Dynamic Changes in the Bacterial Community and Metabolic Profile during Fermentation of Low-Salt Shrimp Paste (Terasi)
Abstract
1. Introduction
2. Results
2.1. Protein Content of Shrimp Paste with Various Salt Concentrations
2.2. Changes in Physicochemical Properties and Culturable Bacterial Population
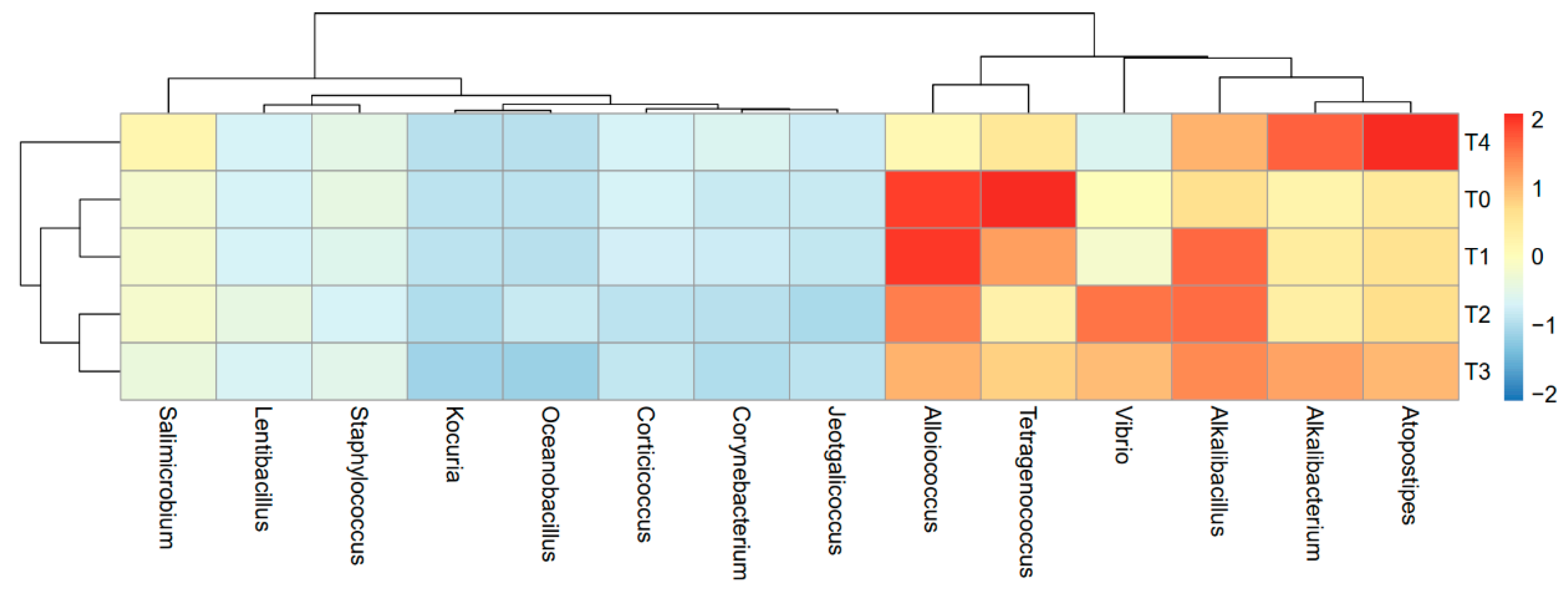
2.3. Bacterial Communities in Shrimp and Low-Salt Shrimp Paste
2.4. Detection of Pathogenic Bacteria (Escherichia coli and Salmonella) Culture-Dependent Method
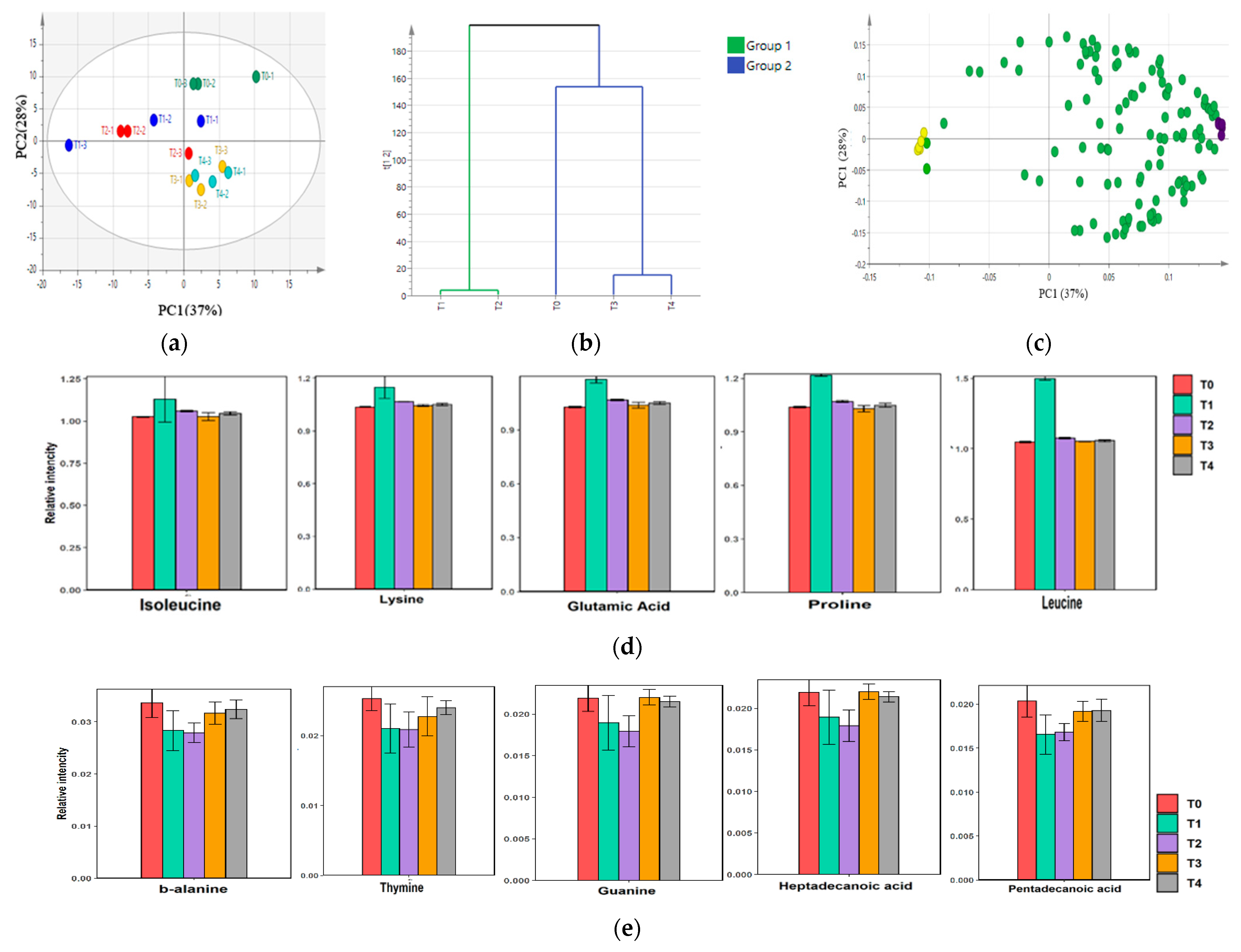
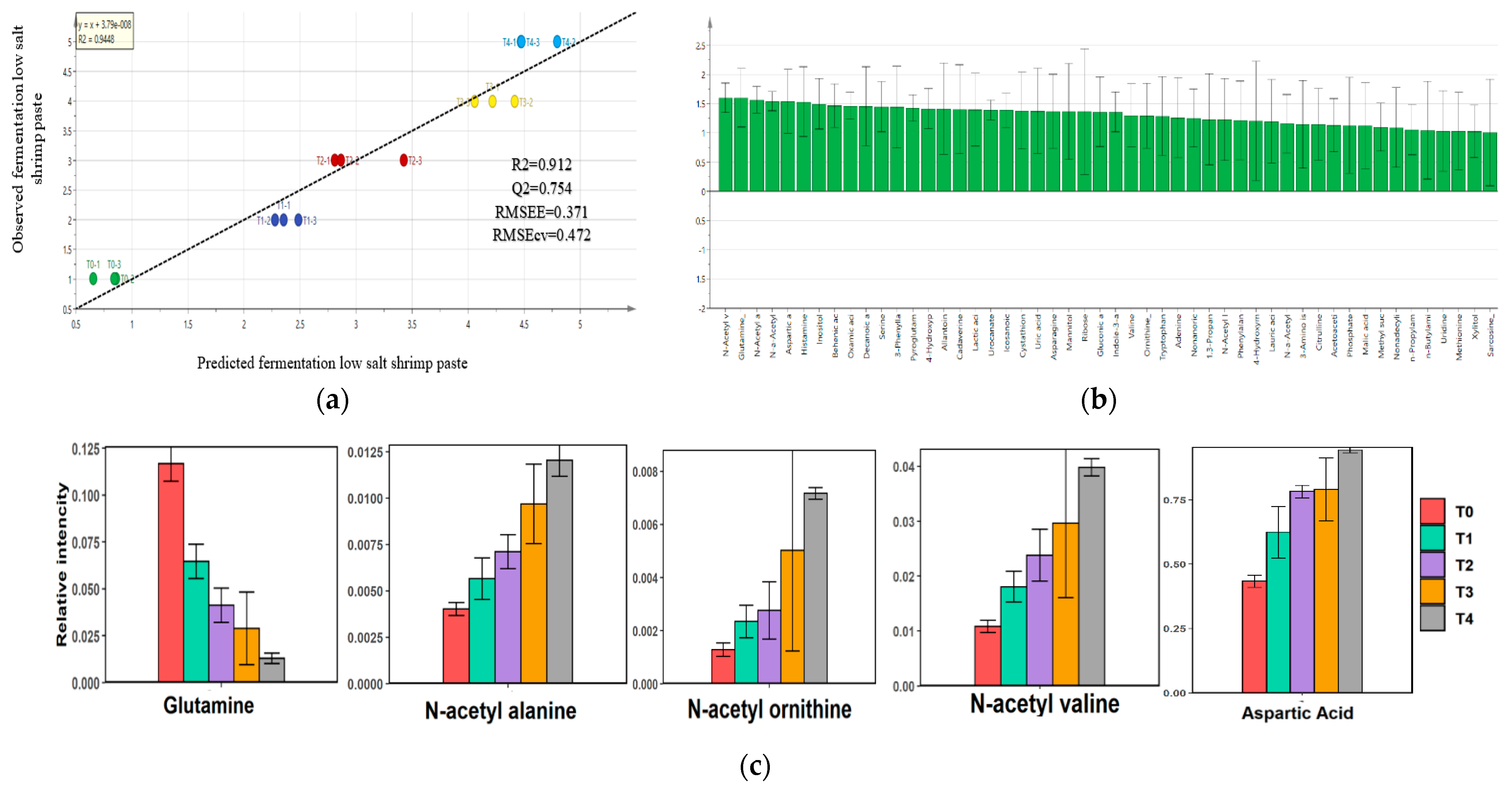
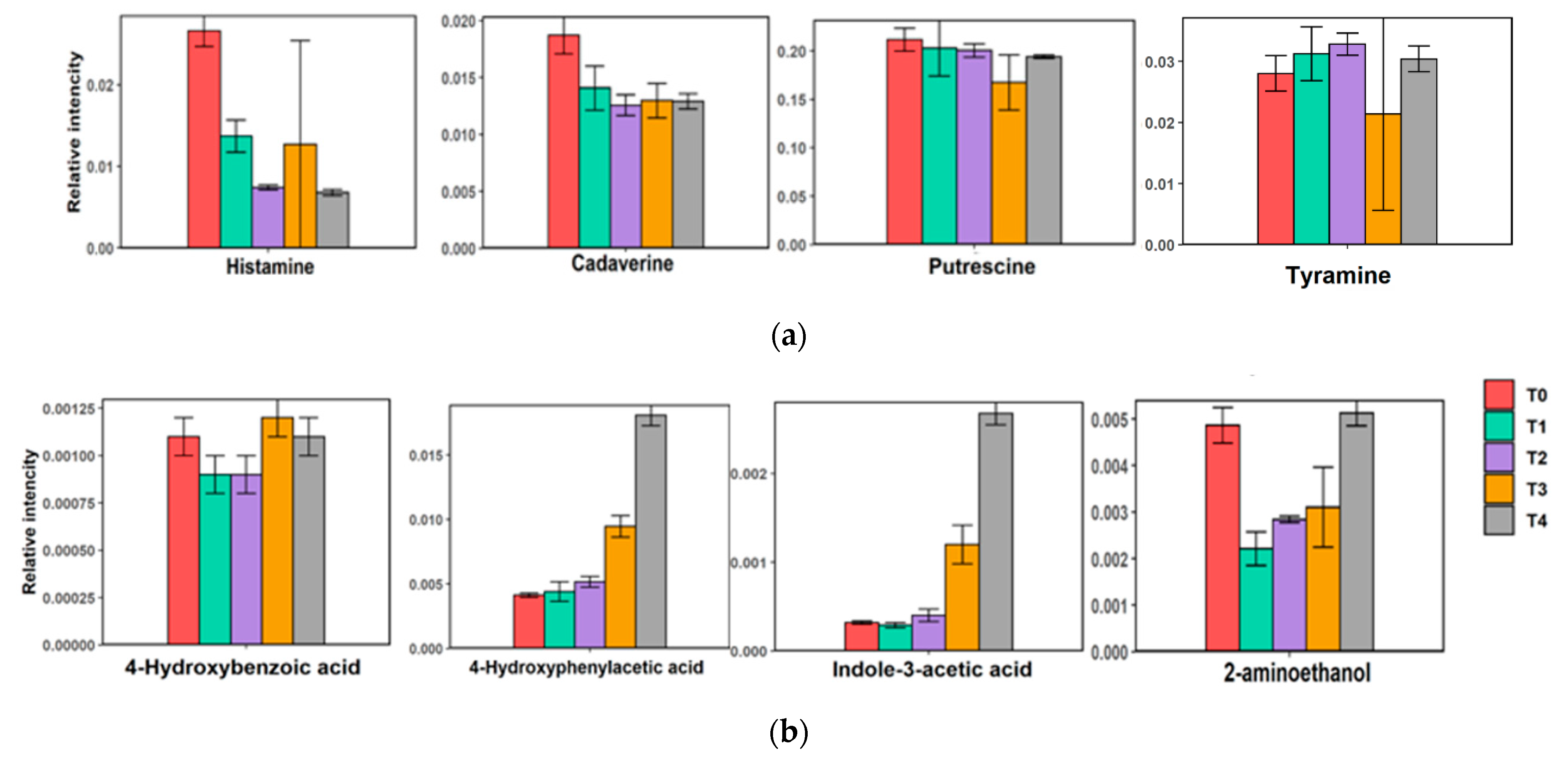
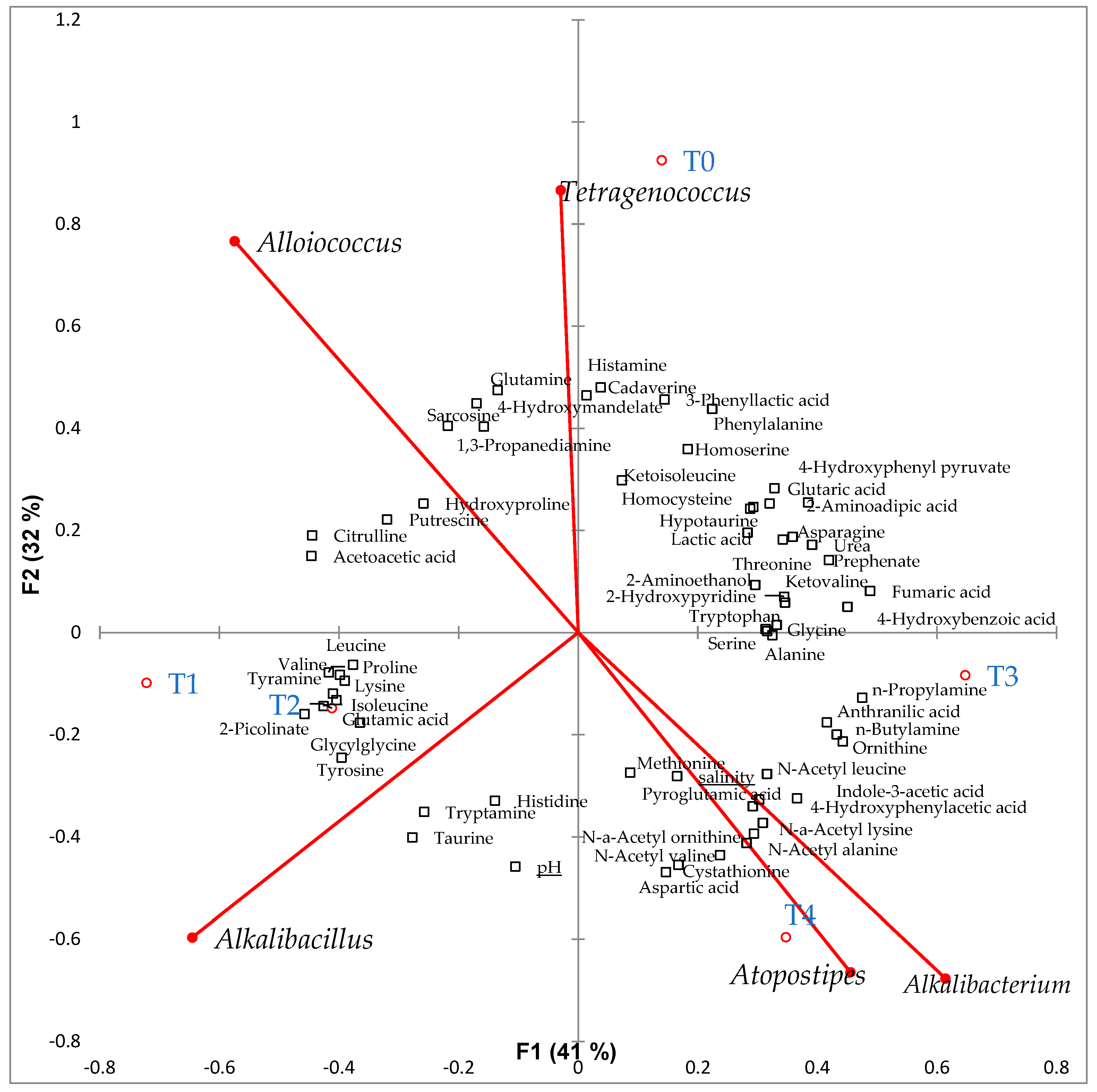
2.5. Metabolite Composition of Low-Salt Terasi
3. Discussion
4. Materials and Methods
4.1. Preparation of Shrimp Paste (Terasi)
4.2. Protein Content of Shrimp Paste
4.3. Physicochemical Analysis and Enumeration of Bacteria
4.4. Detection of Pathogenic Bacteria (E. coli and Salmonella) in Low-Salt Shrimp Paste Qualitatively Used Culture-Dependent Method
4.5. Bacterial Communities Analysis in Shrimp and Shrimp Paste
4.5.1. Extraction of DNA
4.5.2. PCR Amplification and Sequencing
4.5.3. Data Analysis
4.6. Metabolites Detection in Shrimp Paste
4.6.1. Preparation of Samples
4.6.2. GC/MS Analysis
4.6.3. Data Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hajeb, P.; Jinap, S. Fermented Shrimp Products as Source of Umami in Southeast Asia. J. Nutr. Food Sci. 2012, 10, 6. [Google Scholar] [CrossRef]
- Li, W.; Lu, H.; He, Z.; Sang, Y.; Sun, J. Quality characteristics and bacterial community of a Chinese salt-fermented shrimp paste. LWT 2021, 136, 110358. [Google Scholar] [CrossRef]
- Pongsetkul, J.; Benjakul, S.; Sampavapol, P.; Osako, K.; Faithong, N. Chemical composition and physical properties of salted shrimp paste (Kapi) produced in Thailand. Int. Aquat. Res. 2014, 6, 155–166. [Google Scholar] [CrossRef]
- Faithong, N.; Benjakul, S.; Phatcharat, S.; Binsan, W. Chemical composition and antioxidative activity of Thai traditional fermented shrimp and krill products. Food Chem. 2010, 119, 133–140. [Google Scholar] [CrossRef]
- Kim, Y.-B.; Choi, Y.-S.; Ku, S.-K.; Jang, D.-J.; Ibrahim, H.H.; Moon, K.B. Comparison of quality characteristics between belacan from Brunei Darussalam and Korean shrimp paste. J. Ethn. Foods 2014, 1, 19–23. [Google Scholar] [CrossRef]
- Pongsetkul, J.; Benjakul, S.; Sampavapol, P.; Osako, K.; Faithong, N. Chemical compositions, sensory and antioxidative properties of salted shrimp paste (Ka-pi) in Thailand. Int. Food Res. J. 2015, 22, 1454–1465. [Google Scholar]
- Prapasuwannakul, N.; Suwannahong, K. Chemical Composition and Antioxidant Activity of Klongkone Shrimp Paste. Procedia-Soc. Behav. Sci. 2015, 197, 1095–1100. [Google Scholar] [CrossRef]
- Peralta, E.M.; Hatate, H.; Kawabe, D.; Kuwahara, R.; Wakamatsu, S.; Yuki, T.; Murata, H. Improving antioxidant activity and nutritional components of Philippine salt-fermented shrimp paste through prolonged fermentation. Food Chem. 2008, 111, 72–77. [Google Scholar] [CrossRef]
- Phewpan, A.; Phuwaprisirisan, P.; Takahashi, H.; Ohshima, C.; Lopetcharat, K.; Techaruvichit, P.; Keeratipibul, S. Microbial diversity during processing of Thai traditional fermented shrimp paste, determined by next generation sequencing. LWT 2020, 122, 108989. [Google Scholar] [CrossRef]
- Sang, X.; Li, K.; Zhu, Y.; Ma, X.; Hao, H.; Bi, J.; Zhang, G.; Hou, H. The Impact of Microbial Diversity on Biogenic Amines Formation in Grasshopper Sub Shrimp Paste During the Fermentation. Front. Microbiol. 2020, 11, 782. [Google Scholar] [CrossRef]
- Cai, L.; Wang, Q.; Dong, Z.; Liu, S.; Zhang, C.; Li, J. Biochemical, Nutritional, and Sensory Quality of the Low Salt Fermented Shrimp Paste. J. Aquat. Food Prod. Technol. 2017, 26, 706–718. [Google Scholar] [CrossRef]
- Surono, I.S.; Hosono, A. Microflora and Their Enzyme Profile in “Terasi” Starter. Biosci. Biotechnol. Biochem. 1994, 58, 1167–1169. [Google Scholar] [CrossRef]
- Ali, M.; Kusnadi, J.; Aulanni’am, A.; Yunianta, Y. Amino acids, fatty acids and volatile compounds of Terasi Udang, an Indonesian Shrimp paste, during fermentation. AACL Bioflux 2020, 13, 938–950. [Google Scholar]
- Surono, I.S.; Hosono, A. Chemical and Aerobic Bacterial Composition of “Terasi”, a Traditional Fermented Product from Indonesia. J. Food Hyg. Soc. Jpn. 1994, 35, 299. [Google Scholar] [CrossRef]
- Pongsetkul, J.; Benjakul, S.; Vongkamjan, K.; Sumpavapol, P.; Osako, K. Microbiological and chemical changes of shrimp Acetes vulgaris during Kapi production. J. Food Sci. Technol. 2017, 54, 3473–3482. [Google Scholar] [CrossRef]
- Albarracín, W.; Sánchez, I.C.; Grau, R.; Barat, J.M. Salt in food processing; usage and reduction: A review. Int. J. Food Sci. Technol. 2011, 46, 1329–1336. [Google Scholar] [CrossRef]
- Stringer, S.; Pin, C. Microbial Risks Associated with Salt Reduction in Certain Foods and Alternative Options for Preservation; Technical Report; Institute of Food Research: Norwich, UK, 2005. [Google Scholar]
- Xu, W.; Yu, G.; Xue, C.; Xue, Y.; Ren, Y. Biochemical changes associated with fast fermentation of squid processing by-products for low salt fish sauce. Food Chem. 2008, 107, 1597–1604. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Lee, H.J.; Jeon, C.O. Microbial succession and metabolite changes during fermentation of saeu-jeot: Traditional Korean salted seafood. Food Microbiol. 2013, 34, 360–368. [Google Scholar] [CrossRef]
- Lee, S.H.; Jung, J.Y.; Jeon, C.O. Microbial successions and metabolite changes during fermentation of salted shrimp (saeu-jeot) with different salt concentrations. PLoS ONE 2014, 9, e090115. [Google Scholar] [CrossRef]
- Kleekayai, T.; Pinitklang, S.; Laohakunjit, N.; Suntornsuk, W. Volatile components and sensory characteristics of Thai traditional fermented shrimp pastes during fermentation periods. J. Food Sci. Technol. 2016, 53, 1399–1410. [Google Scholar] [CrossRef]
- Zhao, C.J.; Schieber, A.; Gänzle, M.G. Formation of taste-active amino acids, amino acid derivatives and peptides in food fermentations—A review. Food Res. Int. 2016, 89, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Lingying, D.; Limei, W.; Jiang, S.; Lixue, Z.; Bin, Q. Microbial Community Structure and Diversity of Shrimp Paste at Different Fermentation Stages 2. BioRxiv 2018, 334136. [Google Scholar] [CrossRef]
- Padilah, B.; Bahruddin, S.; Fazilah, A.; Ahmad, M.; Gulam, R.R.A. Biogenic amines analysis in shrimp pastes belacan obtained from the Northern States of Peninsular Malaysia. Int. Food Res. J. 2018, 25, 1893–1899. [Google Scholar]
- Lv, X.; Li, Y.; Cui, T.; Sun, M.; Bai, F.; Li, X.; Li, J.; Yi, S. Bacterial community succession and volatile compound changes during fermentation of shrimp paste from Chinese Jinzhou region. LWT 2020, 122, 108998. [Google Scholar] [CrossRef]
- Fernández, M.; Zúñiga, M. Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 2006, 32, 155–183. [Google Scholar] [CrossRef]
- Vanholder, R.; De Smet, R.; Lesaffer, G. p-Cresol: A toxin revealing many neglected but relevant aspects of uraemic toxicity. Nephrol. Dial. Transplant. 1999, 14, 2813–2815. [Google Scholar] [CrossRef]
- Selvaraj, B.; Buckel, W.; Golding, B.T.; Ullmann, G.M.; Martins, B.M. Structure and function of 4-hydroxyphenylacetate decarboxylase and its cognate activating enzyme. J. Mol. Microbiol. Biotechnol. 2016, 26, 76–91. [Google Scholar] [CrossRef]
- Kung, H.F.; Lee, Y.C.; Huang, Y.L.; Huang, Y.R.; Su, Y.C.; Tsai, Y.H. Degradation of histamine by lactobacillus plantarum isolated from MISO products. J. Food Prot. 2017, 80, 1682–1688. [Google Scholar] [CrossRef]
- Knorr, S.; Sinn, M.; Galetskiy, D.; Williams, R.M.; Wang, C.; Müller, N.; Mayans, O.; Schleheck, D.; Hartig, J.S. Widespread bacterial lysine degradation proceeding via glutarate and L-2-hydroxyglutarate. Nat. Commun. 2018, 9, 5071. [Google Scholar] [CrossRef]
- Kobayashi, T.; Kajiwara, M.; Wahyuni, M.; Kitakado, T.; Hamada-Sato, N.; Imada, C.; Watanabe, E. Isolation and characterization of halophilic lactic acid bacteria isolated from “terasi” shrimp paste: A traditional fermented seafood product in Indonesia. J. Gen. Appl. Microbiol. 2003, 49, 279–286. [Google Scholar] [CrossRef]
- Thongsanit, J.; Tanasupawat, S.; Keeratipibul, S.; Jatikavanich, S. Characterization and Identification of Tetragenococcus halophilus and Tetragenococcus muriaticus Strains from Fish Sauce (Nam-pla). Jpn. J. Lact. Acid Bact. 2002, 13, 46–52. [Google Scholar] [CrossRef][Green Version]
- Kim, M.S.; Park, E.J. Bacterial Communities of Traditional Salted and Fermented Seafoods from Jeju Island of Korea Using 16S rRNA Gene Clone Library Analysis. J. Food Sci. 2014, 79, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Kuda, T.; Izawa, Y.; Yoshida, S.; Koyanagi, T.; Takahashi, H.; Kimura, B. Rapid identification of Tetragenococcus halophilus and Tetragenococcus muriaticus, important species in the production of salted and fermented foods, by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Food Control 2014, 35, 419–425. [Google Scholar] [CrossRef]
- Kimura, B.; Konagaya, Y.; Fujii, T. Histamine formation by Tetragenococcus muriaticus, a halophilic lactic acid bacterium isolated from fish sauce. Int. J. Food Microbiol. 2001, 70, 71–77. [Google Scholar] [CrossRef]
- Konagaya, Y.; Kimura, B.; Ishida, M.; Fujii, T. Purification and properties of a histidine decarboxylase from Tetragenococcus muriaticus, a halophilic lactic acid bacterium. J. Appl. Microbiol. 2002, 92, 1136–1142. [Google Scholar] [CrossRef]
- Kim, K.H.; Lee, S.H.; Chun, B.H.; Jeong, S.E.; Jeon, C.O. Tetragenococcus halophilus MJ4 as a starter culture for repressing biogenic amine (cadaverine) formation during saeu-jeot (salted shrimp) fermentation. Food Microbiol. 2019, 82, 465–473. [Google Scholar] [CrossRef]
- Alexander, D.L.J.; Tropsha, A.; Winkler, D.A. Beware of R2: Simple, Unambiguous Assessment of the Prediction Accuracy of QSAR and QSPR Models. J. Chem. Inf. Model. 2015, 55, 1316–1322. [Google Scholar] [CrossRef]
- Ikram, M.M.M.; Ridwani, S.; Putri, S.P.; Fukusaki, E. GC-MS based metabolite profiling to monitor ripening-specific metabolites in pineapple (Ananas comosus). Metabolites 2020, 10, 134. [Google Scholar] [CrossRef]
- Niquet, C.; Tessier, F.J. Free glutamine as a major precursor of brown products and fluorophores in Maillard reaction systems. Amino Acids 2007, 33, 165–171. [Google Scholar] [CrossRef]
- Rössner, J.; Velíšek, J.; Pudil, F.; Davídek, J. Strecker degradation products of aspartic and glutamic acids and their amides. Czech. J. Food Sci. 2018, 19, 41–45. [Google Scholar] [CrossRef]
- Schiffman, S.; Moroch, K.; Dunbar, J. Taste of acetylated amino acids. Chem. Senses 1975, 1, 387–401. [Google Scholar] [CrossRef]
- Schilling, B.; Christensen, D.; Davis, R.; Sahu, A.K.; Hu, L.I.; Walker-Peddakotla, A.; Sorensen, D.J.; Zemaitaitis, B.; Gibson, B.W.; Wolfe, A.J. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol. Microbiol. 2015, 98, 847–863. [Google Scholar] [CrossRef] [PubMed]
- Christensen, D.G.; Baumgartner, J.T.; Xie, X.; Jew, K.M.; Basisty, N.; Schilling, B.; Kuhn, M.L.; Wolfe, A.J. Mechanisms, detection, and relevance of protein acetylation in prokaryotes. MBio 2019, 10, e02708-18. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, T.R.; Cotta, M.A. Characterisation and comparison of microbial populations in swine faeces and manure storage pits by 16S rDNA gene sequence analyses. Anaerobe 2001, 7, 181–187. [Google Scholar] [CrossRef]
- Dabert, P.; Godon, J.; Delgene, J. Characterisation of the microbial diversity in a pig manure storage pit using small subunit rDNA sequence analysis. FEMS Microbiol. Ecol. 2005, 52, 229–242. [Google Scholar] [CrossRef]
- Miller, D.N.; Varel, V.H. Swine manure composition affects the biochemical origins, composition, and accumulation of odorous compounds. J. Anim. Sci. 2003, 81, 2131–2138. [Google Scholar] [CrossRef]
- Cotta, M.A.; Whitehead, T.R.; Collins, M.D.; Lawson, P.A. Atopostipes suicloacale gen. nov., sp. nov., isolated from an underground swine manure storage pit. Anaerobe 2004, 10, 191–195. [Google Scholar] [CrossRef]
- Ishikawa, M.; Tanasupawat, S.; Nakajima, K.; Kanamori, H.; Ishizaki, S.; Kodama, K.; Okamoto-Kainuma, A.; Koizumi, Y.; Yamamoto, Y.; Yamasato, K. Alkalibacterium thalassium sp. nov., Alkalibacterium pelagium sp. nov., Alkalibacterium putridalgicola sp. nov. and Alkalibacterium kapii sp. nov., slightly halophilic and alkaliphilic marine lactic acid bacteria isolated from marine organisms and salted. Int. J. Syst. Evol. Microbiol. 2009, 59, 1215–1226. [Google Scholar] [CrossRef]
- Hanisak, M.D. Nitrogen release from decomposing seaweeds: Species and temperature effects. J. Appl. Phycol. 1993, 5, 175–181. [Google Scholar] [CrossRef]
- Kim, J.; Shahidi, F.; Heu, M. soo Characteristics of salt-fermented sauces from shrimp processing byproducts. J. Agric. Food Chem. 2003, 51, 784–792. [Google Scholar] [CrossRef]
- Geng, J.T.; Takahashi, K.; Kaido, T.; Kasukawa, M.; Okazaki, E.; Osako, K. Relationship among pH, generation of free amino acids, and Maillard browning of dried Japanese common squid Todarodes pacificus meat. Food Chem. 2019, 283, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Peralta, E.M.; Serrano, A.E.J. Activity of naturally occurring antioxidants during heat processing of low-salt fermented shrimp. Anim. Biol. Anim. Husb. Int. J. Bioflux Soc. 2014, 6, 27–33. [Google Scholar]
- Hong, S.I.; Park, W.S.; Pyun, Y.R. Inactivation of Lactobacillus sp. from Kimchi by high pressure carbon dioxide. LWT-Food Sci. Technol. 1997, 30, 681–685. [Google Scholar] [CrossRef]
- Persson, J.A.; Wennerholm, M.; O’Halloran, S. Handbook for Kjeldahl Digestion; FOSS: Hilleroed, Denmark, 2008; pp. 30–36. [Google Scholar]
- AOAC. Official Methods of Analysis, 17th ed.; AOAC International: Gaithersburg, MD, USA, 2000; ISBN 0935584870. [Google Scholar]
- Fukui, Y.; Yoshida, M.; Shozen, K.; Funatsu, Y.; Takano, T.; Oikawa, H.; Yano, Y.; Satomi, M. Bacterial communities in fish sauce mash using culture-dependent and -independent methods. J. Gen. Appl. Microbiol. 2012, 58, 273–281. [Google Scholar] [CrossRef] [PubMed]
- FDA. Bacteriological Analytical Manual. Revision A, 8th ed.; AOAC International: Gaithersburg, MD, USA, 1998. [Google Scholar]
- Huang, X.; Yu, S.; Han, B.; Chen, J. Bacterial community succession and metabolite changes during sufu fermentation. LWT 2018, 97, 537–545. [Google Scholar] [CrossRef]

| Shrimp Paste | Protein Content * |
|---|---|
| 5% salt | 36.03 ± 0.14 |
| 10% salt | 33.56 ± 0.25 |
| 15% salt | 31.23 ± 0.30 |
| 20% salt | 28.89 ± 0.36 |
| Physicochemical and Culturable Bacteria | Shrimp | T0 | T1 | T2 | T3 | T4 |
|---|---|---|---|---|---|---|
| pH | 8.26 ± 0.01 a | 7.68 ± 0.02 b | 7.80 ± 0.02 ab | 7.81 ± 0.01 ab | 7.80 ± 0.04 ab | 7.84 ± 0.02 ab |
| Salinity (%) | 3.78 ± 0.25 c | 6.65 ± 0.49 b | 7.32 ± 0.55 ab | 6.24 ± 0.46 b | 7.20 ± 0.54 ab | 7.91 ± 0.59 a |
| Moisture (%) | 50.46 ± 1.82 a | 43.59 ± 0.48 b | 44.04 ± 0.56 b | 43.28 ± 0.75 b | 43.25 ± 1.06 b | 43.18 ± 1.67 b |
| L * | ND | 44.05 ± 0.54 a | 43.52 ± 1.87 a | 45.39 ± 0.54 a | 44.54 ± 1.58 a | 43.96 ± 1.97 a |
| a * | ND | 5.64 ± 0.21 a | 5.90 ± 0.22 a | 5.39 ± 0.17 a | 6.41 ± 0.60 a | 5.39 ± 0.93 a |
| b * | ND | 5.76 ± 0.54 b | 5.52 ± 0.76 b | 5.51 ± 0.16 b | 7.37 ± 0.69 a | 6.71 ± 0.54 ab |
| Total aerobic bacteria (log CFU/g) | 5.39 ± 0.22 a | 5.25 ± 0.24 a | 4.48 ± 0.33 b | 4.01 ± 0.12 c | 3.73 ± 0.26 d | 3.57 ± 0.14 d |
| Total lactic acid bacteria (log CFU/g) | ND | 5.56 ± 0.13 a | 5.19 ± 0.55 a | 4.96 ± 0.43 ab | 4.74 ± 0.29 ab | 4.03 ± 0.21 b |
| Total halophilic bacteria (log CFU/g) | ND | 6.43 ± 0.02 a | 5.44 ± 0.16 b | 5.18 ± 0.24 bc | 4.92 ± 0.53 c | 4.72 ± 0.25 c |
| Sample (1) | Total Bases (2) | Read Count (3) | N (%) (4) | GC (%) (5) | Q20 (%) (6) | Q30 (%) (7) | OTU(s) (8) | Chao1 (9) | Shannon (10) | Inverse Simpson (11) | Good Coverage (12) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Shrimp | 7.3 × 107 | 1.6 × 105 | 0.0001 | 53.14 | 99.18 | 97.22 | 24 | 29.00 | 1.02 | 0.38 | 0.99 |
| T0 | 9.0 × 107 | 1.9 × 105 | 0.0001 | 52.94 | 99.17 | 97.15 | 108 | 120.05 | 3.23 | 0.85 | 0.99 |
| T1 | 8.5 × 107 | 1.8 × 105 | 0.0001 | 52.97 | 99.19 | 97.19 | 111 | 123.75 | 3.28 | 0.86 | 0.99 |
| T2 | 9.2 × 107 | 2.0 × 105 | 0.0001 | 52.99 | 99.16 | 97.12 | 114 | 121.56 | 3.52 | 0.88 | 0.99 |
| T3 | 8.6 × 107 | 1.8 × 105 | 0.0001 | 53.17 | 99.19 | 97.20 | 105 | 106.50 | 3.67 | 0.89 | 0.99 |
| T4 | 8.1 × 107 | 1.7 × 105 | 0.0001 | 52.75 | 99.20 | 97.22 | 82 | 85.11 | 3.37 | 0.87 | 0.99 |
| Sample | E. coli (Presence/Absence in 25 g) | Salmonella (Presence/Absence in 25 g) |
|---|---|---|
| T0 | - | - |
| T1 | - | - |
| T2 | - | - |
| T3 | - | - |
| T4 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Helmi, H.; Astuti, D.I.; Putri, S.P.; Sato, A.; Laviña, W.A.; Fukusaki, E.; Aditiawati, P. Dynamic Changes in the Bacterial Community and Metabolic Profile during Fermentation of Low-Salt Shrimp Paste (Terasi). Metabolites 2022, 12, 118. https://doi.org/10.3390/metabo12020118
Helmi H, Astuti DI, Putri SP, Sato A, Laviña WA, Fukusaki E, Aditiawati P. Dynamic Changes in the Bacterial Community and Metabolic Profile during Fermentation of Low-Salt Shrimp Paste (Terasi). Metabolites. 2022; 12(2):118. https://doi.org/10.3390/metabo12020118
Chicago/Turabian StyleHelmi, Henny, Dea Indriani Astuti, Sastia Prama Putri, Arisa Sato, Walter A. Laviña, Eiichiro Fukusaki, and Pingkan Aditiawati. 2022. "Dynamic Changes in the Bacterial Community and Metabolic Profile during Fermentation of Low-Salt Shrimp Paste (Terasi)" Metabolites 12, no. 2: 118. https://doi.org/10.3390/metabo12020118
APA StyleHelmi, H., Astuti, D. I., Putri, S. P., Sato, A., Laviña, W. A., Fukusaki, E., & Aditiawati, P. (2022). Dynamic Changes in the Bacterial Community and Metabolic Profile during Fermentation of Low-Salt Shrimp Paste (Terasi). Metabolites, 12(2), 118. https://doi.org/10.3390/metabo12020118








