Phylogenetic Studies and Metabolite Analysis of Sticta Species from Colombia and Chile by Ultra-High Performance Liquid Chromatography-High Resolution-Q-Orbitrap-Mass Spectrometry
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification of Metabolites in 11 Lichen Species
2.1.1. Saturated Organic Acids
2.1.2. Small Phenolic Compounds
2.1.3. Typical Lichenic Phenolic Compounds (Depsides, Depsidones and Anthraquinones)
2.1.4. Terpenes
2.1.5. Lipids
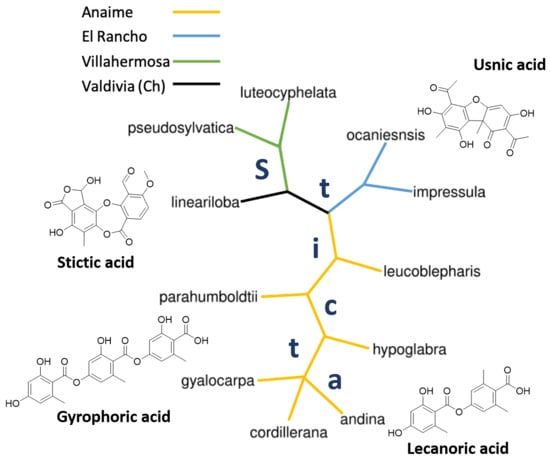
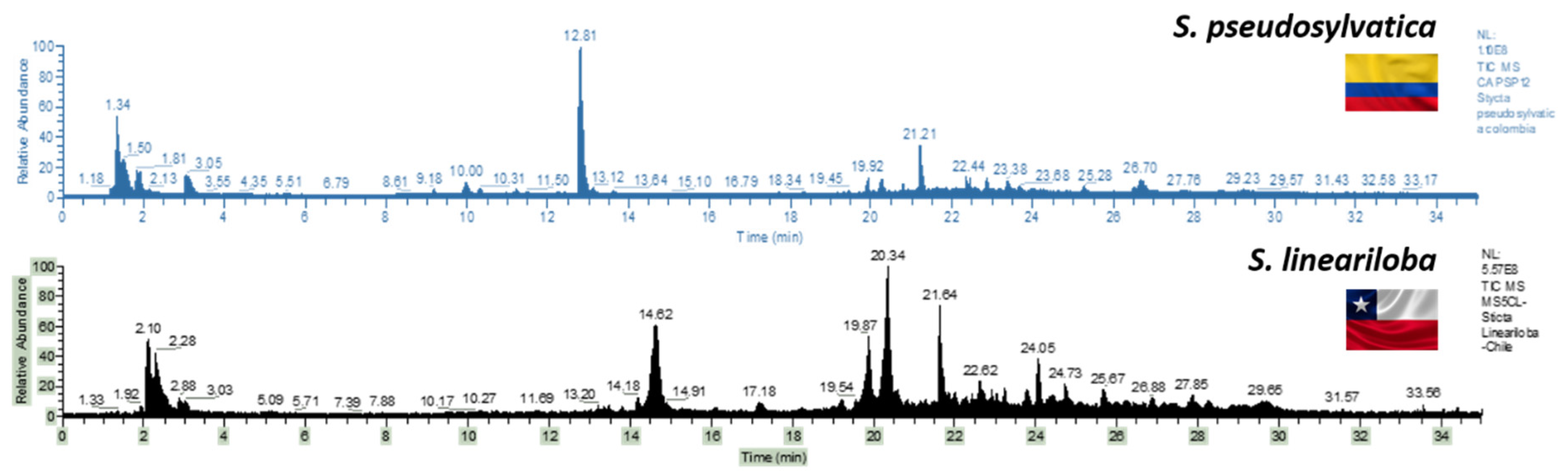
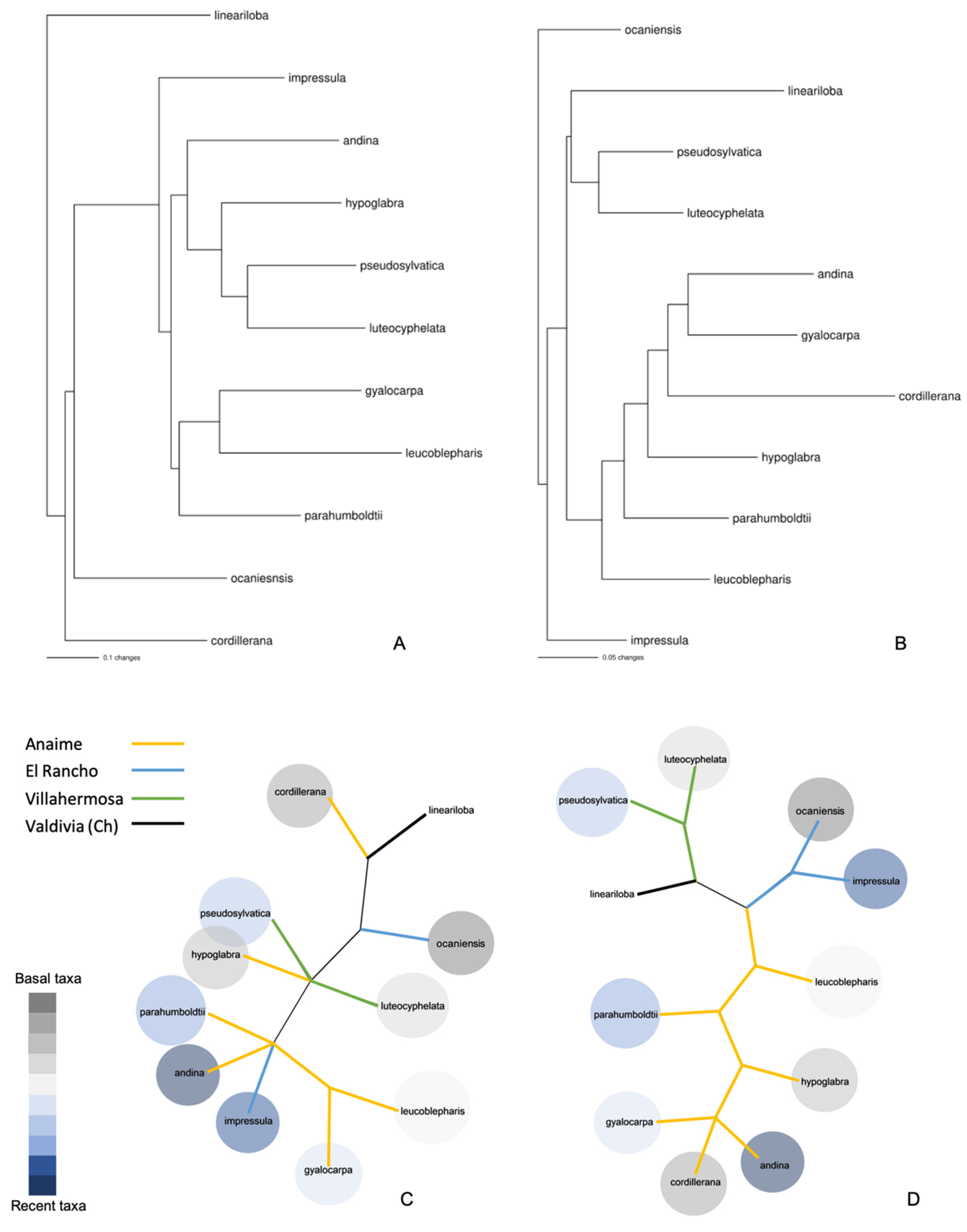
2.1.6. Distance and Phylogenetic Analysis
3. Materials and Methods
3.1. Chemicals
3.2. Lichen Material
3.3. Preparation of the Sample for Analyses
3.4. Instrument
3.5. LC Parameters
3.6. MS Parameters
3.7. Similarity and Phylogenetic Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nash, T. Lichen Biology, 2nd ed.; Cambridge University Press: Cambridge, MA, USA, 2008. [Google Scholar]
- Armaleo, D.; Clerc’, P. Lichen chimeras: DNA analysis suggests that one fungus forms twp morphotypes. Exp. Mycol. 1991, 15, 1–10. [Google Scholar] [CrossRef]
- Sanders, W.B. Composite lichen thalli of sticta sp. from Brazil, with morphologically similar lobes containing either a chlorobiont or a cyanobiont layer. Symbiosis 2001, 31, 47–55. [Google Scholar]
- Henskens, F.L.; Green, T.G.A.; Wilkins, A. Cyanolichens can have both cyanobacteria and green algae in a common layer as major contributors to photosynthesis. Ann. Bot. 2012, 110, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Galloway, D.J. Studies on the lichen genus Sticta (Schreber) Ach.: I. Southern South American species. Lichenol 1994, 26, 223–282. [Google Scholar] [CrossRef]
- Galloway, D.J. Studies on the lichen genus Sticta (Schreber) Ach.: V*. Australian species. Bryophyt. Divers. Evol. 1998, 15, 117–160. [Google Scholar] [CrossRef]
- Galloway, D.J. Studies on the lichen genus Sticta (Schreber) Ach. IV. New Zealand species. Lichenologist 1997, 29, 105–168. [Google Scholar] [CrossRef]
- Moncada, B.; Aguirre, J.; Lücking, R. Ecogeografía del género Sticta (ascomycota liquenizados: Lobariaceae) en Colombia. Rev. Biol. Trop. 2014, 62, 266–281. [Google Scholar] [CrossRef][Green Version]
- Moncada, B.; Luecking, R.K.; Lumbsch, H.T. Rewriting the evolutionary history of the lichen genus Sticta (Ascomycota: Peltigeraceae subfam. Lobarioideae) in the Hawaiian Islands. Plant Fungal Syst. 2020, 65, 95–119. [Google Scholar] [CrossRef]
- Moncada, B.; Lücking, R. Ten new species of Sticta and counting: Colombia as a hot spot for unrecognized diversification in a conspicuous macrolichen genus. Phytotaxa 2012, 74, 1–29. [Google Scholar] [CrossRef]
- Moncada, B.; Mercado-Díaz, J.A.; Magain, N.; Hodkinson, B.P.; Smith, C.W.; Bungartz, F.; Pérez-Pérez, R.E.; Gumboski, E.; Sérusiaux, E.; Lumbsch, H.T.; et al. Phylogenetic diversity of two geographically overlapping lichens: Isolation by distance, environment, or fragmentation? J. Biogeogr. 2021, 48, 676–689. [Google Scholar] [CrossRef]
- Mercado-Díaz, J.A.; Lücking, R.; Moncada, B.; Widhelm, T.J.; Lumbsch, H.T. Elucidating species richness in lichen fungi: The genus Sticta (Ascomycota: Peltigeraceae) in Puerto Rico. Taxon 2020, 69, 851–891. [Google Scholar] [CrossRef]
- Sepahvand, A.; Studzińska-Sroka, E.; Ramak, P.; Karimian, V. Usnea sp.: Antimicrobial potential, bioactive compounds, ethnopharmacological uses and other pharmacological properties; a review article. J. Ethnopharmacol. 2021, 268, 113656. [Google Scholar] [CrossRef] [PubMed]
- Calcott, M.J.; Ackerley, D.F.; Knight, A.; Keyzers, R.A.; Owen, J.G. Secondary metabolism in the lichen symbiosis. Chem. Soc. Rev. 2018, 47, 1730–1760. [Google Scholar] [CrossRef] [PubMed]
- Corbett, B.R.E.; Young, H.J. Lichens and fungi. Part III. Structural elucidation of 15α, 22-dihydroxyhopane from Sticta billardierii Del. Chem. Soc. C Org. 1966, 1564–1567. [Google Scholar] [CrossRef]
- Chin, W.J.; Corbett, R.E.; Heng, C.K.; Wilkins, A.L. Lichens and fungi. Part XI. Isolation and structural elucidation of a new group of triterpenes from Sticta coronata, S. colensoi, and S. flavicans. J. Chem. Soc. Perkin Trans. 1 1973, 14, 1437–1446. [Google Scholar] [CrossRef]
- Piovano, M.; Chamy, M.C.; Garbarino, J.A.; Quilhot, W. Secondary metabolites in the genus Sticta (lichens). Biochem. Syst. Ecol. 2000, 28, 589–590. [Google Scholar] [CrossRef]
- Zhang, H.J.; Guo, H.F.; Lou, H.X. Secondary metabolites from the Chinese lichen Sticta nylanderiana A. Z. Biochem. Syst. Ecol. 2006, 34, 760–762. [Google Scholar] [CrossRef]
- Liang, Z.; Sorribas, A.; Sulzmaier, F.J.; Jiménez, J.I.; Wang, X.; Sauvage, T.; Yoshida, W.Y.; Wang, G.; Ramos, J.W.; Williams, P.G. Stictamides A-C, MMP12 inhibitors containing 4-amino-3-hydroxy-5- phenylpentanoic acid subunits. J. Org. Chem. 2011, 76, 3635–3643. [Google Scholar] [CrossRef]
- Le Pogam, P.; Schinkovitz, A.; Legouin, B.; Le Lamer, A.C.; Boustie, J.; Richomme, P. Matrix-free UV-laser desorption ionization mass spectrometry as a versatile approach for accelerating dereplication studies on lichens. Anal. Chem. 2015, 87, 10421–10428. [Google Scholar] [CrossRef]
- Musharraf, S.G.; Kanwal, N.; Thadhani, V.M.; Choudhary, M.I. Rapid identification of lichen compounds based on the structure-fragmentation relationship using ESI-MS/MS analysis. Anal. Methods 2015, 7, 6066–6076. [Google Scholar] [CrossRef]
- Simirgiotis, M.J.; Quispe, C.; Areche, C.; Sepúlveda, B. Phenolic compounds in chilean mistletoe (quintral, Tristerix tetrandus) analyzed by UHPLC-Q/Orbitrap/MS/MS and its antioxidant properties. Molecules 2016, 21, 245. [Google Scholar] [CrossRef] [PubMed]
- Cornejo, A.; Salgado, F.; Caballero, J.; Vargas, R.; Simirgiotis, M.; Areche, C. Secondary metabolites in Ramalina terebrata detected by UHPLC/ESI/MS/MS and identification of parietin as tau protein inhibitor. Int. J. Mol. Sci. 2016, 17, 1303. [Google Scholar] [CrossRef]
- Castro, O.N.; Benites, J.; Rodilla, J.; Santiago, J.C.; Simirgiotis, M.; Sepulveda, B.; Areche, C. Metabolomic analysis of the lichen Everniopsis trulla using ultra high performance liquid chromatography-quadrupole-orbitrap mass spectrometry (UHPLC-Q-OT-MS). Chromatographia 2017, 80, 967–973. [Google Scholar] [CrossRef]
- Torres-Benítez, A.; Rivera-Montalvo, M.; Sepúlveda, B.; Castro, O.N.; Nagles, E.; Simirgiotis, M.J.; Garciá-Beltrán, O.; Areche, C. Metabolomic analysis of two Parmotrema lichens: P. robustum (Degel.) Hale and P. andinum (Mull. rg.) hale using UHPLC-ESI-OT-MS-MS. Molecules 2017, 22, 1861. [Google Scholar] [CrossRef]
- Salgado, F.; Albornoz, L.; Cortéz, C.; Stashenko, E.; Urrea-Vallejo, K.; Nagles, E.; Galicia-Virviescas, C.; Cornejo, A.; Ardiles, A.; Simirgiotis, M.; et al. Secondary metabolite profiling of species of the genus Usnea by UHPLC-ESI-OT-MS-MS. Molecules 2018, 23, 54. [Google Scholar] [CrossRef]
- Jiménez-González, A.; Quispe, C.; Bórquez, J.; Sepúlveda, B.; Riveros, F.; Areche, C.; Nagles, E.; García-Beltrán, O.; Simirgiotis, M.J. UHPLC-ESI-ORBITRAP-MS analysis of the native Mapuche medicinal plant palo negro (Leptocarpha rivularis DC.–Asteraceae) and evaluation of its antioxidant and cholinesterase inhibitory properties. J. Enzyme Inhib. Med. Chem. 2018, 23, 54. [Google Scholar] [CrossRef] [PubMed]
- Areche, C.; Fernandez-Burgos, R.; Terrones, T.C.D.; Simirgiotis, M.; García-Beltrán, O.; Borquez, J.; Sepulveda, B. Mulinum crassifolium Phil; two new mulinanes, gastroprotective activity and metabolomic analysis by UHPLC-orbitrap mass spectrometry. Molecules 2019, 24, 1673. [Google Scholar] [CrossRef]
- Parrot, D.; Jan, S.; Baert, N.; Guyot, S.; Tomasi, S. Comparative metabolite profiling and chemical study of Ramalina siliquosa complex using LC-ESI-MS/MS approach. Phytochemistry 2013, 89, 114–124. [Google Scholar] [CrossRef]
- Ly, H.D.; Vo, T.N.; Duong, T.H.; Nguyen, T.D.; Nguyen, K.P.P. A new depside and two new diphenyl ether compounds from the lichen Ramalina farinacea (L.). Ach. Phytochem. Lett. 2015, 11, 146–150. [Google Scholar] [CrossRef]
- Alam, M.A.; Khatoon, R.; Huda, S.; Ahmad, N.; Sharma, P.K. Biotechnological Applications of Lichens. In Lichen-Derived Products; John Wiley and Sons: Hoboken, NJ, USA, 2020; pp. 203–219. [Google Scholar]
- Nguyen, T.T.; Nallapaty, S.; Rao, G.S.N.K.; Koneru, S.T.; Annam, S.S.P.; Tatipamula, V.B. Evaluating the in vitro activity of depsidones from Usnea subfloridana Stirton as key enzymes involved in inflammation and gout. Pharm. Sci. 2021, 27, 291–296. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, M.; Xu, B. A comprehensive review on secondary metabolites and health-promoting effects of edible lichen. J. Funct. Foods 2020, 80, 104283. [Google Scholar] [CrossRef]
- Nguyen, T.T.H.; Dinh, M.H.; Chi, H.T.; Wang, S.L.; Nguyen, Q.V.; Tran, T.D.; Nguyen, A.D. Antioxidant and cytotoxic activity of lichens collected from Bidoup Nui Ba National Park, Vietnam. Res. Chem. Intermed. 2019, 45, 33–49. [Google Scholar] [CrossRef]
- Aoussar, N.; Laasri, F.E.; Bourhia, M.; Manoljovic, N.; Mhand, R.A.; Rhallabi, N.; Ullah, R.; Shahat, A.A.; Noman, O.M.; Nasr, F.A.; et al. Phytochemical analysis, cytotoxic, antioxidant, and antibacterial activities of lichens. Evid. Based Complement. Altern. Med. 2020, 2020, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Elix, J.; Elix, J.; Venables, D.; Venables, D.; Lumbsch, H.; Lumbsch, H.; Brako, L.; Brako, L. Further new metabolites from lichens. Aust. J. Chem. 1994, 47, 1619. [Google Scholar] [CrossRef]
- Sultana, N.; Afolayan, A.J. A new depsidone and antibacterial activities of compounds from Usnea undulata Stirton. J. Asian Nat. Prod. Res. 2011, 13, 1158–1164. [Google Scholar] [CrossRef]
- Le Pogam, P.; Le Lamer, A.C.; Siva, B.; Legouin, B.; Bondon, A.; Graton, J.; Jacquemin, D.; Rouaud, I.; Ferron, S.; Obermayer, W.; et al. Minor pyranonaphthoquinones from the apothecia of the lichen Ophioparma ventosa. J. Nat. Prod. 2016, 79, 1005–1011. [Google Scholar] [CrossRef]
- Goel, M.; Dureja, P.; Rani, A.; Uniyal, P.L.; Laatsch, H. Isolation, characterization and antifungal activity of major constituents of the himalayan Lichen Parmelia reticulata tayl. J. Agric. Food Chem. 2011, 59, 2299–2307. [Google Scholar] [CrossRef]
- Asplund, J.; Gauslaa, Y. Content of secondary compounds depends on thallus size in the foliose lichen Lobaria pulmonaria. Lichenologist 2007, 39, 273–278. [Google Scholar] [CrossRef]
- Elix, J.A.; Jariangprasert, S.; Archer, A.W. New Pertusaria (lichenized Ascomycota) from Australia and Thailand. Telopea 2008, 12, 263–272. [Google Scholar] [CrossRef]
- Brodo, I.M.; Tønsberg, T. Opegrapha halophila (Opegraphaceae), a new lichen species from coastal British Columbia, Canada, and Alaska, USA. Bryologist 2019, 122, 457–462. [Google Scholar] [CrossRef]
- Xiang, W.J.; Wang, Q.Q.; Ma, L.; Hu, L.H. β-Orcinol-type depsides from the lichen Thamnolia vermicularis. Nat. Prod. Res. 2013, 27, 804–808. [Google Scholar] [CrossRef] [PubMed]
- Nallasivam, J.L.; Fernandes, R.A. A protecting-group-free synthesis of (+)-nephrosteranic, (+)-protolichesterinic, (+)-nephrosterinic, (+)-phaseolinic, (+)-rocellaric acids and (+)-methylenolactocin. Org. Biomol. Chem. 2017, 15, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Bogo, D.; Honda, N.K.; Alcantara, G.B.; Brandão, L.F.G.; Aléssio, G.F.; Guimarães, R.D.C.A.; Matos, M.D.F.C. Cytotoxic activity of compounds from lichens of the cerrado biome in Brazil. Orbital 2020, 12, 7–16. [Google Scholar] [CrossRef]
- Dieu, A.; Mambu, L.; Champavier, Y.; Chaleix, V.; Sol, V.; Gloaguen, V.; Millot, M. Antibacterial activity of the lichens Usnea florida and Flavoparmelia caperata (Parmeliaceae). Nat. Prod. Res. 2020, 34, 3358–3362. [Google Scholar] [CrossRef] [PubMed]
- Simirgiotis, M.J.; Ramirez, J.E.; Schmeda, H.G.; Kennelly, E.J. Bioactive coumarins and HPLC-PDA-ESI-ToF-MS metabolic profiling of edible queule fruits (Gomortega keule), an endangered endemic Chilean species. Food Res. Int. 2013, 54, 532–543. [Google Scholar] [CrossRef]
- Brahmi-Chendouh, N.; Piccolella, S.; Nigro, E.; Hamri-Zeghichi, S.; Madani, K.; Daniele, A.; Pacifico, S. Urtica dioica L. leaf chemical composition: A never-ending disclosure by means of HR-MS/MS techniques. J. Pharm. Biomed. Anal. 2021, 195, 113892. [Google Scholar] [CrossRef]
- Gouveia-Figueira, S.; Danielsson, K.; Fowler, C.J. Changes in proportions of linoleic acid-derived oxylipins in oral lichen planus. Acta Derm. Venereol. 2019, 99, 1051–1052. [Google Scholar] [CrossRef]
- Moncada, B.; Lücking, R.; Suárez, A. Molecular phylogeny of the genus Sticta (lichenized Ascomycota: Lobariaceae) in Colombia. Fungal Divers. 2014, 64, 205–231. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods); Version 4; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Culberson, W.L. Chemosystematics and Ecology of Lichen-Forming Fungi. Annu. Rev. Ecol. Syst 1970, 1, 153–170. [Google Scholar] [CrossRef]
- Mittermeier, V.K.; Schmitt, N.; Volk, L.P.M.; Suárez, J.P.; Beck, A.; Eisenreich, W. Metabolic profiling of alpine and ecuadorian lichens. Molecules 2015, 20, 18047–18065. [Google Scholar] [CrossRef]
- Farkas, E.; Biró, B.; Varga, N.; Sinigla, M.; Lőkös, L. Analysis of lichen secondary chemistry doubled the number of Cetrelia W.L. Culb. & C.F. Culb. species (Parmeliaceae, lichenised Ascomycota) in Hungary. Cryptogamie. Mycologie 2021, 42, 1–16. [Google Scholar] [CrossRef]
- Quilhot, W.; Piovano, M.; Arancibia, H. Studies on Chilean Lichens, XII. Chemotaxonomy of the Genus Psoroma. J. Nat. Prod. 1989, 52, 191–192. [Google Scholar] [CrossRef]
- Konoreva, L.; Prokopiev, I.; Frolov, I.; Chesnokov, S.; Rozhina, S.; Poryadina, L.; Shavarda, A. Metabolite profiling of the Cladonia lichens using gas chromatography-mass spectrometry. Biochem. Syst. Ecol. 2019, 85, 3–12. [Google Scholar] [CrossRef]
- Vondrák, J.; Frolov, I.; Košnar, J.; Arup, U.; Veselská, T.; Halıcı, G.; Malíček, J.; Søchting, U. Substrate switches, phenotypic innovations and allopatric speciation formed taxonomic diversity within the lichen genus Blastenia. J. Syst. Evol. 2020, 58, 295–330. [Google Scholar] [CrossRef]
- Frolov, I.; Vondrák, J.; Košnar, J.; Arup, U. Phylogenetic relationships within Pyrenodesmia sensu lato and the role of pigments in its taxonomic interpretation. J. Syst. Evol. 2021, 59, 454–474. [Google Scholar] [CrossRef]
- Le Corvec, M.; Boussard-Plédel, C.; Charpentier, F.; Fatih, N.; Le Dare, B.; Massart, F.; Rojas, F.; Tariel, H.; Loréal, O.; Bureau, B.; et al. Chemotaxonomic discrimination of lichen species using an infrared chalcogenide fibre optic sensor: A useful tool for on-field biosourcing. RSC Adv. 2016, 6, 108187–108195. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Andersen, B.; Thrane, U. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol. Res. 2008, 112, 231–240. [Google Scholar] [CrossRef]
- Xu, M.; Heidmarsson, S.; Olafsdottir, E.S.; Buonfiglio, R.; Kogej, T.; Omarsdottir, S. Secondary metabolites from cetrarioid lichens: Chemotaxonomy, biological activities and pharmaceutical potential. Phytomedicine 2016, 23, 441–459. [Google Scholar] [CrossRef]
- Vu, T.H.; Catheline, D.; Delmail, D.; Boustie, J.; Legrand, P.; Lohezic-Le Devehat, F. Gas chromatographic analysis to compare the fatty acid composition of fifteen lichen species, with a focus on Stereocaulon. Lichenologist 2016, 48, 323–337. [Google Scholar] [CrossRef]


| Peak | Tentative Identification | [M-H]- | Retention Time (min) | Theoretical Mass (m/z) | Measured Mass (m/z) | Accuracy (ppm) | Metabolite Type ** | MS Ions (m/z) | Lichen Species * |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Gluconic Acid | C6H11O7 | 1.28 | 195.0509 | 195.0505 | 2.0 | Acid | 165.0401; | 2; 3; 7; 9; 10; 11 |
| 2 | Unknown | C11H5O5N | 1.28 | 231.0184 | 231.0195 | −4.8 | - | --- | 6 |
| 3 | Manitol | C6H13O6 | 1.31 | 181.0712 | 181.0714 | −1.1 | Carbohidrate | 181.0717; 163.0606 | 7; 9; 10; 11 |
| 4 | Arabic acid | C5H9O6 | 1.29 | 165.0399 | 165.0401 | −1.2 | Acid | 147.0293; 113.0237; 129.0196 | 6 |
| 5 | Citric Acid | C6H7O7 | 1.38 | 191.0196 | 191.0192 | 2.0 | Acid | 111.0080 | 1; 2; 3; 4; 5; 6; 7; 8; 9; 11 |
| 6 | Unknown | C15H5O3N2 | 1.38 | 261.0289 | 261.0300 | −4.2 | -- | --- | 1; 7; 8; 10 |
| 7 | 4-ethyl-2-Ethylisophthalic acid | C10H9O4 | 1.44 | 193.0504 | 193.0501 | 1.6 | A | 133.0288 | 1; 2; 3; 4; 6 |
| 8 | Unknown | C8H10O5N | 1.56 | 200.0563 | 200.0559 | 2.0 | - | -- | 1; 3; 4; 6 |
| 9 | Unknown | C15H5O3N2 | 1.64 | 261.0247 | 261.0278 | −11.8 | - | --- | 7; 8; 10 |
| 10 | Isocitric Acid | C6H7O7 | 1.77 | 191.0195 | 191.0192 | 1.6 | Acid | 111.0079; | 1; 2; 3; 4; 6; 7 |
| 11 | Unknown | C7H9O7 | 1.91 | 205.0352 | 205.0348 | 2.0 | - | 187.0245; 173.0087; 121.1131 | 2 |
| 12 | Unknown | C7H7O6 | 2.24 | 187.0246 | 187.0243 | 1.6 | - | 125.0237; 137.2503; | 2 |
| 13 | 2-Ethylisophthalic acid | C10H9O4 | 2.85 | 193.0505 | 193.0501 | 2.0 | A | 161.0240; 133.0290 | 1; 2; 3; 4; 6; 7; 8; 9; 10 |
| 14 | Trihydroxy benzaledehyde | C7H5O4 | 4.81 | 153.0188 | 153.0189 | −0.7 | A | 137.0238 | 1; 2; 3; 4 |
| 15 | 2-Hydroxyisophthalic acid | C8H5O5 | 7.63 | 181.0137 | 181.0141 | −2.2 | A | 137.0238 | 4 |
| 16 | 2,4-dihydroxy benzaldehyde | C7H5O3 | 8.02 | 137.0238 | 137.0239 | −0.7 | A | 121.0289 | 1; 2; 3; 4 |
| 17 | 4-O-Demethylglomellic acid | C24H25O9 | 8.65 | 457.1476 | 457.1499 | −5.0 | d | --- | 5; 8 |
| 18 | Unknow | C22H23O7 | 8.77 | 399.1444 | 399.1413 | 7.8 | - | --- | 8 |
| 19 | Unknow | C7H11O5 | 8.86 | 175.0606 | 175.0611 | −2.9 | - | --- | 5 |
| 20 | Grayanic acid | C23H25O7 | 8.91 | 413.1600 | 413.1569 | 7.5 | d | 181.06503 | 8 |
| 21 | Unknown | C13H16O7N | 8.98 | 298.0940 | 298.0927 | 4.4 | - | 175.0609; 283.0210; 202.0696; | 5 |
| 22 | Unknown | C11H9O7 | 9.33 | 253.0361 | 253.0348 | 5.1 | - | 137.0603; 181,0505; 249.3808; 173.4203; | 5 |
| 23 | Atranol | C8H7O3 | 9.44 | 151.0395 | 151.0396 | −0.7 | A | 123.0445; 135.0445 | 3 |
| 24 | Unknown | C24H23 O8 | 9.48 | 439.1371 | 439.1393 | −5.0 | - | 151.0397; 105.0948; 121.4871; | 5 |
| 25 | 5,7-Dihydroxy-6-methylphthalide | C9H7O4 | 9.64 | 179.0344 | 179.0347 | −1.6 | A | 135.0444; 107.0494 | 4; 5; 7; 8; 10 |
| 26 | Unknown | C16H15O10 | 9.73 | 367.0665 | 367.0639 | 7.1 | - | --- | 1; 4 |
| 27 | Unknown | C30H27O13N | 10.03 | 609.1475 | 609.1482 | −1.1 | - | --- | 1; 4; 6 |
| 28 | Unknow | C18H15O4 | 10.31 | 295.0970 | 295.0935 | 11.8 | - | --- | 9 |
| 29 | Unknow | C17H13O6 | 11.31 | 313.0712 | 313.0724 | −3.8 | -- | --- | 1; 2; 3; 4; 5 |
| 30 | 1,5-Pentanedicarboxylic acid | C7H11O4 | 10.56 | 159.0657 | 159.0660 | −1.9 | L | 115.0758 | 5 |
| 31 | Unknown | C29H25O13N | 10.54 | 595.1316 | 595.1326 | −1.7 | - | --- | 3 |
| 32 | Didechlorolecideoidin | C17 H13O7 | 10.64 | 329.0661 | 329.0676 | −4.6 | D | 209.0456; 285.0776; 151.0396; 179.0347; 123.0443 | 3 |
| 33 | Decahydroxyoxopentacosanoic acid | C25H47O13 | 10.71 | 555.3017 | 555.3047 | −5.4 | L | --- | 8 |
| 34 | Orsellinic acid | C8H7O4 | 11.04 | 167.0347 | 167.0344 | 1.8 | A | 123.0442 | 2; 3; 4; 5; 6 |
| 35 | Unknow | C21H19O12 | 11.04 | 463.0877 | 463.0893 | −3.4 | - | --- | 3 |
| 36 | Unknow | C10H9O5 | 11.07 | 209.0450 | 209.0458 | −3.8 | - | --- | 5; 6 |
| 37 | Nor 8′-methylconstictic acid | C21H19O11 | 11.11 | 447.0927 | 447.0942 | −3.4 | d | 209.0455 | 2 |
| 38 | Unknow | C17H13O6 | 11.20 | 313.0712 | 313.0720 | −2.5 | - | --- | 7 |
| 39 | Metil-2,6-dihidroxibenzoate | C8H7O4 | 11.21 | 167.0344 | 167.0346 | −1.1 | A | 109.0287; 137.0238 | 9 |
| 40 | Hypostictic acid isomer | C19H15O8 | 11.36 | 371.0778 | 371.0782 | −1.1 | D | 195.0665; 327.0885; 341.0679; 179.0347 | 1; 3; 4; 7 |
| 41 | Unknow | C19H16O9 N | 11.69 | 402.0825 | 402.0841 | −3.98 | - | --- | 11 |
| 42 | Fumarprotocetraric acid derivative | C17H11O6 | 11,70 | 311.0556 | 311.0564 | −2.5 | d | --- | 7; 10 |
| 43 | 4,5-Dihydroxy-2-nonenoic acid | C9H15O4 | 12.11 | 187.0974 | 187.0977 | −1.6 | L | 171.1025; 143.1072 | 1; 2; 3; 4; 9; 10 |
| 44 | 2,4-Dicarboxy-3-hydroxy-5-methoxytoluene | C10H9O6 | 12.14 | 225.0407 | 225.0399 | 3.5 | A | 181.0503; 167.0346; 149.0240 | 4 |
| 45 | Unknown | C17H13O6 | 12.16 | 313.0724 | 313.0727 | −1.0 | - | --- | 4 |
| 46 | Unknown | C18H15O7 | 12.37 | 343.0818 | 343.0826 | −2.3 | - | --- | 7; 9 |
| 47 | Unknown | C21H17O12 | 12.40 | 461.0737 | 461.0720 | 3.7 | - | --- | 6 |
| 48 | Unknown | C20H17O8 | 12.52 | 385.0939 | 385.0923 | 4.1 | - | --- | 1; 2; 3 |
| 49 | Unknow | C10H9O4 | 12.81 | 193.0501 | 193.0502 | −0.5 | - | --- | 1; 2; 3; 4; 5; 6; 7; 8; 9; 10 |
| 50 | Unknow | C27H45O6 | 13.04 | 465.3216 | 465.3231 | −3.2 | - | --- | 3 |
| 51 | 2,4-dihydroxy benzaldehyde | C7H5O3 | 13.07 | 137.0237 | 137.0239 | −1.5 | A | 121.0288 | 5 |
| 52 | Consalizinic acid derivative I | C19H13O11 | 13.36 | 417.0458 | 417.0474 | −3.8 | D | 373.0573; 387.0373; 225.0406, 177.0193 | 11 |
| 53 | 4-Ethoxy-3-formyl-2-hydroxy-6-methylbenzoic acid | C11H11O5 | 13.41 | 223.0614 | 223.0606 | 3.6 | A | 177.0190; 133.0296; | 1; 2; 3; 4; 7; 9; 10 |
| 54 | Unknow | C20H15O8 | 13.47 | 383.0767 | 383.0781 | −3.6 | - | --- | 1 |
| 55 | Consalizinic acid derivative II | C20H17O11 | 13,47 | 433.0771 | 433.0787 | −3.7 | D | 401.0524; 417.0474; 373.0574 | 11 |
| 56 | Cynodontin or Citreorosein isomer, | C15H9O6 | 13.71 | 285.0399 | 285.0410 | −3.9 | Anthraquinone | 151.0396; 137.0237 | 2 |
| 57 | Consalizinic acid derivative I isomer | C19H13O11 | 13.78 | 417.0458 | 417.0474 | −3.84 | D | 373.0573; 343.0467, 77.0190; 401.0523 | 10 |
| 58 | Unknow | C30H47O7 | 13.79 | 519.3322 | 519.3337 | −2.9 | - | --- | 3 |
| 59 | 1,4,5,6,8-Pentahydroxy-3-ethylanthraquinone | C15H9O7 | 13.87 | 301.0348 | 301.0361 | −4.3 | Anthraquinone | --- | 4 |
| 60 | Unknow | C19H15O4 | 13.87 | 307.0970 | 307.0939 | 10.0 | - | --- | 8 |
| 61 | Unknow | C14H13O7 | 13.99 | 293.0661 | 293.0674 | −4.4 | - | --- | 3 |
| 62 | Haemathamnolic acid isomer | C19H15O10 | 14.18 | 403.0665 | 403.0681 | −3.97 | D | 359.0781; 371.0414; 209.0455 | 11 |
| 63 | Fumarprotocetraric acid derivative | C17H11O6 | 14.78 | 311.0567 | 311.0569 | −0.6 | d | --- | 1; 2; 3; 7; 10 |
| 64 | Constictic acid | C19H13O10 | 14.61 | 401.0509 | 401.0528 | −4.74 | D | 357.0625; 313.0726; 283.0619; 255.0670; 121.0289 | 11 |
| 65 | Hypostictic acid isomer | C19H15O8 | 14.98 | 371.0781 | 371.0767 | 3.8 | D | 327.0883; 195.0664; 179.0347 | 5 |
| 66 | Thelephoric acid | C18H7O8 | 14.81 | 351.0141 | 351.0154 | −3.7 | Terphenylquinones | --- | 3 |
| 67 | Methylstictic acid | C20H15O9 | 15.26 | 399.0716 | 399.0728 | −3.0 | D | 371.0779; 193.0504 | 2; 7 |
| 68 | Nor 8′-metilconstictic acid | C21H19O11 | 15.28 | 447.0927 | 447.0944 | −3.80 | D | 401.0524; 209.0455 | 11 |
| 69 | Protocetraric acid | C18H13O9 | 17.17 | 373.0560 | 373.0574 | −3.75 | D | 355.0468; 329.0674; 311.0568; 227.0352; 267.0669; 285.0777 | 11 |
| 70 | Hypoconstictic acid | C19H15O9 | 17.27 | 387.0716 | 387.0729 | −3.4 | D | 267.0673; 311.0552; 149.0238; 343.0827;167.0345 | 3 |
| 71 | Unknow | C14H13O6 | 17.84 | 277.0712 | 277.0724 | −4.3 | - | -- | 3; 6 |
| 72 | 12,13,15-Trihydroxy-9-octadecenoic acid | C18H15O5 | 18.11 | 329.2328 | 329.2340 | −3.6 | L | 285.1716; | 2; 6 |
| 73 | Unknow | C20H13O8 | 18.19 | 381.0610 | 381.0626 | −4.2 | - | --- | 3; 4 |
| 74 | Salazinic acid | C18H11O10 | 18.21 | 387.0352 | 387.0368 | −4.13 | D | 343.0468; 269.0458; 241.0507; 325.0365; 299.0569 | 11 |
| 75 | Unknown | C10H9O4 | 18.45 | 193.0505 | 193.0501 | 2.1 | - | --- | 3; 4 |
| 76 | Unknown | C30H47O7 | 18.61 | 519.3322 | 519.3319 | 0.6 | - | -- | 3 |
| 77 | Menegazziaic acid | C18H13O9 | 18.79 | 373.0560 | 373.0575 | −4.0 | D | 311,0570; 255.0666; 329.0679 | 3 |
| 78 | Norstictic acid | C18H11O9 | 18.86 | 371.0403 | 371.0417 | −3.8 | D | 327.0526; 151.0396; 123.0444; | 3 |
| 79 | Unknow | C22H19O10 | 18.88 | 443.0978 | 443.0996 | −4.1 | - | --- | 3 |
| 80 | Physodalic acid | C20H15O10 | 18.99 | 415.0665 | 415.0681 | −3.85 | D | 359.0417; 315.0520; 343.0832; 387,0367; 373.0573; 401.0525 | 11 |
| 81 | Unknow | C26H19O10 | 19.00 | 491.0978 | 491.0997 | −3.9 | - | --- | 3 |
| 82 | Derivative methyl 8-hydroxy-4-0-demethylbarbatate | C19H19O9 | 19.05 | 391.1045 | 391.1029 | 4.1 | d | 359.0788 | 3 |
| 83 | 12,13,15-Trihydroxy-9-octadecenoic acid | C18H33O5 | 19.05 | 329.2328 | 329.2336 | −2.4299 | L | --- | 7 |
| 84 | Haemoventosin | C15H11O7 | 19.16 | 303,0519 | 303.0505 | 4.7 | Naphthaquinone | 259.0619; 231.0667; 189.0560; | 3 |
| 85 | α-acetilconstictic acid derivative I | C21H17O11 | 19.22 | 445.0771 | 445.0786 | −3.3 | D | 415.0680; 371.0780; 427.0676; 343.0830;193.0504, 401.0522 | 11 |
| 86 | Conhypoprotocetraric acid or Convirensic acid | C18H15O8 | 19.25 | 359.0781 | 359.0767 | 3.9 | D | 344.0545; 302.0442 | 3 |
| 87 | 4-O-dimethylbaemycesic acid | C18H15O8 | 19.27 | 359.0781 | 359.0767 | 3.9 | d | 181.0714; 163.0397; 137.0236 | 1; 2; 3;5; 6 |
| 88 | Orsellinic acid Isomer | C8H7O4 | 19.45 | 167.0344 | 167.0346 | −1.1974 | A | 123.0440; 149.0235 | 9; 10 |
| 89 | Lecanoric acid | C16H13O7 | 19.51 | 317.0661 | 317.0671 | 0.6 | d | 167.0345; 123.0443; 149.0238; | 1; 2; 3; 4; 5; 6 |
| 90 | Constictic acid isomer | C19H13O10 | 19.56 | 401.0509 | 401.0524 | −3.74 | D | 357,0626; 313.0726; 343.0831; 255.0622 | 11 |
| 91 | Pentahydroxytetracosanoic acid | C24H47O7 | 19.67 | 447.3322 | 447.3336 | −3.1 | L | --- | 1; 3; 7; 9; 10 |
| 92 | 2-Methyl-5-hydroxy-6-hydroxymethyl-7-Methoxychromone | C12H11O5 | 19.73 | 235.0606 | 235.0615 | −3.8 | C | 181.0504 | 3 |
| 93 | Unknown | C20H17O8 | 19.79 | 385.0939 | 385.0923 | 4.1 | - | --- | 1; 4; 5; 10 |
| 94 | Heptahydroxytrioxooctadecanoic acid | C18H29O12 | 19.86 | 437.1664 | 437.1645 | 4.3 | L | --- | 1; 4; 5; 6; 7; 9; 10 |
| 95 | 5,7-Dihydroxy-6-methylphthalide derivative | C9H7O3 | 19.88 | 163.0395 | 163.0392 | 1.8 | A | 119.0492 | 10 |
| 96 | Criptostictic acid derivative | C18H11O8 | 20.04 | 355.0454 | 355.0462 | −2.2 | D | 133.0288; 239.0715; 311.0572; 179.0345; | 7 |
| 97 | Unknow | C18H17O6 | 20.08 | 329.1025 | 329.1032 | −2.1 | - | --- | 9 |
| 98 | Unknow | C20H15O8 | 20.16 | 383.0767 | 383.0775 | −2.0 | - | --- | 9; 10 |
| 99 | Unknow | C21H19O9 | 20.18 | 415.1045 | 415.1029 | 3.9 | - | --- | 3; 5; 9; 10 |
| 100 | Unknow | C28H23O11 | 20.12 | 535.1240 | 535.1257 | −3.1 | - | --- | 3 |
| 101 | Unknow | C15H13O3 | 20.13 | 241.0872 | 241.0874 | −0.8 | - | ---- | 1; 2 |
| 102 | Heptahydroxytetraoxoicosanoic acid | C20H31O13 | 21.19 | 479.1765 | 479.1746 | 3.9 | L | --- | 7 |
| 103 | Tetrahydroxytricosanoic acid | C23H45O6 | 20.26 | 417.3232 | 417.3216 | 3.9 | L | 403.3073 | 1; 3; 4; 7 |
| 104 | Tetrahydroxytrioxoundecanoic acid | C11H15O9 | 20.30 | 291.0716 | 291.0699 | 5.8 | L | --- | 8 |
| 105 | Stictic acid | C19H13O9 | 20.34 | 385.0560 | 385.0576 | −4.1 | D | 341.0674; 357.0622; 297.0774; 313.0721; 193.0504; 269.0826 | 11 |
| 106 | Parietin | C16H11O5 | 20.39 | 283.0606 | 283.0617 | −3.9 | Antraquinone | 179.0345 | 1; 2; 6 |
| 107 | Unknow | C24H47O11N2 | 20.39 | 539.3157 | 539.3180 | −4.3 | - | --- | 3 |
| 108 | Evernic acid isomer | C17H15O7 | 20.46 | 331.0818 | 331.0830 | −3.6 | d | 167.0347; 123.0447; 149.0240 | 1 |
| 109 | Hypoconstictic acid | C19H15O9 | 20.50 | 387.0716 | 387.0732 | −4.1 | D | 149.0238; 343.0836; 167.0345 | 4 |
| 110 | Cryptostictic acid | C19H15O9 | 20.50 | 387.0716 | 387.0725 | −2.3 | D | 267,0661; 343,0825; 311.05067; 239.0710 | 7; 8 |
| 111 | Retigeric acid derivative | C30H43O7 | 20.51 | 515.3009 | 515.3025 | −3.1 | Triterpene | --- | 3 |
| 112 | Retigeric acid B | C30H45O6 | 20.56 | 501.3216 | 501.3236 | −4.0 | Triterpene | --- | 3 |
| 113 | Salazinic acid isomer | C18H11O10 | 20.58 | 387.0352 | 387.0368 | −4.13 | D | 343.0468; 299.0565 | 11 |
| 114 | Unknown | C23H22O10N | 20.68 | 472.1244 | 472.1259 | −3.2 | - | ----- | 6 |
| 115 | 9,10-dihydroxyoctadecatrienoic acid | C18H29O4 | 20.69 | 309.2081 | 309.2066 | 4.9 | L | 291.1975 | 1; 2; 4 |
| 116 | Unknow | C17H13O6 | 20.84 | 313.0712 | 313.0727 | −4.8 | - | ---- | 1; 2; 6 |
| 117 | Pulvinic acid derivative I | C18H11O6 | 20.97 | 323.0556 | 323.0556 | 0.0 | Pulvinic acid y derivates | 133.0286; 117.0335 | 10 |
| 118 | 9,10,12 Trihydroxytriacontaheptaenoic acid | C30H45O5 | 20.99 | 485.3284 | 485.3267 | 3.5 | L | --- | 3 |
| 119 | 4-0-Demethylbarbatic acid | C18H17O7 | 20.99 | 345.0974 | 345.0989 | −4.3 | d | 181.0505; 163.0396; 137.0603 | 4 |
| 120 | Unknow | C24H23O10N | 21.02 | 485.1322 | 485.1319 | 0.6 | - | --- | 1; 5 |
| 121 | Methyl orsellinate | C9H9O4 | 21.05 | 181.0502 | 181.0501 | 0.5 | A | 163.0389 | 1 |
| 122 | Heptahydroxyetraoxoicosanoic acid | C20H31O13 | 21.17 | 479.1752 | 479.1765 | −2.7 | L | --- | 1; 2; 3; 4; 5; 6; 9 |
| 123 | Gyrophoric Acid | C24H19O10 | 21.25 | 467.0991 | 467.0978 | 2.8 | d | 167.0346; 317.0673; 123.0445; 149.0238; | 1; 3; 4; 6; 11 |
| 124 | Galbinic acid | C20H13O11 | 21.27 | 429.0458 | 429.0474 | −3.73 | D | 403.0681; 371.0417; 401.0524; 327.0518; 149.0239 | 11 |
| 125 | Hyposalazinic acid | C18H13O8 | 21.28 | 357.0610 | 357.0623 | −3.6 | D | 313.0723; 135.0444; 179.0348 | 1 |
| 126 | Hydroxytetracosapentaenoic acid | C24H37O3 | 21.42 | 373.2743 | 373.2743 | 0.0 | L | --- | 10 |
| 127 | Orsellinic acid isomer | C8H7O4 | 21.47 | 167.0347 | 167.0344 | 1.8 | A | 149.0239; 123.0443 | 1; 4; 6 |
| 128 | Dihydroxyoctadecenoic acid | C18H33O4 | 21.48 | 313.2390 | 313.2395 | −1.6 | L | ---- | 2; 5; 6 |
| 129 | Norstictic acid | C18H11O9 | 21.64 | 371.0403 | 371.0417 | −3.77 | D | 27.0517; 227.0716; 151.0390; 243.0297 | 11 |
| 130 | Dihydroxyoctadec-6-enoic acid | C18H33O4 | 21.58 | 313.2379 | 313.2379 | 0.0 | L | --- | 10 |
| 131 | Loxodinol isomer | C25H29O9 | 21.64 | 473.1812 | 473.1818 | −1.2 | DE | 429.1919 | 9 |
| 132 | EthyI 2,4-dihydroxy-6-n-nonylbenzoate | C18H27O4 | 21.65 | 307.1909 | 307.1922 | −4.2 | A | 263.1659 | 1; 2; 3; 4 |
| 133 | Evernic Acid | C17H15O7 | 21.81 | 331.0828 | 331.0818 | 3.0 | d | 167.0345; 123.0444; 149,0238; | 1; 2; 3; 4; 5 |
| 134 | Protocetraric acid Isomer | C18H13O9 | 21.85 | 373.0560 | 373.0573 | −3.48 | D | 355.0460; 329.0674; 285.0780, 311.0567; 255.,0672 | 11 |
| 135 | Unknown | C22H22O8N | 21.83 | 428.1360 | 428.1345 | 3.5 | - | ---- | 6 |
| 136 | Strepsilin | C15H9O5 | 21.89 | 269.0450 | 269.0462 | −4.5 | DBF | 225.0554 | 2; 5 |
| 137 | Unknown | C30H29O4 | 21.97 | 453.2066 | 453.2061 | 1.1 | - | --- | 3 |
| 138 | Unknown | C18H11O6 | 22.01 | 323.0556 | 323.0570 | −4.33 | - | --- | 11 |
| 139 | Hexahydroxytrioxooctacosatrienoic acid | C28H43O11 | 22.12 | 555.2805 | 555.2841 | −6.4832 | L | --- | 9 |
| 140 | Nonahydroxyoctacosatetraenoic acid | C28 H47O11 | 22.26 | 559.3124 | 559.3132 | −1.4 | L | --- | 2 |
| 141 | Unknow | C28H41O9N2 | 22.26 | 549.2849 | 549.2812 | 6.8 | - | --- | 3 |
| 142 | Norsolorinic acid | C20H17O7 | 22.44 | 369.0974 | 369.0989 | −4.06 | - | --- | 11 |
| 143 | Unknow | C25H33O13 | 22.46 | 541.1921 | 541.1909 | 2.2 | - | --- | 1; 2; 3; 6; 10 |
| 144 | Hydroxytetracosapentaenoic acid derivative | C24H37O3 | 22.61 | 373.2743 | 373.2741 | 0.5 | - | --- | 10 |
| 145 | Hydroxytrioxotricosanoic acid | C23H39O6 | 22.53 | 411.2747 | 411.2757 | −2.4 | L | --- | 8 |
| 146 | Squamatic acid | C19H17O9 | 22.89 | 389.0873 | 389.0886 | −3.3 | d | 343.0836; 163.0396; 193.0139; 149.0238; 121.0286 | 1; 3; 4 |
| 147 | Picrolichenic acid | C25H29O7 | 22.72 | 441.1913 | 441.1926 | −3.0 | Depsones | --- | 1 |
| 148 | Heptahydroxydioxohexacosanoic acid | C26H47O11 | 22.76 | 535.3118 | 535.3134 | −3.0 | L | --- | 6 |
| 149 | Unknow | C30H45O4 | 22.83 | 469.3319 | 469.3335 | −3.4 | - | --- | 3 |
| 150 | 2,2′-Di-O-methylanziaic acid | C26H33O7 | 22.85 | 457.2226 | 457.2244 | −3.9 | d | 413.2345; | 4 |
| 151 | Dihydroxytetracosahexaenoic acid | C24H35O4 | 22.85 | 387.2535 | 387.2552 | −4.4 | L | --- | 5 |
| 152 | Hydroxyoctadecadienoic acid | C18H31O3 | 22.90 | 295.2273 | 295.2273 | 0.0 | L | --- | 10 |
| 153 | Orsellinic acid Isomer | C8H7O4 | 22.92 | 167.0344 | 167.0348 | −2.3 | A | 149.0240; 123.0445 | 11 |
| 154 | Pulvinic acid | C18H11O5 | 22.98 | 307.0606 | 307.0613 | −2.2 | Pulvinic acid y derivates | 117.0338; 263.0713 | 9 |
| 155 | 4-0-Demethylbarbatic acid | C18H17O7 | 23.02 | 345.0974 | 345.0986 | −3.5 | d | 123.0443; 137.0237; 181.0502 | 1 |
| 156 | Psoromic acid | C18H13O8 | 23.06 | 357.0610 | 357.0626 | −4.4 | D | 313.0726; 181.0502; 179.0347; 327.0520; 269.0826; 285.0776 | 11 |
| 157 | Methylgyrophoric acid | C25H21O10 | 23.15 | 481.1135 | 481.1147 | −2.5 | d | 149.0238; 123.0442; 167.0346; 317.0671 | 1; 4 |
| 158 | Evernic acid isomer | C17H15O7 | 23.22 | 331.0818 | 331.0832 | −4.2 | d | 149.0239; 123.0443; 167.0346; 105.0337 | 11 |
| 159 | Skyrin | C30H17O10 | 23.28 | 537.0822 | 537.0840 | −3.4 | Anthraquinones | ---- | 3 |
| 160 | Angardianic acid | C19H35O4 | 23.36 | 327.2543 | 327.2547 | −1.2 | Acids | 283.2649; 309.2081 | 2; 4 |
| 161 | Pentadecatetraenoic acid | C15H21O2 | 23.38 | 233.1542 | 233.1545 | −1.2 | L | --- | 9; 10 |
| 162 | 9-hydroxyoctadecatrienoic acid | C18 H29O3 | 23.45 | 293.2117 | 293.2130 | −4.4 | L | 277.2180 | 6 |
| 163 | Unknow | C18H15O7 | 23.53 | 343.0818 | 343.0824 | −1.7 | - | --- | 9 |
| 164 | Pulvinic acid derivative II | C19H13O5 | 23.68 | 321.0763 | 321.0770 | −2.1 | Pulvinic acid y derivates | 117.0337 | 9; 10; 11 |
| 165 | Pulvinic acid | C18H11O5 | 23.77 | 307.0606 | 307.0620 | −4.5 | Pulvinic acid y derivates | 263.0720; 117.0339 | 11 |
| 166 | Furfuric acid isomer | C28H23O12 | 23.82 | 551.1190 | 551.1197 | −1.2 | D | 371.0784; 193.0504; 179.0347; 207.0297; 193.0504 | 8 |
| 167 | Unknow | C26H47O5N2 | 23.82 | 467.3485 | 467.3492 | −3,9 | - | --- | 3 |
| 168 | Unknow | C30H27O6 | 23.89 | 483.1808 | 483.1820 | −2,5 | - | --- | 1 |
| 169 | Unknow | C30H25O6 | 24.01 | 481.1651 | 481.1663 | −2,5 | - | --- | 1 |
| 170 | Unknow | C15H13O3 | 24.02 | 241.0872 | 241.0872 | 0,0 | - | --- | 1 |
| 171 | Trihydroxyheptacosa pentaenoic acid | C27H43O5 | 24.05 | 447.3110 | 447.3127 | −3.8 | L | --- | 8 |
| 172 | Barbatic Acid | C19H19O7 | 24.26 | 359.1141 | 359.1131 | 2.8 | d | 137.0603; 163.0396; 181.0509 | 1; 4 |
| 173 | Hydroxytrioxodocosanoic acid | C22H37O6 | 24.29 | 397.2590 | 397.2601 | −2.8 | L | --- | 8 |
| 174 | Thamnolic acid isomer | C19H15O11 | 24.41 | 419.0614 | 419.0630 | −3.8 | d | 375.0730; 167.0344; 209.0455; 181.0503 | 11 |
| 175 | Orsenillic acid derivated II | C8H7O4 | 24.73 | 167.0344 | 167.0347 | −1.80 | - | 149.0239; 1230444; | 11 |
| 176 | Unknow | C26H33O8 | 24.73 | 473.2190 | 473.2175 | 3.2 | - | --- | 2 |
| 177 | Lobaric acid | C25H27O8 | 24.81 | 455.1706 | 455.1718 | −2.6 | D | 411.1824; 367.1811 | 1 |
| 178 | Unknow | C22H27O7 | 24.97 | 403.1770 | 403.1757 | 3.2 | - | 2 | |
| 179 | Unknow | C30H41O8 | 25.27 | 529.2819 | 529.2801 | 3.4 | - | --- | 4 |
| 180 | Hypothamnolic acid | C19H17O10 | 25.49 | 405.0822 | 405.0832 | −2.5 | d | 209.0456; 181.0499 | 1 |
| 181 | Unknow | C25H11O7 | 25.53 | 423.0505 | 423.0497 | 1.9 | - | --- | 4 |
| 182 | Pulvinic acid derivative III | C19H13O5 | 25.67 | 321.0763 | 321.0777 | −4.3 | Pulvinic acid y derivates | 117.0338 | 11 |
| 183 | Dihydroxyicosahexaenoic acid | C20H27O4 | 26.02 | 331.1909 | 331.1925 | −4.8 | L | ----- | 11 |
| 184 | Usnic acid | C18H15O7 | 26.05 | 343.0818 | 343.0831 | −3.8 | DBF | 231.0658; 328.0585; 259.0604 | 3; 4; 6; 7; 8 |
| 185 | Nephromopsic acid orRoccellaric acid | C19H33O4 | 26.32 | 325.2392 | 325.2379 | 4.0 | Acids | 281.2494 | 3; 4 |
| 186 | Unknow | C28H25O5N | 26.87 | 455.1733 | 455.1723 | 2.20 | - | --- | 11 |
| 187 | Perlatolic acid | C25H31O7 | 26.98 | 443.2070 | 443.2078 | −1.8 | d | 205.0867; 179.1073; 223.0973 | 7; 8 |
| 188 | Caperatic acid | C21H37O7 | 28.14 | 401.2539 | 401.2549 | −2.4 | Acids | 255.2327 | 8 |
| 189 | Atranorin | C19H17O8 | 29.64 | 373.0923 | 373.0937 | −3.75 | d | 177.0192; 163.0397 | 9; 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albornoz, L.; Torres-Benítez, A.; Moreno-Palacios, M.; Simirgiotis, M.J.; Montoya-Serrano, S.A.; Sepulveda, B.; Stashenko, E.; García-Beltrán, O.; Areche, C. Phylogenetic Studies and Metabolite Analysis of Sticta Species from Colombia and Chile by Ultra-High Performance Liquid Chromatography-High Resolution-Q-Orbitrap-Mass Spectrometry. Metabolites 2022, 12, 156. https://doi.org/10.3390/metabo12020156
Albornoz L, Torres-Benítez A, Moreno-Palacios M, Simirgiotis MJ, Montoya-Serrano SA, Sepulveda B, Stashenko E, García-Beltrán O, Areche C. Phylogenetic Studies and Metabolite Analysis of Sticta Species from Colombia and Chile by Ultra-High Performance Liquid Chromatography-High Resolution-Q-Orbitrap-Mass Spectrometry. Metabolites. 2022; 12(2):156. https://doi.org/10.3390/metabo12020156
Chicago/Turabian StyleAlbornoz, Laura, Alfredo Torres-Benítez, Miguel Moreno-Palacios, Mario J. Simirgiotis, Saúl A. Montoya-Serrano, Beatriz Sepulveda, Elena Stashenko, Olimpo García-Beltrán, and Carlos Areche. 2022. "Phylogenetic Studies and Metabolite Analysis of Sticta Species from Colombia and Chile by Ultra-High Performance Liquid Chromatography-High Resolution-Q-Orbitrap-Mass Spectrometry" Metabolites 12, no. 2: 156. https://doi.org/10.3390/metabo12020156
APA StyleAlbornoz, L., Torres-Benítez, A., Moreno-Palacios, M., Simirgiotis, M. J., Montoya-Serrano, S. A., Sepulveda, B., Stashenko, E., García-Beltrán, O., & Areche, C. (2022). Phylogenetic Studies and Metabolite Analysis of Sticta Species from Colombia and Chile by Ultra-High Performance Liquid Chromatography-High Resolution-Q-Orbitrap-Mass Spectrometry. Metabolites, 12(2), 156. https://doi.org/10.3390/metabo12020156