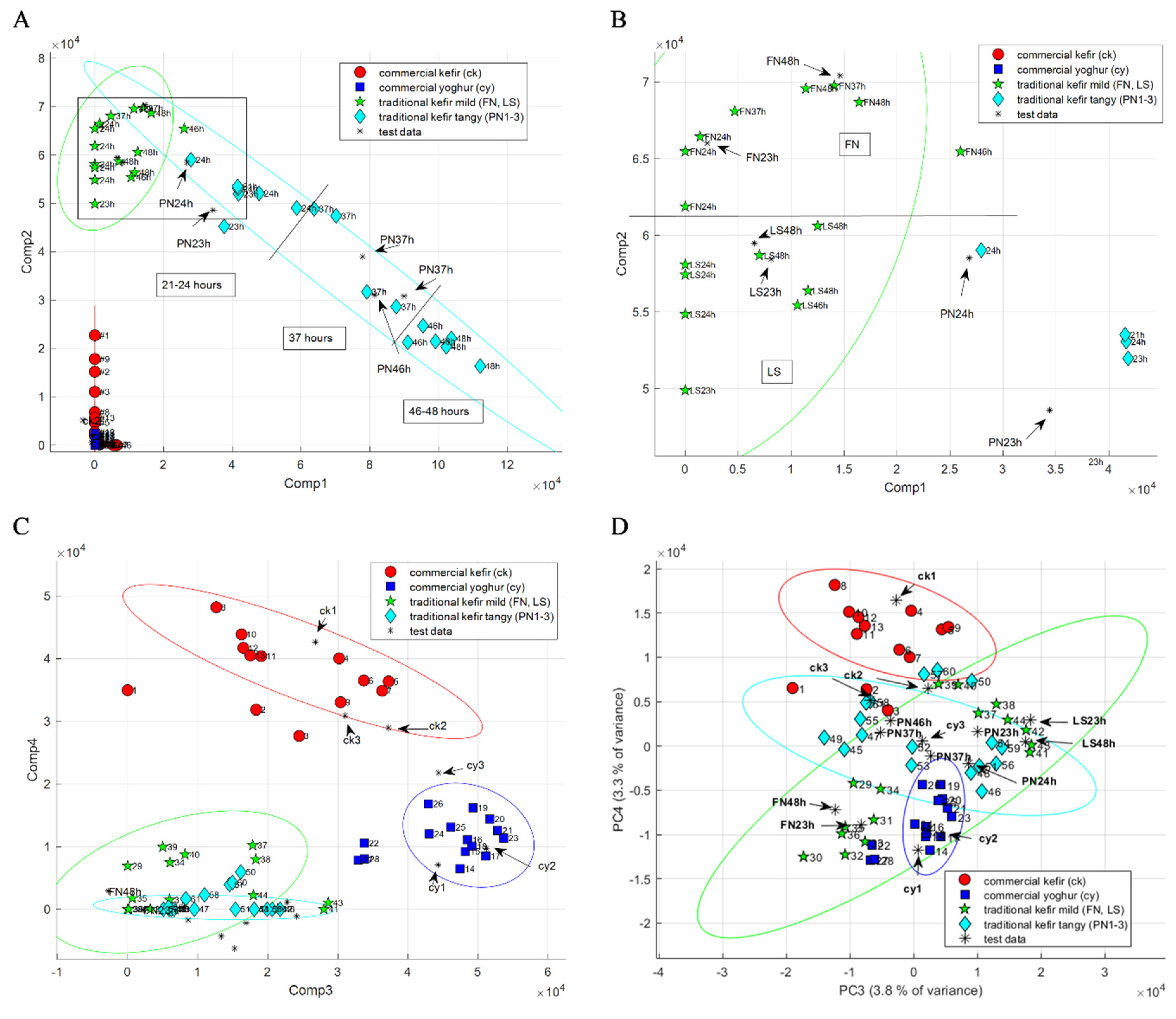
2.2. Kefir Discrimination by PCA and NNMF
2.3. Comparison of PCA and NNMF for Kefir Classification Based on HS-GC-IMS Data
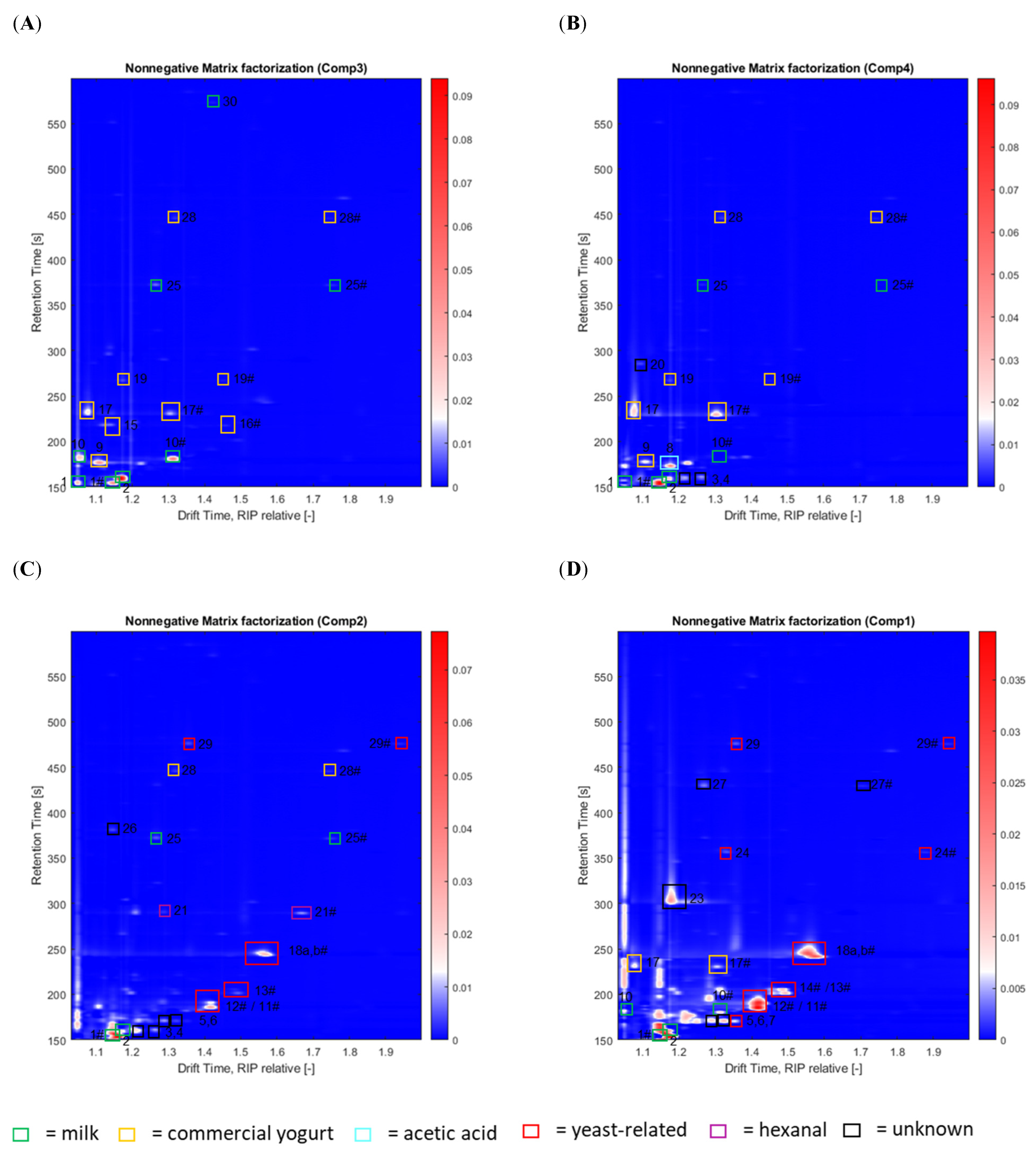
2.4. Backward Projection of Loadings Using Four NNMF Components and Substance Identification
2.5. Identification of Microorganisms Using qPCR
2.6. Further Investigation of the Origin of Hexanal in Kefir Samples
3. Materials and Methods
3.1. Reagents and Fermented Dairy Samples
3.2. Analysis of Microbial Composition of Kefirs
3.3. Metabolite Analysis (Acetic Acid Quantification)
3.4. HS-GC-MS System
3.5. Reference HS-GC-MS/IMS System Based on Standard OEM-IMS Cell
3.6. GC-HTIMS Prototype with Adjustable Drift Tube Temperature
3.7. Ambient Pressure Measurements
3.8. Data Preprocessing
3.9. Chemometric Data Analysis and Software
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sample Name | Manufacturer | Fat Content | Organic | Supplementary Cultures |
|---|---|---|---|---|
| ck01 | Alnatura | 1.50% | x | |
| ck03 | Andechser expired | 1.50% | x | |
| ck02 | Andechser (M3) frozen | 1.50% | x | |
| ck04 | Berchtesgadener | 1.50% | x | |
| ck05 | Berchtesgadener | 1.50% | ||
| ck06 | Brandenburg (M1) frozen | 3.50% | ||
| ck07 | Gut & Günstig | 1.50% | ||
| ck08 | GutBio | 1.50% | x | |
| ck09 | Müller (M2) frozen | 1.50% | ||
| ck10 | Müller | 1.50% | ||
| ck11 | Quarki | 2.00% | ||
| ck12 | Schrozberger | 1.50% | x | AB |
| ck13 | Starter_culture (23h) | 3.50% | x | |
| ck15 | Starter_culture (48h) | 3.50% | x | |
| ck14 | Starter_culture (23h) frozen | 3.50% | x | |
| ck16 | Starter_culture (48h) frozen | 3.50% | x |
| # | Kefir Grain | Harvest Time (h) | Run | Data Obtained in |
|---|---|---|---|---|
| FN_R2_23h | FN | 23 | R2 | 02/2021 |
| FN_R1_24h | FN | 24 | R1 | 12/2020 |
| FN_R2_46h | FN | 46 | R2 | 02/2021 |
| FN_R1_48h | FN | 48 | R1 | 12/2020 |
| FN_R3_24h | FN | 24 | R3 | 04/2021 |
| FN_R3_37h | FN | 37 | R3 | 04/2021 |
| FN_R3_48h | FN | 48 | R3 | 04/2021 |
| FN_R4_24h | FN | 24 | R4 | 04/2021 |
| FN_R4_37h | FN | 37 | R4 | 04/2021 |
| FN_R4_48h | FN | 48 | R4 | 04/2021 |
| LS_R2_23h | LS | 23 | R2 | 02/2021 |
| LS_R1_24h | LS | 24 | R1 | 12/2020 |
| LS_R2_46h | LS | 46 | R2 | 02/2021 |
| LS_R1_48h | LS | 48 | R1 | 12/2020 |
| LS_R3_24h | LS | 24 | R3 | 04/2021 |
| LS_R3_37h | LS | 37 | R3 | 04/2021 |
| LS_R3_48h | LS | 48 | R3 | 04/2021 |
| LS_R4_24h | LS | 24 | R4 | 04/2021 |
| LS_R4_37h | LS | 37 | R4 | 04/2021 |
| LS_R4_48h | LS | 48 | R4 | 04/2021 |
| PN1_R2_23h | PN1 | 23 | R2 | 02/2021 |
| PN1_R2_46h | PN1 | 46 | R2 | 02/2021 |
| PN1_R3_24h | PN1 | 24 | R3 | 04/2021 |
| PN1_R3_37h | PN1 | 37 | R3 | 04/2021 |
| PN1_R3_48h | PN1 | 48 | R3 | 04/2021 |
| PN1_R4_24h | PN1 | 24 | R4 | 04/2021 |
| PN1_R4_37h | PN1 | 37 | R4 | 04/2021 |
| PN1_R4_48h | PN1 | 48 | R4 | 04/2021 |
| PN2_R2_23h | PN2 | 23 | R2 | 02/2021 |
| PN2_R2_46h | PN2 | 46 | R2 | 02/2021 |
| PN2_R3_24h | PN2 | 24 | R3 | 04/2021 |
| PN2_R3_37h | PN2 | 37 | R3 | 04/2021 |
| PN2_R4_24h | PN2 | 24 | R4 | 04/2021 |
| PN2_R4_37h | PN2 | 37 | R4 | 04/2021 |
| PN3_R2_23h | PN3 | 23 | R2 | 02/2021 |
| PN3_R2_46h | PN3 | 46 | R2 | 02/2021 |
| PN3_R3_24h | PN3 | 24 | R3 | 04/2021 |
| PN3_R3_37h | PN3 | 37 | R3 | 04/2021 |
| PN3_R3_48h | PN3 | 48 | R3 | 04/2021 |
| PN3_R4_24h | PN3 | 24 | R4 | 04/2021 |
| PN3_R4_37h | PN3 | 37 | R4 | 04/2021 |
| PN3_R4_48h | PN3 | 48 | R4 | 04/2021 |
| Sample Name | Manufacturer | Fat Content | Organic | Supplementary Cultures |
|---|---|---|---|---|
| cy1 | Andechser | 0.10% | x | AB |
| cy2 | Andechser | 3.80% | x | AB |
| cy3 | Berchtesgadener | 3.50% | x | AB |
| cy4 | Berchtesgadener | 3.90% | AB | |
| cy5 | Gut & Günstig | 1.80% | C | |
| cy6 | GutBio | 3.80% | x | |
| cy7 | Ja! | 1.50% | ||
| cy8 | Milsani | 1.80% | C | |
| cy9 | Schrozberger | 1.80% | x | AB |
| cy10 | Schrozberger | 3.50% | x | AB |
| cy11 | Schrozberger_ABC | 3.50% | x | ABC |
| cy12 | Schwalbenhof | 3.50% | x | |
| cy15 | Söbbeke_1 | 1.50% | x | |
| cy16 | Söbbeke_2 | 1.50% | x | |
| cy18 | Söbbeke_ABC | 3.80% | x | ABC |
| cy17 | Söbbeke | 3.80% | x | |
| cy13 | Starter_culture (13h) | 3.50% | x | |
| cy14 | Starter_culture (13h) frozen | 3.50% | x |
References
- Gilliland, S.E. Health and nutritional benefits from lactic acid bacteria. FEMS Microbiol. Rev. 1990, 7, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.F.; Shahani, K.M.; Amer, M.A. Therapeutic role of dietary lactobacilli and lactobacillic fermented dairy products. FEMS Microbiol. Lett. 1987, 46, 343–356. [Google Scholar] [CrossRef]
- García Fontán, M.C.; Martínez, S.; Franco, I.; Carballo, J. Microbiological and chemical changes during the manufacture of Kefir made from cows’ milk, using a commercial starter culture. Int. Dairy J. 2006, 16, 762–767. [Google Scholar] [CrossRef]
- Li, J.; Yuan, H.; Yao, Y.; Hua, J.; Yang, Y.; Dong, C.; Deng, Y.; Wang, J.; Li, H.; Jiang, Y.; et al. Rapid volatiles fingerprinting by dopant-assisted positive photoionization ion mobility spectrometry for discrimination and characterization of Green Tea aromas. Talanta 2019, 191, 39–45. [Google Scholar] [CrossRef]
- Gerhardt, N.; Birkenmeier, M.; Schwolow, S.; Rohn, S.; Weller, P. Volatile-Compound Fingerprinting by Headspace-Gas-Chromatography Ion-Mobility Spectrometry (HS-GC-IMS) as a Benchtop Alternative to 1H NMR Profiling for Assessment of the Authenticity of Honey. Anal. Chem. 2018, 90, 1777–1785. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Maecker, R.; Vyhmeister, E.; Meisen, S.; Rosales Martinez, A.; Kuklya, A.; Telgheder, U. Identification of terpenes and essential oils by means of static headspace gas chromatography-ion mobility spectrometry. Anal. Bioanal. Chem. 2017, 409, 6595–6603. [Google Scholar] [CrossRef]
- Wang, S.; Chen, H.; Sun, B. Recent progress in food flavor analysis using gas chromatography-ion mobility spectrometry (GC-IMS). Food Chem. 2020, 315, 126158. [Google Scholar] [CrossRef]
- Hernández-Mesa, M.; Ropartz, D.; García-Campaña, A.M.; Rogniaux, H.; Dervilly-Pinel, G.; Le Bizec, B. Ion Mobility Spectrometry in Food Analysis: Principles, Current Applications and Future Trends. Molecules 2019, 24, 2706. [Google Scholar] [CrossRef] [Green Version]
- Cohen, G.; Rudnik, D.D.; Laloush, M.; Yakir, D.; Karpas, Z. A Novel Method for Determination of Histamine in Tuna Fish by Ion Mobility Spectrometry. Food Anal. Methods 2015, 8, 2376–2382. [Google Scholar] [CrossRef]
- Borsdorf, H.; Eiceman, G.A. Ion Mobility Spectrometry: Principles and Applications. Appl. Spectrosc. Rev. 2006, 41, 323–375. [Google Scholar] [CrossRef]
- Cumeras, R.; Figueras, E.; Davis, C.E.; Baumbach, J.I.; Gràcia, I. Review on ion mobility spectrometry. Part 2: Hyphenated methods and effects of experimental parameters. Analyst 2015, 140, 1391–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, C.; Jaros, D.; Rohm, H. Ion Mobility Spectrometry as a Potential Tool for Flavor Control in Chocolate Manufacture. Foods 2019, 8, 460. [Google Scholar] [CrossRef] [Green Version]
- Karpas, Z.; Guamán, A.V.; Calvo, D.; Pardo, A.; Marco, S. The potential of ion mobility spectrometry (IMS) for detection of 2,4,6-trichloroanisole (2,4,6-TCA) in wine. Talanta 2012, 93, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Tzschoppe, M.; Haase, H.; Höhnisch, M.; Jaros, D.; Rohm, H. Using ion mobility spectrometry for screening the autoxidation of peanuts. Food Control 2016, 64, 17–21. [Google Scholar] [CrossRef]
- Rearden, P.; Harrington, P.B. Rapid screening of precursor and degradation products of chemical warfare agents in soil by solid-phase microextraction ion mobility spectrometry (SPME–IMS). Anal. Chim. Acta 2005, 545, 13–20. [Google Scholar] [CrossRef]
- Cook, G.W.; LaPuma, P.T.; Hook, G.L.; Eckenrode, B.A. Using gas chromatography with ion mobility spectrometry to resolve explosive compounds in the presence of interferents. J. Forensic Sci. 2010, 55, 1582–1591. [Google Scholar] [CrossRef]
- Zalewska, A.; Pawłowski, W.; Tomaszewski, W. Limits of detection of explosives as determined with IMS and field asymmetric IMS vapour detectors. Forensic Sci. Int. 2013, 226, 168–172. [Google Scholar] [CrossRef]
- Wu, C.; Siems, W.F.; Hill, J.H.H.; Hannan, R.M. Analytical determination of nicotine in tobacco by supercritical fluid chromatography–ion mobility detection. J. Chromatogr. A 1998, 811, 157–161. [Google Scholar] [CrossRef]
- Griffin, G.W.; Dzidic, I.; Carroll, D.I.; Stillwell, R.N.; Horning, E.C. Ion mass assignments based on mobility measuremets. Validity of plasma chromatographic mass mobility correlations. Anal. Chem. 1973, 45, 1204–1209. [Google Scholar] [CrossRef]
- Pomareda, V.; Guamán, A.V.; Mohammadnejad, M.; Calvo, D.; Pardo, A.; Marco, S. Multivariate curve resolution of nonlinear ion mobility spectra followed by multivariate nonlinear calibration for quantitative prediction. Chemom. Intell. Lab. Syst. 2012, 118, 219–229. [Google Scholar] [CrossRef]
- Borsdorf, H.; Rudolph, M. Gas-phase ion mobility studies of constitutional isomeric hydrocarbons using different ionization techniques. Int. J. Mass Spectrom. 2001, 208, 67–72. [Google Scholar] [CrossRef]
- Johnson, P.V.; Beegle, L.W.; Kim, H.I.; Eiceman, G.A.; Kanik, I. Ion mobility spectrometry in space exploration. Int. J. Mass Spectrom. 2007, 262, 1–15. [Google Scholar] [CrossRef]
- Brendel, R.; Schwolow, S.; Rohn, S.; Weller, P. Comparison of PLSR, MCR-ALS and Kernel-PLSR for the quantification of allergenic fragrance compounds in complex cosmetic products based on nonlinear 2D GC-IMS data. Chemom. Intell. Lab. Syst. 2020, 205, 104128. [Google Scholar] [CrossRef]
- Gómez-Ramos, M.M.; Ferrer, C.; Malato, O.; Agüera, A.; Fernández-Alba, A.R. Liquid chromatography-high-resolution mass spectrometry for pesticide residue analysis in fruit and vegetables: Screening and quantitative studies. J. Chromatogr. A 2013, 1287, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Márquez-Sillero, I.; Cárdenas, S.; Sielemann, S.; Valcárcel, M. On-line headspace-multicapillary column-ion mobility spectrometry hyphenation as a tool for the determination of off-flavours in foods. J. Chromatogr. A 2014, 1333, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Shuai, Q.; Zhang, L.; Li, P.; Zhang, Q.; Wang, X.; Ding, X.; Zhang, W. Rapid adulteration detection for flaxseed oil using ion mobility spectrometry and chemometric methods. Anal. Methods 2014, 6, 9575–9580. [Google Scholar] [CrossRef]
- Garrido-Delgado, R.; Dobao-Prieto, M.M.; Arce, L.; Aguilar, J.; Cumplido, J.L.; Valcárcel, M. Ion mobility spectrometry versus classical physico-chemical analysis for assessing the shelf life of extra virgin olive oil according to container type and storage conditions. J. Agric. Food Chem. 2015, 63, 2179–2188. [Google Scholar] [CrossRef]
- Denawaka, C.J.; Fowlis, I.A.; Dean, J.R. Evaluation and application of static headspace-multicapillary column-gas chromatography-ion mobility spectrometry for complex sample analysis. J. Chromatogr. A 2014, 1338, 136–148. [Google Scholar] [CrossRef] [Green Version]
- Krisilova, E.V.; Levina, A.M.; Makarenko, V.A. Determination of the volatile compounds of vegetable oils using an ion-mobility spectrometer. J. Anal. Chem. 2014, 69, 371–376. [Google Scholar] [CrossRef]
- Danezis, G.P.; Tsagkaris, A.S.; Camin, F.; Brusic, V.; Georgiou, C.A. Food authentication: Techniques, trends & emerging approaches. TrAC Trends Anal. Chem. 2016, 85, 123–132. [Google Scholar] [CrossRef] [Green Version]
- Karpas, Z. Applications of ion mobility spectrometry (IMS) in the field of foodomics. Food Res. Int. 2013, 54, 1146–1151. [Google Scholar] [CrossRef]
- Hernández-Mesa, M.; Escourrou, A.; Monteau, F.; Le Bizec, B.; Dervilly-Pinel, G. Current applications and perspectives of ion mobility spectrometry to answer chemical food safety issues. TrAC Trends Anal. Chem. 2017, 94, 39–53. [Google Scholar] [CrossRef]
- Cristoni, S.; Bernardi, L.R.; Malvandi, A.M.; Larini, M.; Longhi, E.; Sortino, F.; Conti, M.; Pantano, N.; Puccio, G. A case of personalized and precision medicine: Pharmacometabolomic applications to rare cancer, microbiological investigation, and therapy. Rapid Commun. Mass Spectrom. 2021, 35, e8976. [Google Scholar] [CrossRef] [PubMed]
- Cristoni, S.; Rossi Bernardi, L.; Larini, M.; Natale, G.; Didomenico, N.; Varelli, M.; Conti, M.; Dorna, I.; Puccio, G. Predicting and preventing intestinal dysbiosis on the basis of pharmacological gut microbiota metabolism. Rapid Commun. Mass Spectrom. 2019, 33, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Baduel, C.; Mueller, J.F.; Tsai, H.; Gomez Ramos, M.J. Development of sample extraction and clean-up strategies for target and non-target analysis of environmental contaminants in biological matrices. J. Chromatogr. A 2015, 1426, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Schwolow, S.; Gerhardt, N.; Rohn, S.; Weller, P. Data fusion of GC-IMS data and FT-MIR spectra for the authentication of olive oils and honeys-is it worth to go the extra mile? Anal. Bioanal. Chem. 2019, 411, 6005–6019. [Google Scholar] [CrossRef]
- Kessler, W. Multivariate Datenanalyse: Für Die Pharma-, Bio- und Prozessanalytik: Ien Lehrbuch; Wiley-VCH: Weinheim, Germany, 2007; ISBN 978-3-527-31262-7. [Google Scholar]
- Maimon, O.; Rokach, L. Data Mining and Knowledge Discovery Handbook; Springer: Boston, MA, USA, 2005; ISBN 978-0-387-24435-8. [Google Scholar]
- Granato, D.; Santos, J.S.; Escher, G.B.; Ferreira, B.L.; Maggio, R.M. Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical perspective. Trends Food Sci. Technol. 2018, 72, 83–90. [Google Scholar] [CrossRef]
- Robinson, R.K.; Lucey, J.A.; Tamime, A.Y. Manufacture of Yoghurt. In Fermented Milks; Tamime, A., Ed.; Blackwell Publishing Ltd.: Oxford, UK, 2006; pp. 53–75. ISBN 9780470995501. [Google Scholar]
- Dan, T.; Wang, D.; Wu, S.; Jin, R.; Ren, W.; Sun, T. Profiles of Volatile Flavor Compounds in Milk Fermented with Different Proportional Combinations of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus. Molecules 2017, 22, 1633. [Google Scholar] [CrossRef] [Green Version]
- Dan, T.; Ren, W.; Liu, Y.; Tian, J.; Chen, H.; Li, T.; Liu, W. Volatile Flavor Compounds Profile and Fermentation Characteristics of Milk Fermented by Lactobacillus delbrueckii subsp. bulgaricus. Front. Microbiol. 2019, 10, 2183. [Google Scholar] [CrossRef]
- Spietelun, A.; Pilarczyk, M.; Kloskowski, A.; Namieśnik, J. Current trends in solid-phase microextraction (SPME) fibre coatings. Chem. Soc. Rev. 2010, 39, 4524–4537. [Google Scholar] [CrossRef]
- Souza Silva, E.A.; Risticevic, S.; Pawliszyn, J. Recent trends in SPME concerning sorbent materials, configurations and in vivo applications. TrAC Trends Anal. Chem. 2013, 43, 24–36. [Google Scholar] [CrossRef]
- Wouters, J.T.; Ayad, E.H.; Hugenholtz, J.; Smit, G. Microbes from raw milk for fermented dairy products. Int. Dairy J. 2002, 12, 91–109. [Google Scholar] [CrossRef]
- Prado, M.R.; Blandón, L.M.; Vandenberghe, L.P.S.; Rodrigues, C.; Castro, G.R.; Thomaz-Soccol, V.; Soccol, C.R. Milk kefir: Composition, microbial cultures, biological activities, and related products. Front. Microbiol. 2015, 6, 1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diniz, R.; Garla, L.; Schneedorf, J.; Carvalho, J. Study of anti-inflammatory activity of Tibetan mushroom, a symbiotic culture of bacteria and fungi encapsulated into a polysaccharide matrix. Pharmacol. Res. 2003, 47, 49–52. [Google Scholar] [CrossRef]
- Huseini, H.F.; Rahimzadeh, G.; Fazeli, M.R.; Mehrazma, M.; Salehi, M. Evaluation of wound healing activities of kefir products. Burns 2012, 38, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Nalbantoglu, U.; Cakar, A.; Dogan, H.; Abaci, N.; Ustek, D.; Sayood, K.; Can, H. Metagenomic analysis of the microbial community in kefir grains. Food Microbiol. 2014, 41, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wu, F.; Wang, X.; Sui, Y.; Yang, L.; Wang, J. Characterization of Lactobacillus plantarum Lp27 isolated from Tibetan kefir grains: A potential probiotic bacterium with cholesterol-lowering effects. J. Dairy Sci. 2013, 96, 2816–2825. [Google Scholar] [CrossRef] [Green Version]
- Garrote, G.L.; Abraham, A.G.; De Antoni, G.L. Chemical and microbiological characterisation of kefir grains. J. Dairy Res. 2001, 68, 639–652. [Google Scholar] [CrossRef]
- La Rivière, J.W.; Kooiman, P. Kefiran, a novel polysaccharide produced in the kefir grain by Lactobacillus brevis. Arch. Mikrobiol. 1967, 59, 269–278. [Google Scholar] [CrossRef]
- Wszolek, M.; Kupiec-Teahan, B.; Skov Guldager, H.; Tamime, A.Y. Production of Kefir, Koumiss and other Related Products. In Fermented Milks; Tamime, A., Ed.; Blackwell Publishing Ltd.: Oxford, UK, 2006; pp. 174–216. ISBN 9780470995501. [Google Scholar]
- Simova, E.; Beshkova, D.; Angelov, A.; Hristozova, T.; Frengova, G.; Spasov, Z. Lactic acid bacteria and yeasts in kefir grains and kefir made from them. J. Ind. Microbiol. Biotechnol. 2002, 28, 1–6. [Google Scholar] [CrossRef]
- Rea, M.C.; Lennartsson, T.; Dillon, P.; Drinan, F.D.; Reville, W.J.; Heapes, M.; Cogan, T.M. Irish kefir-like grains: Their structure, microbial composition and fermentation kinetics. J. Appl. Bacteriol. 1996, 81, 83–94. [Google Scholar] [CrossRef]
- Güzel-Seydim, Z.B.; Seydim, A.C.; Greene, A.K.; Bodine, A.B. Determination of Organic Acids and Volatile Flavor Substances in Kefir during Fermentation. J. Food Compos. Anal. 2000, 13, 35–43. [Google Scholar] [CrossRef]
- Battcock, M.; Azam-Ali, S. Fermented Fruits and Vegetables: A Global Perspective; Food and Agriculture Organization of the United Nations: Rome, Italy, 1998; ISBN 92-5-104226-8. [Google Scholar]
- Cogan, T.M.; Hill, C. Cheese Starter Cultures. In Cheese: Chemistry, Physics and Microbiology; Fox, P.F., Ed.; Springer: Boston, MA, USA, 1993; pp. 193–255. ISBN 978-1-4613-6138-1. [Google Scholar]
- Tamime, A.Y.; Marshall, V.M.E. Microbiology and technology of fermented milks. In Microbiology and Biochemistry of Cheese and Fermented Milk; Law, B.A., Ed.; Springer: Boston, MA, USA, 1997; pp. 57–152. ISBN 978-1-4612-8427-7. [Google Scholar]
- Tomasini, A.; Bustillo, G.; Lebeault, J.-M. Production of blue cheese flavour concentrates from different substrates supplemented with lipolyzed cream. Int. Dairy J. 1995, 5, 247–257. [Google Scholar] [CrossRef]
- Berger, C.; Khan, J.A.; Molimard, P.; Martin, N.; Spinnler, H.E. Production of sulfur flavors by ten strains of Geotrichum candidum. Appl. Environ. Microbiol. 1999, 65, 5510–5514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Kranenburg, R.; Kleerebezem, M.; van Hylckama Vlieg, J.; Ursing, B.M.; Boekhorst, J.; Smit, B.A.; Ayad, E.H.; Smit, G.; Siezen, R.J. Flavour formation from amino acids by lactic acid bacteria: Predictions from genome sequence analysis. Int. Dairy J. 2002, 12, 111–121. [Google Scholar] [CrossRef]
- Tamime, A.Y.; Skriver, A.; Nilsson, L.-E. Starter Cultures. In Fermented Milks; Tamime, A., Ed.; Blackwell Publishing Ltd.: Oxford, UK, 2006; pp. 11–52. ISBN 9780470995501. [Google Scholar]
- Brendel, R.; Schwolow, S.; Rohn, S.; Weller, P. Volatilomic Profiling of Citrus Juices by Dual-Detection HS-GC-MS-IMS and Machine Learning-An Alternative Authentication Approach. J. Agric. Food Chem. 2021, 69, 1727–1738. [Google Scholar] [CrossRef]
- Gerhardt, N.; Birkenmeier, M.; Sanders, D.; Rohn, S.; Weller, P. Resolution-optimized headspace gas chromatography-ion mobility spectrometry (HS-GC-IMS) for non-targeted olive oil profiling. Anal. Bioanal. Chem. 2017, 409, 3933–3942. [Google Scholar] [CrossRef]
- Blanca, M.J.; Alarcón, R.; Arnau, J.; Bono, R.; Bendayan, R. Effect of variance ratio on ANOVA robustness: Might 1.5 be the limit? Behav. Res. Methods 2018, 50, 937–962. [Google Scholar] [CrossRef]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE 2019, 14, e0224365. [Google Scholar] [CrossRef]
- Walsh, A.M.; Crispie, F.; Kilcawley, K.; O’Sullivan, O.; O’Sullivan, M.G.; Claesson, M.J.; Cotter, P.D. Microbial Succession and Flavor Production in the Fermented Dairy Beverage Kefir. mSystems 2016, 1, e00052-16. [Google Scholar] [CrossRef] [Green Version]
- Ruth, J.H. Odor thresholds and irritation levels of several chemical substances: A review. Am. Ind. Hyg. Assoc. J. 1986, 47, A142–A151. [Google Scholar] [CrossRef]
- Lide, D.R. CRC Handbook of Chemistry and Physics: A Ready-Reference Book of Chemical and Physical Data, 87th ed.; CRC Taylor & Francis: Boca Raton, FL, USA, 2006; ISBN 0849304873. [Google Scholar]
- Jarabek, A.M. Inhalation RfC Methodology: Dosimetric Adjustments and Dose-Response Estimation of Noncancer Toxicity in the Upper Respiratory Tract. Inhal. Toxicol. 1994, 6, 301. Available online: https://hero.epa.gov/hero/index.cfm/reference/details/reference_id/6487 (accessed on 26 January 2022).
- Gossauer, A. Struktur und Reaktivität der Biomoleküle: Eine Einführung in Die Organische Chemie; Verl. Helvetica Chimica Acta: Zürich, Switzerland, 2006; ISBN 978-3-906390-29-1. [Google Scholar]
- Kubitschke, J.; Lange, H.; Strutz, H. Carboxylic Acids, Aliphatic. Ullmann’s Encyclopedia of Industrial Chemistry; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2000; pp. 1–18. ISBN 9783527306732. [Google Scholar]
- Wang, H.; Sun, X.; Song, X.; Guo, M. Effects of kefir grains from different origins on proteolysis and volatile profile of goat milk kefir. Food Chem. 2021, 339, 128099. [Google Scholar] [CrossRef] [PubMed]
- Mallikarjuna, N.; Yellamma, K. Genetic and Metabolic Engineering of Microorganisms for the Production of Various Food Products. In Recent Developments in Applied Microbiology and Biochemistry; Elsevier: Cambride, MA, USA, 2019; pp. 167–182. ISBN 9780128163283. [Google Scholar]
- Cheng, H. Volatile flavor compounds in yogurt: A review. Crit. Rev. Food Sci. Nutr. 2010, 50, 938–950. [Google Scholar] [CrossRef]
- Beshkova, D.M.; Simova, E.D.; Frengova, G.I.; Simov, Z.I.; Dimitrov, Z. Production of volatile aroma compounds by kefir starter cultures. Int. Dairy J. 2003, 13, 529–535. [Google Scholar] [CrossRef]
- García-Llatas, G.; Lagarda, M.J.; Romero, F.; Abellán, P.; Farré, R. A headspace solid-phase microextraction method of use in monitoring hexanal and pentane during storage: Application to liquid infant foods and powdered infant formulas. Food Chem. 2007, 101, 1078–1086. [Google Scholar] [CrossRef] [Green Version]
- Buttery, R.G.; Turnbaugh, J.G.; Ling, L.C. Contribution of volatiles to rice aroma. J. Agric. Food Chem. 1988, 36, 1006–1009. [Google Scholar] [CrossRef]
- Nejati, F.; Junne, S.; Kurreck, J.; Neubauer, P. Quantification of Major Bacteria and Yeast Species in Kefir Consortia by Multiplex TaqMan qPCR. Front. Microbiol. 2020, 11, 1291. [Google Scholar] [CrossRef]
- De Luca, L.; Aiello, A.; Pizzolongo, F.; Blaiotta, G.; Aponte, M.; Romano, R. Volatile Organic Compounds in Breads Prepared with Different Sourdoughs. Appl. Sci. 2021, 11, 1330. [Google Scholar] [CrossRef]
- Marini, F. Chemometrics in Food Chemistry, 1st ed.; Elsevier Science: Burlington, NJ, USA, 2013; ISBN 9780444595294. [Google Scholar]
- Varmuza, K.; Filzmoser, P. Introduction to Multivariate Statistical Analysis in Chemometrics; CRC Taylor & Francis: Boca Raton, FL, USA, 2016; ISBN 9780429145049. [Google Scholar]
- SDBSWeb: National Institute of Advanced Industrial Science and Technology. Available online: https://sdbs.db.aist.go.jp (accessed on 16 September 2021).


| Pattern Recognition and Data Reduction Technique | Supervised Method | CV Error Rate (%) | Prediction Accuracy (Posterior) (%) |
|---|---|---|---|
| PCA (PC 1 to 4) | LDA | 0 | 100 |
| kNN (k = 5) | 0 | 100 | |
| SVM | 3.3 (2 out of 61) | 100 | |
| PLS (PLS 1 to 4) | DA | 1.6 (1 out of 61) | 100 |
| PLS (PLS 1 to 5) | DA | 0 | 100 |
| NNMF (C 1 to 4) | LDA | 3.3 (2 out of 61) | 87 (13 out of 15) |
| kNN (k = 5) | 0 | 100 | |
| SVM | 5.0 (3 out of 61) | 100 |
| Pattern Recognition and Data Reduction Technique | Supervised Method | CV Error Rate (8-Fold CV) (%) | Prediction Accuracy (Posterior) (%) |
|---|---|---|---|
| PCA (PC 1 to 4) | LDA | 1.6 (1 out of 61) | 0 |
| kNN (k = 5) | 0 | 0 | |
| SVM | 6.6 (4 out of 61) | 0 | |
| PCA (PC 1 to 5) | LDA | 0 | 100 |
| kNN (k = 5) | 0 | 100 | |
| SVM | 9.8 (6 out of 61) | 93 (14 out of 15) | |
| PLS (PLS 1 to 4) | DA | 0 | 100 |
| PLS (PLS 1 to 5) | DA | 0 | 100 |
| NNMF (C 1 to 5) | LDA | 5.0 (3 out of 61) | 93 (14 out of 15) |
| kNN (k = 5) | 0 | 100 | |
| SVM | 8.2 (5 out of 61) | 87 (13 out of 15) |
| # | Compound | Retention Time Start (s) | Molar Mass (g mol−1) | Odor Descriptor | Identification Method |
|---|---|---|---|---|---|
| 1 | Ethanol | 150.1 | 46 | Dry, dust [68] | RS |
| 2 | Acetone | 155.6 | 58 | Earthy, fruity, wood pulp, hay [68] | RS, MS |
| 3 | Tentatively pentane | 155.6 | 72 | Faint gasoline-like [69] | RS |
| 4 | Unknown | 155.9 | |||
| 5 | Tentatively propanol | 167.6 | 60 | Mild, alcohol-like [70] | RS |
| 6 | Unknown | 167.6 | |||
| 7 | 2-methyl propanal (isobutyraldehyde) | 167.6 | 58 | Faint gasoline-like, natural gas [71] | RS, MS |
| 8 | Acetic acid | 169.9 | 88 | Vinegar, peppers, green, fruity, floral, sour [68] | RS, MS |
| 9 | Butane-2,3-dione (diacetyl) | 174.0 | 86 | Buttery, strong [68] | RS, MS |
| 10 | 2-butanone | 178.0 | 80 | Buttery, sour milk, etheric [68] | RS |
| 11 | Ethyl acetate | 182.2 | 60 | Solvent, pineapple, fruity, apples [68] | RS, MS |
| 12 | 2-methyl-1-propanol (isobutanol) | 188.1 | 74 | Malty [68] | RS, MS |
| 13 | 3-methylbutanal | 198.7 | 86 | Malty, cheesy, green, dark chocolate, cocoa [68] | RS, MS |
| 14 | 2-methylbutanal | 203.3 | 86 | Malty, dark chocolate, almond, cocoa, coffee [68] | RS |
| 15 | 2,3-pentandione | 216.3 | 100 | Creamy, cheesy, oily, sweet buttery, caramellic [68] | RS |
| 16 | 2-pentanone | 214.2 | 86 | Orange peel, sweet, fruity [68] | RS |
| 17 | 3-hydroxybutan-2-one (acetoin) | 226.7 | 88 | Bland, yogurt-like [72] | RS |
| 18 | 2-methyl-1-butanol | 244.4 | 88 | Penetrating, alcohol, wine-like, plastic [68] | RS |
| 19 | 3-methyl-1-butanol (isoamyl alcohol) | 239.4 | 88 | Fresh cheese, breathtaking, alcoholic, fruity, grainy, solvent-like, floral, malty [68] | RS, MS |
| 20 | Butyric acid | 264.3 | 88 | Unpleasant, similar to vomit or body odor [73] | RS |
| 21 | Unknown | 282.1 | |||
| 22 | Hexanal | 286.3 | 100 | Green, slightly fruity, lemon, herbal, grassy, tallow [68] | RS |
| 23 | Unknown | 299.8 | |||
| 24 | Unknown | 320.5 | |||
| 25 | 3-methylbutyl acetate (isoamyl acetate) | 355.1 | 130 | Fruity, banana, candy, sweet, apple peel [68] | RS |
| 26 | 2-heptanone | 369.2 | 114 | Blue cheese, spicy, Roquefort [68] | RS |
| 27 | Unknown | 378.7 | |||
| 28 | Unknown | 427.3 | |||
| 29 | Hexanoic acid (caproic acid) | 443.7 | 116 | Sweaty, cheesy, sharp, goaty, bad breath, acidic [68] | RS |
| 30 | Ethyl hexanoate | 474.0 | 144 | Fruity, malty, young cheese, moldy, apple, green, orange, pineapple, banana [68] | RS |
| 31 | 2-nonanone | 570.8 | 142 | Malty, fruity, hot milk, smoked cheese, lipid metabolism [68] | RS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capitain, C.C.; Nejati, F.; Zischka, M.; Berzak, M.; Junne, S.; Neubauer, P.; Weller, P. Volatilomics-Based Microbiome Evaluation of Fermented Dairy by Prototypic Headspace-Gas Chromatography–High-Temperature Ion Mobility Spectrometry (HS-GC-HTIMS) and Non-Negative Matrix Factorization (NNMF). Metabolites 2022, 12, 299. https://doi.org/10.3390/metabo12040299
Capitain CC, Nejati F, Zischka M, Berzak M, Junne S, Neubauer P, Weller P. Volatilomics-Based Microbiome Evaluation of Fermented Dairy by Prototypic Headspace-Gas Chromatography–High-Temperature Ion Mobility Spectrometry (HS-GC-HTIMS) and Non-Negative Matrix Factorization (NNMF). Metabolites. 2022; 12(4):299. https://doi.org/10.3390/metabo12040299
Chicago/Turabian StyleCapitain, Charlotte C., Fatemeh Nejati, Martin Zischka, Markus Berzak, Stefan Junne, Peter Neubauer, and Philipp Weller. 2022. "Volatilomics-Based Microbiome Evaluation of Fermented Dairy by Prototypic Headspace-Gas Chromatography–High-Temperature Ion Mobility Spectrometry (HS-GC-HTIMS) and Non-Negative Matrix Factorization (NNMF)" Metabolites 12, no. 4: 299. https://doi.org/10.3390/metabo12040299
APA StyleCapitain, C. C., Nejati, F., Zischka, M., Berzak, M., Junne, S., Neubauer, P., & Weller, P. (2022). Volatilomics-Based Microbiome Evaluation of Fermented Dairy by Prototypic Headspace-Gas Chromatography–High-Temperature Ion Mobility Spectrometry (HS-GC-HTIMS) and Non-Negative Matrix Factorization (NNMF). Metabolites, 12(4), 299. https://doi.org/10.3390/metabo12040299








