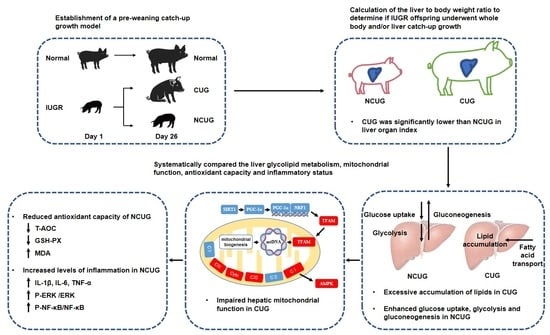
Altered Liver Metabolism, Mitochondrial Function, Oxidative Status, and Inflammatory Response in Intrauterine Growth Restriction Piglets with Different Growth Patterns before Weaning
Abstract
1. Introduction
2. Materials and Methods
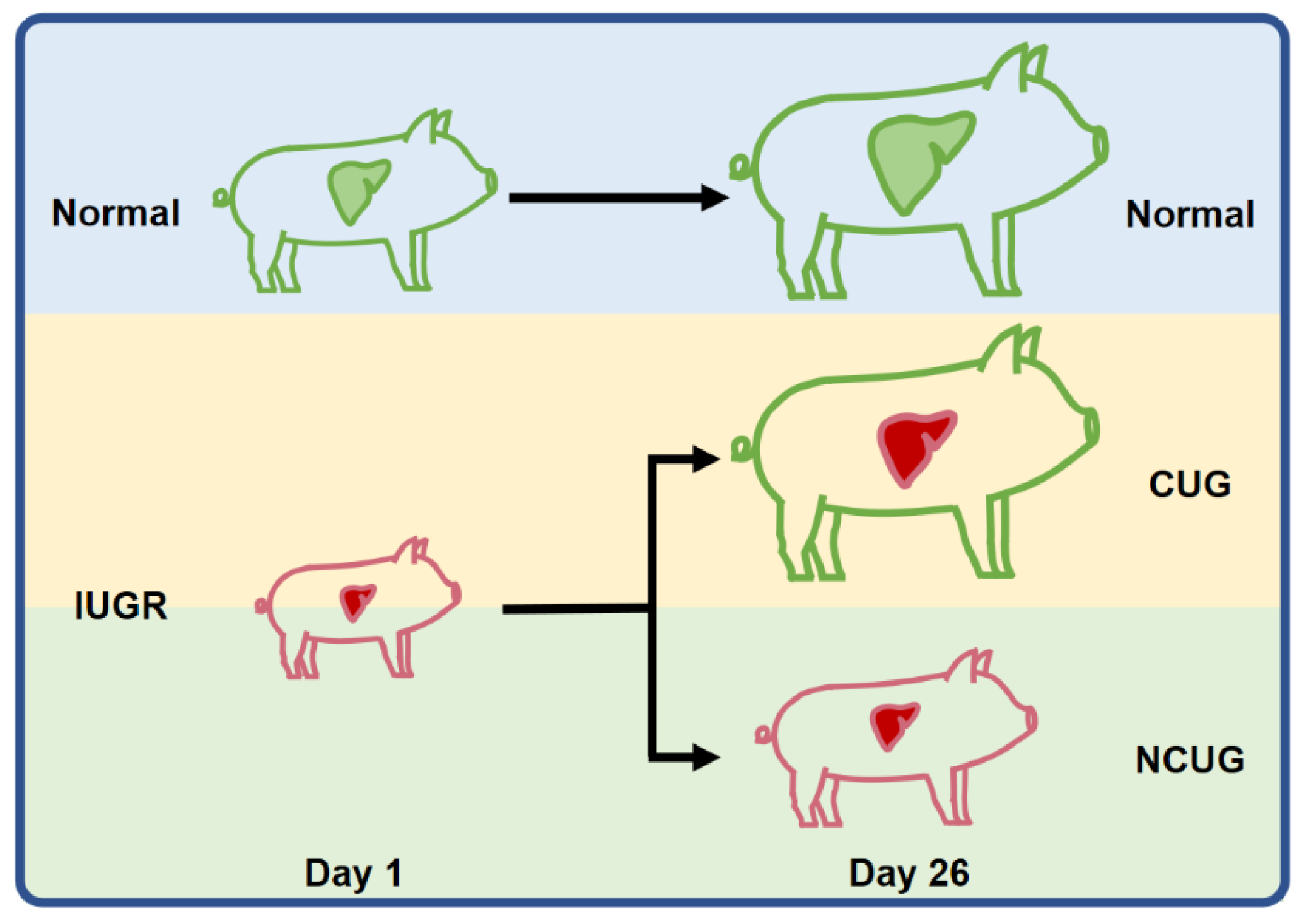
2.1. Animals, Experimental Diets, and Sample Collection
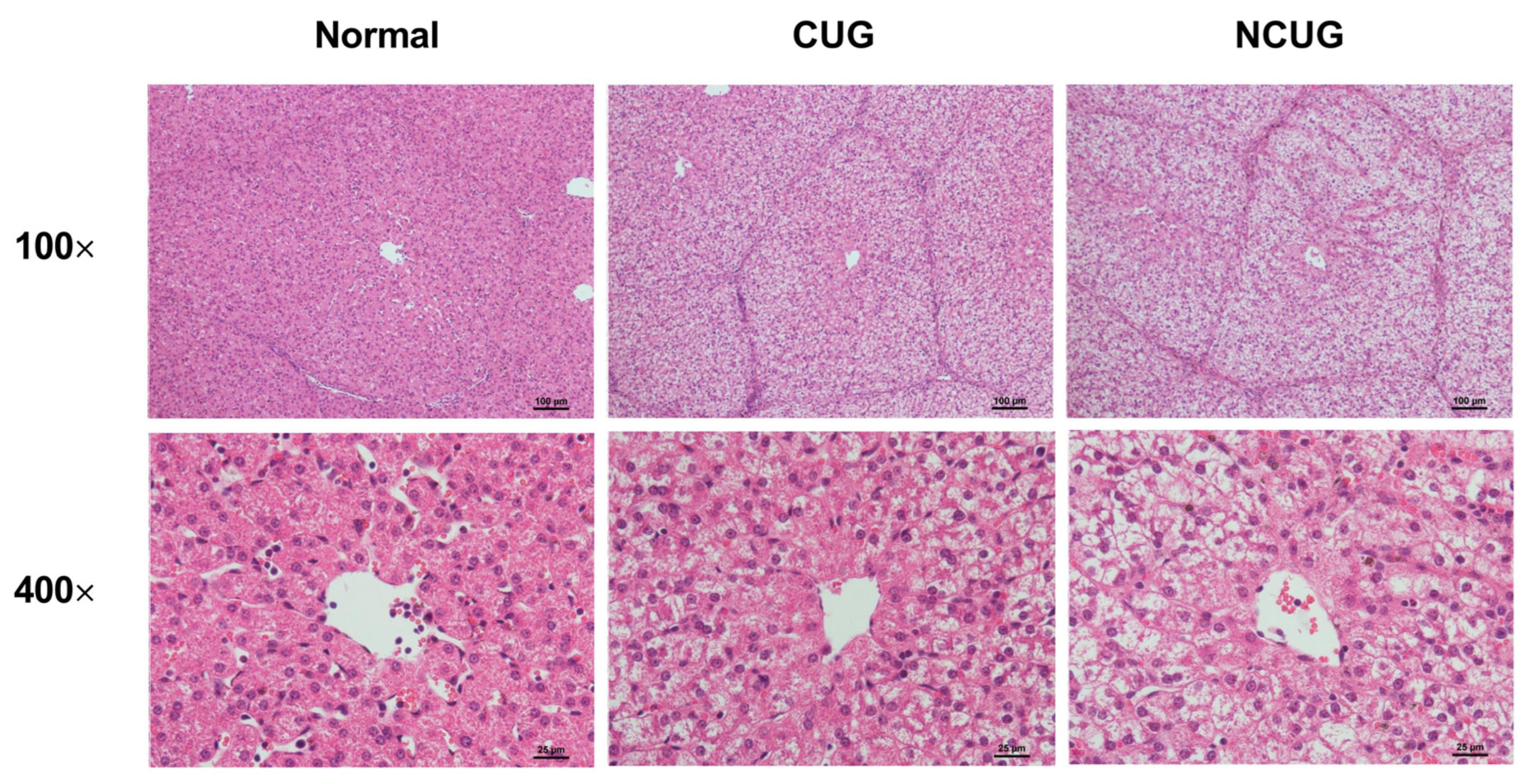
2.2. Liver Biopsy Staining
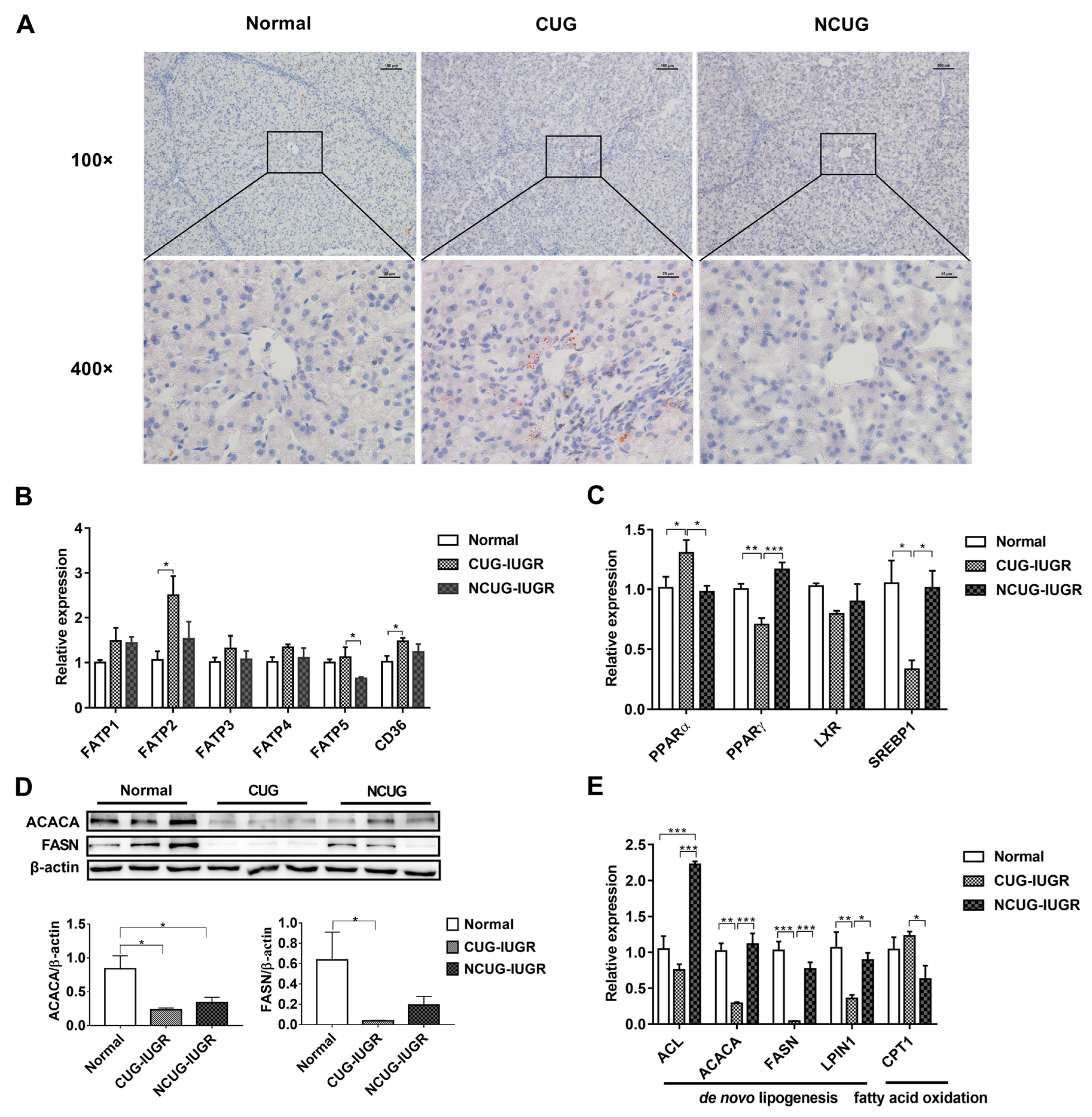
2.3. Oil Red O Staining of Liver Sections
2.4. Hepatic Oxidative Stress
2.5. Total RNA Isolation and Real-Time PCR
2.6. Western Blot Analysis
2.7. Statistical Analysis
2.8. Ethics Approval and Consent to Participate
3. Results
3.1. Growth Performance and Organ Index of Piglets with Different Growth Patterns
3.2. Liver Histology Section
3.3. Hepatic Lipid Metabolism in Piglets with Different Growth Patterns
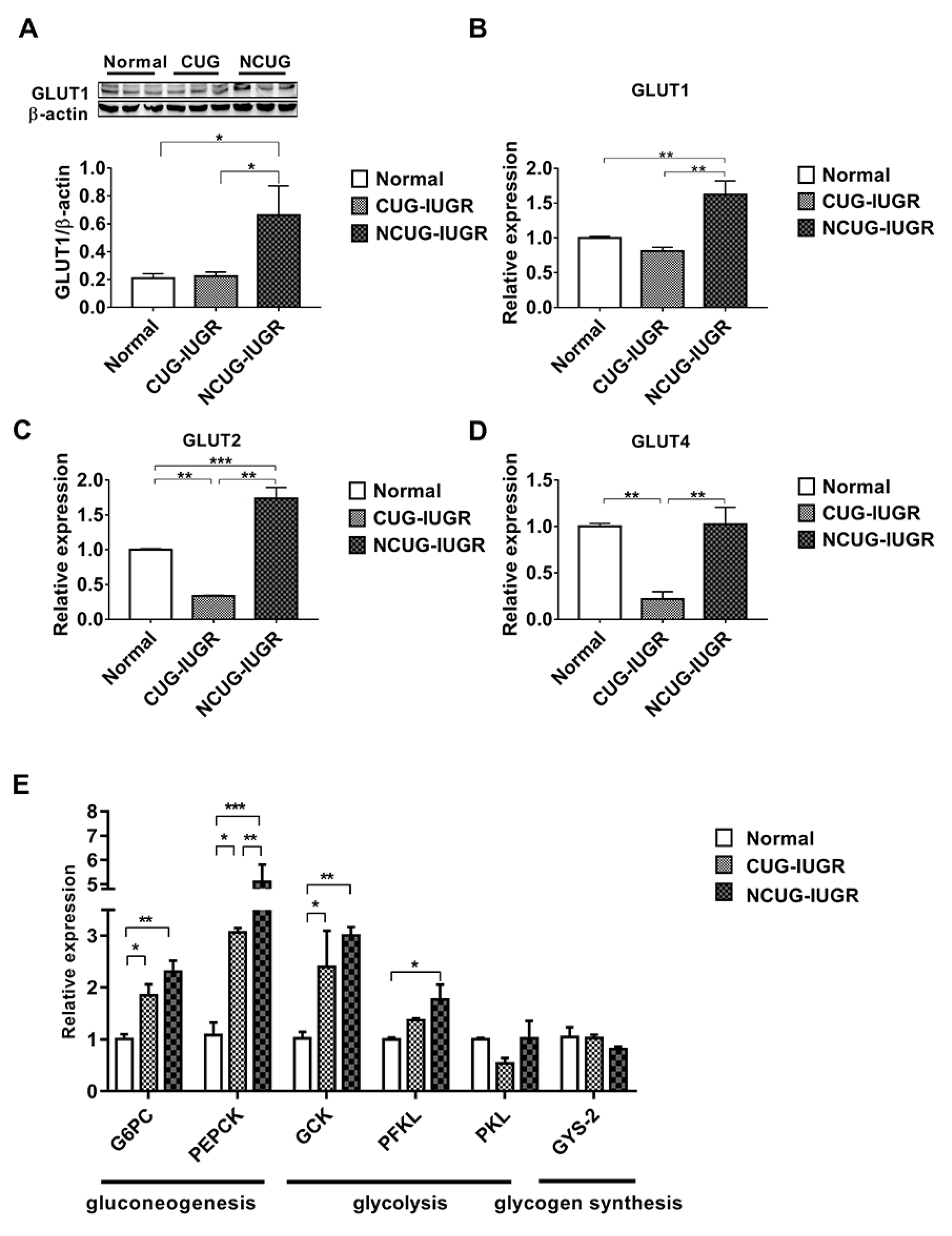
3.4. Hepatic Glucose Metabolism in Piglets with the Different Growth Patterns
3.5. Mitochondrial Electronic Transmission Chain (ETC) and AMPK Signaling Pathway in the Liver among Different Growth Patterns of Piglets
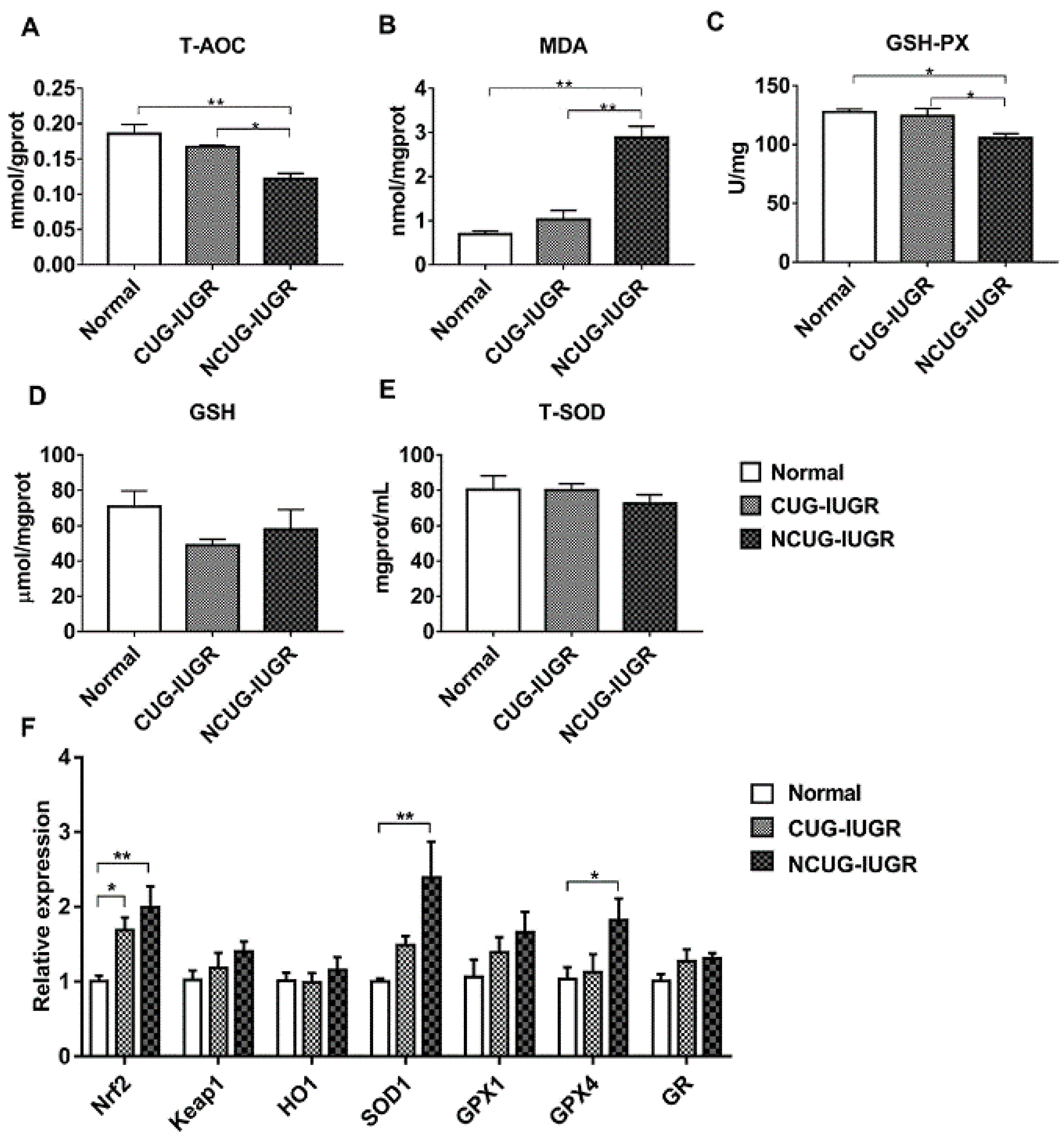
3.6. Hepatic Oxidative Stress Comparison among Different Growth Patterns of Piglets
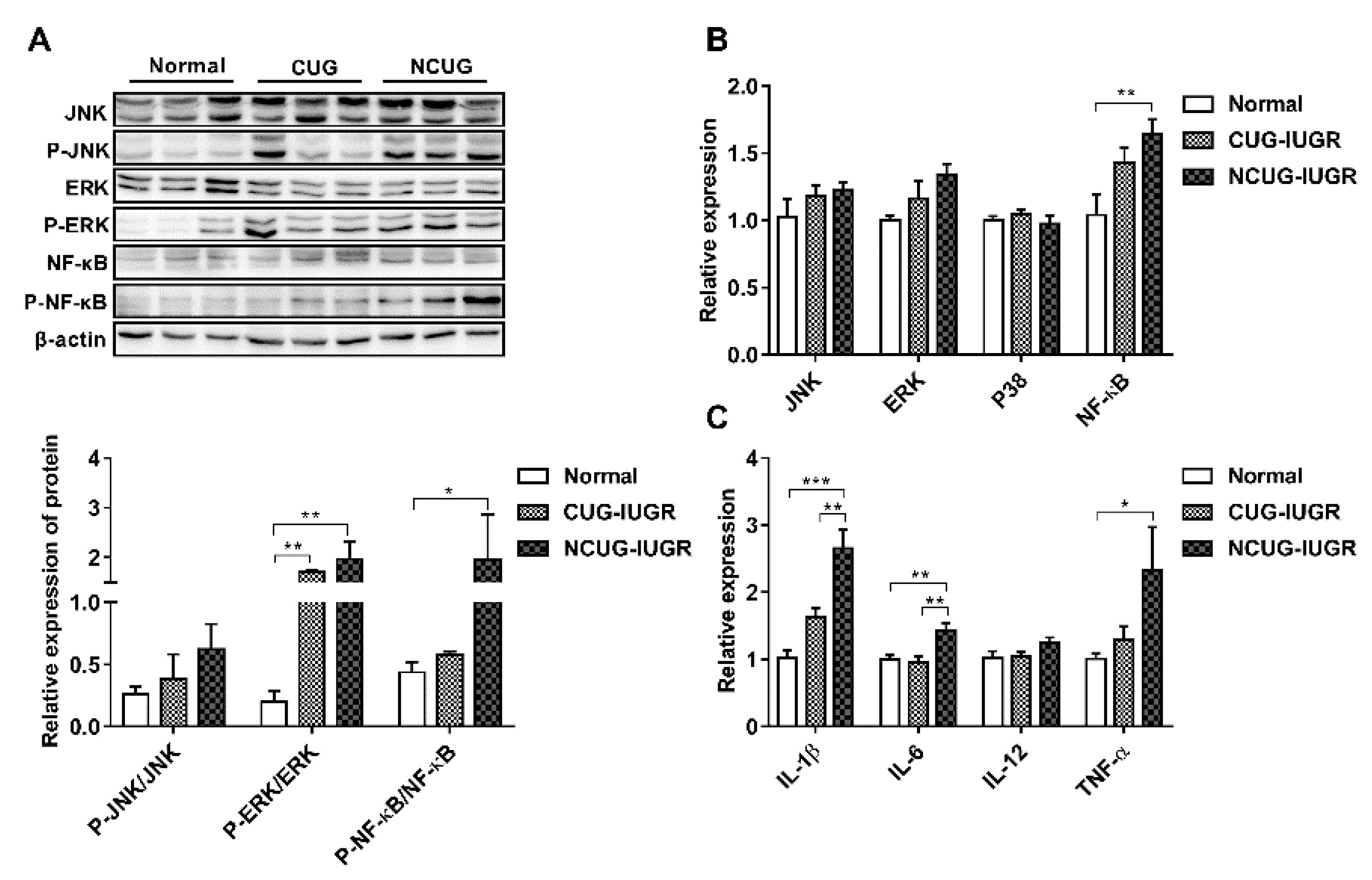
3.7. Hepatic Inflammatory State and NF-κB/MAPK Signaling Pathway
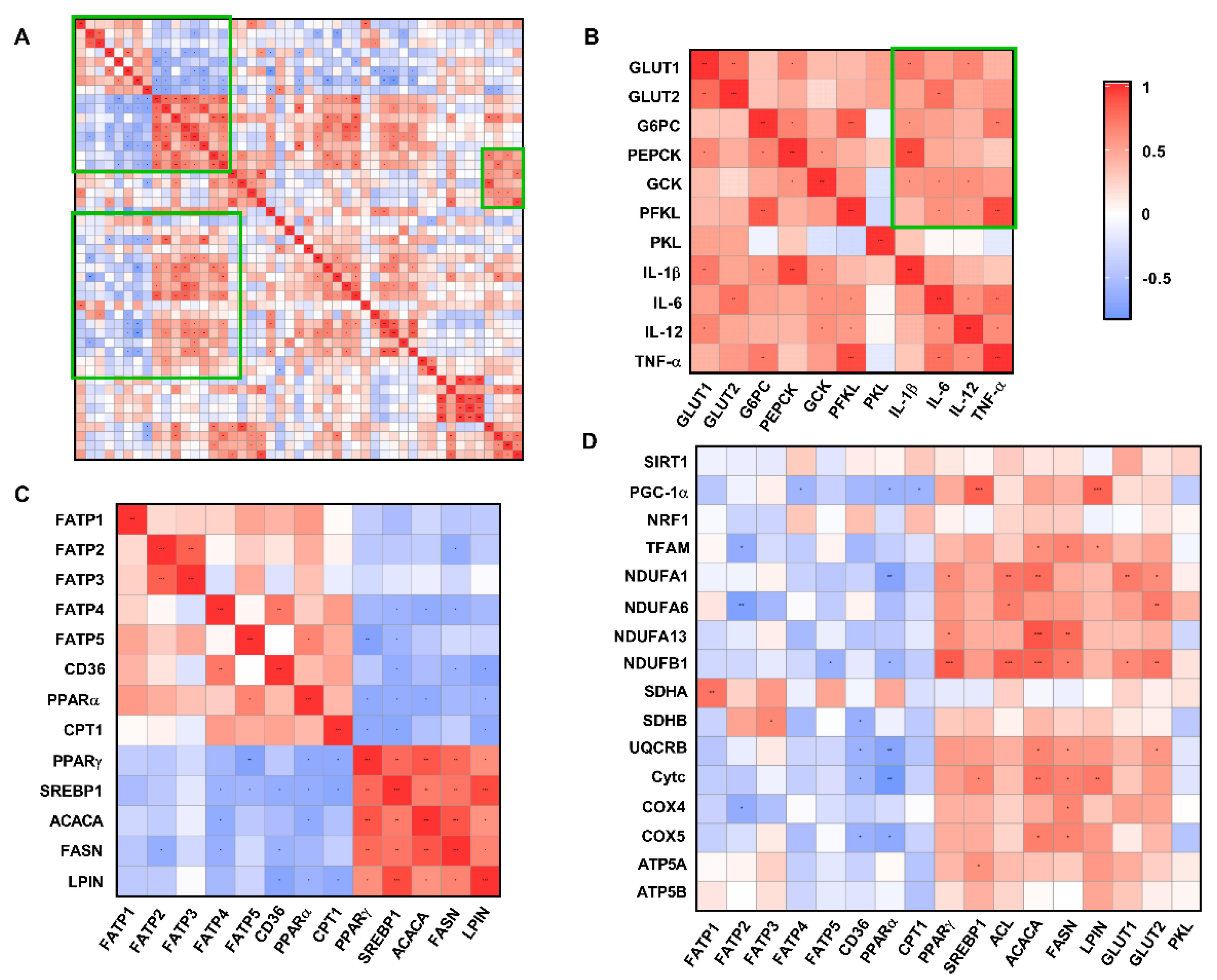
3.8. Correlation Analysis of Liver Metabolism-Related Indexes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Wu, G.; Bazer, F.W.; Wallace, J.M.; Spencer, T.E. Board-invited review: Intrauterine growth retardation: Implications for the animal sciences. J. Anim. Sci. 2006, 84, 2316–2337. [Google Scholar] [CrossRef] [PubMed]
- Soo, J.Y.; Wiese, M.D.; Berry, M.J.; McMillen, I.C.; Morrison, J.L. Intrauterine growth restriction may reduce hepatic drug metabolism in the early neonatal period. Pharmacol. Res. 2018, 134, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Poore, K.R.; Fowden, A.L. The effects of birth weight and postnatal growth patterns on fat depth and plasma leptin concentrations in juvenile and adult pigs. J. Physiol. 2004, 558, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Boersma, B.; Wit, J.M. Catch-up growth. Endocr. Rev. 1997, 18, 646–661. [Google Scholar] [CrossRef] [PubMed]
- Jain, V.; Singhal, A. Catch up growth in low birth weight infants: Striking a healthy balance. Rev. Endocr. Metab. Disord. 2012, 13, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Posont, R.; Yates, D.T. Postnatal Nutrient Repartitioning due to Adaptive Developmental Programming. Veter-Clin. N. Am. Food Anim. Pr. 2019, 35, 277–288. [Google Scholar] [CrossRef]
- Cianfarani, S.; Agostoni, C.; Bedogni, G.; Canani, R.B.; Brambilla, P.; Nobili, V.; Pietrobelli, A. Effect of intrauterine growth retardation on liver and long-term metabolic risk. Int. J. Obes. 2012, 36, 1270–1277. [Google Scholar] [CrossRef]
- Bechmann, L.P.; Hannivoort, R.A.; Gerken, G.; Hotamisligil, G.S.; Trauner, M.; Canbay, A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 2012, 56, 952–964. [Google Scholar] [CrossRef]
- A Gruppuso, P.; A Sanders, J. Regulation of liver development: Implications for liver biology across the lifespan. J. Mol. Endocrinol. 2016, 56, R115–R125. [Google Scholar] [CrossRef]
- Morrison, J.L.; Duffield, J.A.; Muhlhausler, B.; Gentili, S.; McMillen, I.C. Fetal growth restriction, catch-up growth and the early origins of insulin resistance and visceral obesity. Pediatr. Nephrol. 2010, 25, 669–677. [Google Scholar] [CrossRef]
- Hales, C.N.; Barker, D.J. Type 2 (non-insulin-dependent) diabetes mellitus: The thrifty phenotype hypothesis. Diabetologia 1992, 35, 595–601. [Google Scholar] [CrossRef]
- Pendleton, A.L.; Wesolowski, S.R.; Regnault, T.R.H.; Lynch, R.M.; Limesand, S.W. Dimming the powerhouse: Mitochondrial dysfunction in the liver and skeletal muscle of intrauterine growth restricted fetuses. Front. Endocrinol. 2021, 12, 612888. [Google Scholar] [CrossRef]
- Shen, L.; Gan, M.; Zhang, S.; Ma, J.; Tang, G.; Jiang, Y.; Li, M.; Wang, J.; Li, X.; Che, L.; et al. Transcriptome analyses reveal adult metabolic syndrome with intrauterine growth restriction in pig models. Front. Genet. 2018, 9, 291. [Google Scholar] [CrossRef]
- Fernandez-Twinn, D.; Ozanne, S. Mechanisms by which poor early growth programs type-2 diabetes, obesity and the metabolic syndrome. Physiol. Behav. 2006, 88, 234–243. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Su, W.; Ying, Z.; Zhou, L.; Zhang, L.; Wang, T. Resveratrol attenuates mitochondrial dysfunction in the liver of intrauterine growth retarded suckling piglets by improving mitochondrial biogenesis and redox status. Mol. Nutr. Food Res. 2017, 61, 1600653. [Google Scholar] [CrossRef]
- Saget, S.; Cong, R.; Decourtye, L.; Endale, M.-L.; Martinerie, L.; Girardet, C.; Perret, C.; Clemessy, M.; Leneuve, P.; Dinard, L.; et al. Changes in circulating miRNA19a-3p precede insulin resistance programmed by intra-uterine growth retardation in mice. Mol. Metab. 2020, 42, 101083. [Google Scholar] [CrossRef]
- Cui, C.; Wu, C.; Wang, J.; Ma, Z.; Zheng, X.; Zhu, P.; Wang, N.; Zhu, Y.; Guan, W.; Chen, F. Restored intestinal integrity, nutrients transporters, energy metabolism, antioxidative capacity and decreased harmful microbiota were associated with IUGR piglet’s catch-up growth before weanling. J. Anim. Sci. Biotechnol. 2022, 13, 129. [Google Scholar] [CrossRef]
- Ju, L.; Tong, W.; Qiu, M.; Shen, W.; Sun, J.; Zheng, S.; Chen, Y.; Liu, W.; Tian, J. Antioxidant MMCC ameliorates catch-up growth related metabolic dysfunction. Oncotarget 2017, 8, 99931–99939. [Google Scholar] [CrossRef][Green Version]
- Kopec, G.; Shekhawat, P.S.; Mhanna, M.J. Prevalence of diabetes and obesity in association with prematurity and growth restriction. Diabetes Metab. Syndr. Obes. 2017, 10, 285–295. [Google Scholar] [CrossRef]
- Ipsen, D.H.; Lykkesfeldt, J.; Tveden-Nyborg, P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell. Mol. Life Sci. 2018, 75, 3313–3327. [Google Scholar] [CrossRef]
- Duarte, J.A.G.; Carvalho, F.; Pearson, M.; Horton, J.D.; Browning, J.D.; Jones, J.G.; Burgess, S.C. A high-fat diet suppresses de novo lipogenesis and desaturation but not elongation and triglyceride synthesis in mice. J. Lipid Res. 2014, 55, 2541–2553. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, K.L.; Smith, C.I.; Schwarzenberg, S.J.; Jessurun, J.; Boldt, M.D.; Parks, E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Investig. 2005, 115, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Ten Hove, M.; Pater, L.; Storm, G.; Weiskirchen, S.; Weiskirchen, R.; Lammers, T.; Bansal, R. The hepatic lipidome: From basic science to clinical translation. Adv. Drug Deliv. Rev. 2020, 159, 180–197. [Google Scholar] [CrossRef] [PubMed]
- Limesand, S.W.; Rozance, P.J.; Smith, D.; Hay, W.W., Jr. Increased insulin sensitivity and maintenance of glucose utilization rates in fetal sheep with placental insufficiency and intrauterine growth restriction. Am. J. Physiol. Metab. 2007, 293, E1716–E1725. [Google Scholar] [CrossRef] [PubMed]
- Lynegaard, J.; Hansen, C.; Kristensen, A.; Amdi, C. Body composition and organ development of intra-uterine growth restricted pigs at weaning. Animal 2020, 14, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Pericuesta, E.; Gutierrez-Arroyo, J.L.; Sanchez-Calabuig, M.J.; Gutierrez-Adan, A. Postnatal catch-up growth programs telomere dynamics and glucose intolerance in low birth weight mice. Int. J. Mol. Sci. 2021, 22, 3657. [Google Scholar] [CrossRef]
- Houin, S.S.; Rozance, P.J.; Brown, L.D.; Hay, W.W., Jr.; Wilkening, R.B.; Thorn, S.R. Coordinated changes in hepatic amino acid metabolism and endocrine signals support hepatic glucose production during fetal hypoglycemia. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E306–E314. [Google Scholar] [CrossRef]
- Jones, A.K.; Brown, L.D.; Rozance, P.J.; Serkova, N.J.; Hay, W.W., Jr.; Friedman, J.E.; Wesolowski, S.R. Differential effects of intrauterine growth restriction and a hypersinsulinemic-isoglycemic clamp on metabolic pathways and insulin action in the fetal liver. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2019, 316, R427–R440. [Google Scholar] [CrossRef]
- Ying, Z.; Ge, X.; Zhang, H.; Su, W.; Li, Y.; Zhou, L.; Zhang, L.; Wang, T. Effects of dietary methionine restriction on postnatal growth, insulin sensitivity, and glucose metabolism in intrauterine growth retardation pigs at 49 and 105 d of age. J. Anim. Sci. 2018, 97, 610–619. [Google Scholar] [CrossRef]
- Rui, L. Energy metabolism in the liver. Compr. Physiol. 2014, 4, 177–197. [Google Scholar]
- Wang, J.; Cao, M.; Zhuo, Y.; Che, L.; Fang, Z.; Xu, S.; Lin, Y.; Feng, B.; Wu, D. Catch-up growth following food restriction exacerbates adulthood glucose intolerance in pigs exposed to intrauterine undernutrition. Nutrition 2016, 32, 1275–1284. [Google Scholar] [CrossRef]
- Duan, C.; Liu, M.; Xu, H.; Tang, W.; Liu, J.; Hou, L.; Li, L. Decreased expression of GLUT4 in male CG-IUGR rats may play a vital role in their increased susceptibility to diabetes mellitus in adulthood. Acta Biochim. et Biophys. Sin. 2016, 48, 872–882. [Google Scholar] [CrossRef]
- Brown, L.D.; Rozance, P.J.; Bruce, J.L.; Friedman, J.E.; Hay, W.W., Jr.; Wesolowski, S.R. Limited capacity for glucose oxidation in fetal sheep with intrauterine growth restriction. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2015, 309, R920–R928. [Google Scholar] [CrossRef]
- Guo, R.; Gu, J.; Zong, S.; Wu, M.; Yang, M. Structure and mechanism of mitochondrial electron transport chain. Biomed. J. 2018, 41, 9–20. [Google Scholar] [CrossRef]
- Cheng, K.; Jia, P.; Ji, S.; Song, Z.; Zhang, H.; Zhang, L.; Wang, T. Improvement of the hepatic lipid status in intrauterine growth retarded pigs by resveratrol is related to the inhibition of mitochondrial dysfunction, oxidative stress and inflammation. Food Funct. 2020, 12, 278–290. [Google Scholar] [CrossRef]
- Oke, S.L.; Sohi, G.; Hardy, D.B. Perinatal protein restriction with postnatal catch-up growth leads to elevated p66Shc and mitochondrial dysfunction in the adult rat liver. Reproduction 2020, 159, 27–39. [Google Scholar] [CrossRef]
- Xie, X.; Lin, T.; Zhang, M.; Liao, L.; Yuan, G.; Gao, H.; Ning, Q.; Luo, X. IUGR with infantile overnutrition programs an insulin-resistant phenotype through DNA methylation of peroxisome proliferator-activated receptor-gamma coactivator-1alpha in rats. Pediatr. Res. 2015, 77, 625–632. [Google Scholar] [CrossRef][Green Version]
- Ruderman, N.B.; Xu, X.J.; Nelson, L.; Cacicedo, J.M.; Saha, A.K.; Lan, F.; Ido, Y. AMPK and SIRT1: A long-standing partnership? Am. J. Physiol.-Endocrinol. Metab. 2010, 298, E751–E760. [Google Scholar] [CrossRef]
- Wu, S.; Zou, M.H. AMPK, mitochondrial function, and cardiovascular disease. Int. J. Mol. Sci. 2020, 21, 4987. [Google Scholar] [CrossRef]
- Zi, Y.; Ma, C.; Li, H.; Shen, S.; Liu, Y.; Li, M.; Gao, F. Effects of intrauterine growth restriction during late pregnancy on the ovine fetal renal function and antioxidant capacity. Anim. Sci. J. 2021, 92, e13613. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Wang, T. Antioxidant capacity and concentration of redox-active trace mineral in fully weaned intra-uterine growth retardation piglets. J. Anim. Sci. Biotechnol. 2015, 6, 48. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Che, L.; Xuan, Y.; Hu, L.; Liu, Y.; Xu, Q.; Fang, Z.; Lin, Y.; Xu, S.; Wu, D.; Zhang, K.; et al. Effect of Postnatal Nutrition Restriction on the Oxidative Status of Neonates with Intrauterine Growth Restriction in a Pig Model. Neonatology 2014, 107, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yao, Y.; Yu, B.; Mao, X.; Huang, Z.; Chen, D. Effect of folic acid supplementation on hepatic antioxidant function and mitochondrial-related gene expression in weanling intrauterine growth retarded piglets. Livest. Sci. 2012, 146, 123–132. [Google Scholar] [CrossRef]
- Chen, Q.M.; Maltagliati, A.J. Nrf2 at the heart of oxidative stress and cardiac protection. Physiol. Genom. 2018, 50, 77–97. [Google Scholar] [CrossRef] [PubMed]
- Sajadimajd, S.; Khazaei, M. Oxidative Stress and Cancer: The role of Nrf2. Curr. Cancer Drug Target. 2018, 18, 538–557. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Ren, W.; Liu, G.; Duan, J.; Yang, G.; Wu, L.; Li, T.; Yin, Y. Birth oxidative stress and the development of an antioxidant system in newborn piglets. Free Radic. Res. 2013, 47, 1027–1035. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef]
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef]
- Matulewicz, N.; Karczewska-Kupczewska, M. Insulin resistance and chronic inflammation. Postep. Hig. I Med. Dosw. (Online) 2016, 70, 1245–1258. [Google Scholar]
- Oeckinghaus, A.; Hayden, M.S.; Ghosh, S. Crosstalk in NF-kappaB signaling pathways. Nat. Immunol. 2011, 12, 695–708. [Google Scholar] [CrossRef]
- Liu, C.; Lin, G.; Wang, X.; Wang, T.; Wu, G.; Li, D.; Wang, J. Intrauterine growth restriction alters the hepatic proteome in fetal pigs. J. Nutr. Biochem. 2013, 24, 954–959. [Google Scholar] [CrossRef]
- Tarry-Adkins, J.L.; Fernandez-Twinn, D.S.; Hargreaves, I.P.; Neergheen, V.; E Aiken, C.; Martin-Gronert, M.S.; McConnell, J.M.; E Ozanne, S. Coenzyme Q10 prevents hepatic fibrosis, inflammation, and oxidative stress in a male rat model of poor maternal nutrition and accelerated postnatal growth. Am. J. Clin. Nutr. 2016, 103, 579–588. [Google Scholar] [CrossRef]
- Niu, Y.; He, J.; Zhao, Y.; Shen, M.; Zhang, L.; Zhong, X.; Wang, C.; Wang, T. Effect of Curcumin on Growth Performance, Inflammation, Insulin level, and Lipid Metabolism in Weaned Piglets with IUGR. Animals 2019, 9, 1098. [Google Scholar] [CrossRef]


| Item | Content | |
|---|---|---|
| Late Gestation | Lactation | |
| Ingredients | ||
| High quality northeast corn | 47.90 | 57.67 |
| Barley | 30.00 | 10.00 |
| Bran | 5.00 | |
| Soybean meal (43%) | 12.10 | 22.60 |
| Soybeans | 2.00 | |
| Fish meal | 1.00 | 1.50 |
| Soybean oil | 1.63 | |
| Limestone | 1.00 | 1.40 |
| CaHPO4 | 1.20 | 1.20 |
| Salt | 0.50 | 0.60 |
| Lys | 0.15 | 0.20 |
| DL-Met | 0.15 | 0.20 |
| Premix 1 | 1.00 | 1.00 |
| Total | 100 | 100 |
| Nutrient composition 2 | ||
| DE, MJ/kg | 3150.00 | 3400.00 |
| CP | 13.50 | 17.50 |
| EE | 2.50 | 4.80 |
| CF | 3.24 | 2.82 |
| Ash | 5.18 | 5.49 |
| Ca | 0.95 | 1.00 |
| TP | 0.60 | 0.62 |
| AP | 0.35 | 0.45 |
| TLys | 0.70 | 1.20 |
| Gene Name | Gene Accession | Primer Sequences (5′ → 3′) | Size, bp |
|---|---|---|---|
| FATP1 | NM_001083931.1 | F: GGCAACAGACGTGATCTATGAC | 125 |
| R: AGCGGCTGGCTGAAAACT | |||
| FATP2 | JX092264.1 | F: TCTAACACGGATGGGGTCG | 107 |
| R: AGGGCAGGAGTGGAAAAGT | |||
| FATP3 | XM_001929591.2 | F: AGGTCTCAGCCGAAGTGGAT | 206 |
| R: TGGGAGGGCGAGGTAGAT | |||
| FATP4 | XM_003353676.1 | F: AGCCGCATCCTGTCCTTT | 213 |
| R: GACATCCTTGGCGATCTTTT | |||
| FATP5 | AK233227.1 | F: GCGAGCAGACGGAAAAGAAG | 199 |
| R: TGCAGGAAGTCCACGAGTGA | |||
| CD36 | DQ192230.1 | F: GGACTCATTGCTGGTGCTGT | 169 |
| R: GTCTGTAAACTTCCGTGCCTGT | |||
| CPT-1 | NM_001129805 | F: GGTGGTGTCAGCGTAGCA | 391 |
| R: CCTTGTTGTCAGTTTGGGTAA | |||
| PPAR-α | DQ437887.1 | F: CAGCGTGGCACTGAACATC | 144 |
| R: CTCCGATCACATTTGTCATAGAC | |||
| FASN | NM_001099930.1 | F: GCTTGTCCTGGGAAGAGTGTA | 115 |
| R: AGGAACTCGGACATAGCGG | |||
| ACACA | NM_001114269.1 | F: ACATCCCCACGCTAAACA | 186 |
| R: AGCCCATCACTTCATCAAAG | |||
| ACL | NM_001105302 | F: AGCGAGCAGCAGACCTATGAC | 143 |
| R: GGCCACGTTGGTGAAGTTTG | |||
| LPIN1 | NM_001130734.1 | F: CACATTTTGCCCACCCTT | 164 |
| R: GTGCCACGCTCGTTGACC | |||
| SREBP1 | NM_214157.1 | F: AGCGGACGGCTCACAATG | 121 |
| R: CGCAAGACGGCGGATTTA | |||
| PPAR-γ | NM_214379.1 | F: AGCCCTTTGGTGACTT | 213 |
| R: AGGACTCTGGGTGGTT | |||
| LXR | AB254405.1 | F: TTCCGTCGCAGTGTCATC | 184 |
| R: CTTGCCGCTTCAGTTTCTTCA | |||
| GLUT1 | NW_003610563.1 | F: GATGAAGGAGGAGTGCCG | 106 |
| R: CAGCACCACGGCGATGAGGAT | |||
| GLUT2 | NM_001097417.1 | F: ATTCTTTGGTGGGATGCTTG | 118 |
| R: ATGAGATGGTCCCAATTTCG | |||
| GLUT4 | NM_0011288433 | F: TATGTTGCGGATGCTATGGG | 396 |
| R: CTCGGGTTTCAGGCACTTTT | |||
| G6PC | NM_001113445 | F: GGAAATGAGCAGCAAGGT | 176 |
| R: TCGGTGCCACTGATAAAG | |||
| PEPCK | NM_001161753 | F: CACAAGGGCAAAGTGATTATGC | 238 |
| R: GGAACCAGTTGACGTGGAAGA | |||
| GCK | XM_013985832.2 | F: GTGGTGGCAATGGTGAATGAC | 184 |
| R: TCGGCGGTCTTCATAGTAGCA | |||
| PFKL | XM_021071510.1 | F: GAAACGAGAAGTGCCACGAA | 182 |
| R: TACCGTAGTTCCGGTCGAAG | |||
| PKL | XM_021089721.1 | F: AGACTGCCAAGGGTCACTTT | 117 |
| R: CAGCTCCTCAAAGAGTTGCC | |||
| GYS2 | NM_001195511 | F: CATCACCACCAACGACGGA | 193 |
| R: ACACGGCCCAGAGAAAAGG | |||
| NDUFA1 | XM_003135339.5 | F: GCTTCCGGGGAAGGAATCAA | 101 |
| R: CCGGGGAGAATTTCGAACCA | |||
| NDUFA6 | NM_001185178.1 | F: TCTCAGAGCCTTGCATGTCG | 85 |
| R: AAGCCATCCAGCATCGTACC | |||
| NDUFA13 | NM_001244646.1 | F: ATGAAGGATGTGCCGGACTG | 125 |
| R: CCATAGGTGGCGCTGAGAAT | |||
| NDUFB1 | XM_003482306.4 | F: TGCCTTCCGGAACAAGAGTC | 88 |
| R: GCAATTCAGCCACAGCCTTT | |||
| SDHA | XM_021076930.1 | F: CAATAAGAGGTCGTCGGCCA | 127 |
| R: AGAGAGACCAAACGCAGCTC | |||
| SDHB | NM_001104953.1 | F: TCCTATGGTGTTGGATGCGT | 124 |
| R: AGTGTTGCCTCCGTTGATGT | |||
| UQCRB | NM001185172.1 | F: CATCAGGCAACGCTTCTGTC | 81 |
| R: TATACCCTCCAGCCACTTGC | |||
| CytC | NM_001129970.1 | F: CTGGGGAGAGGAGACACTGAT | 158 |
| R: AGGCGGTGGCCAACTTTTAC | |||
| COX IV | XM_021093705.1 | F: CCAAGTGGGACTACGACAAGAAC | 131 |
| R: CCTGCTCGTTTATTAGCACTGG | |||
| COX V | NM_001007517.1 | F: ATCTGGAGGTGGTGTTCCTACTG | 160 |
| R: GTTGGTGATGGAGGGGACTAAA | |||
| ATP5A1 | NM_001185142.1 | F: ACGCCATTGATGGAAAGGGT | 98 |
| R: TGGTTCCCGCACAGAGATTC | |||
| ATP5B1 | XM_001929410.5 | F: CATGTTGGGCTTTGTGGGTC | 139 |
| R: ATAGTCTCTGGCAGGCTGGA | |||
| SIRT1 | NM_001145750.2 | F: TTGCAACAGCATCTTGCCTG | 91 |
| R: GGACATCGAGGAACCACCTG | |||
| PGC-1α | NM_213963.2 | F: GCTTGACGAGCGTCATTCAG | 100 |
| R: GGTCTTCACCAACCAGAGCA | |||
| NRF1 | XM_021078993.1 | F: GAAGCTGTCCAGGGGCTTTA | 116 |
| R: ATCCATGCTCTGCTACTGGG | |||
| TFAM | NM_001130211.1 | F: AGCGAGGTCTGAAGAGTTGC | 114 |
| R: TTGCACCCGTAGACAAAGCA | |||
| POLG | XM_001927064.5 | F: CTGTCAGATGAGGGCGAGTG | 133 |
| R: ACTTCTTCCGTCGTGACTTTCT | |||
| Nrf2 | XM_013984303.2 | F: ATCCAGCGGATTGCTCGTAG | 155 |
| R: TCAAATCCATGTCCTTGGCG | |||
| Keap1 | NM_001114671.1 | F: TCTGCTTAGTCATGGTGACCT | 158 |
| R: GGGGTTCCAGATGACAAGGG | |||
| HO-1 | NM_001004027.1 | F: TGATGGCGTCCTTGTACCAC | 71 |
| R: GACCGGGTTCTCCTTGTTGT | |||
| SOD1 | NM_214127.2 | F: CAAGAAGGGGCACCACGTT | 70 |
| R: CTCAGGGGACGCAAGAACTG | |||
| GPX1 | NM_214201.1 | F: CCTCAAGTACGTCCGACCAG | 85 |
| R: GTGAGCATTTGCGCCATTCA | |||
| GPX4 | NM_214407.1 | F: TGTGTGAATGGGGACGATGC | 135 |
| R: CTTCACCACACAGCCGTTCT | |||
| GR | AY368271.1 | F: GTGAGCCGACTGAACACCAT | 141 |
| R: CAGGATGTGAGGAGCTGTGT | |||
| TNF-α | NM_214022.1 | F: GCCCTTCCACCAACGTTTTC | 97 |
| R: CAAGGGCTCTTGATGGCAGA | |||
| IL-1β | NM_214055.1 | F: ATTCAGGGACCCTACCCTCTC | 92 |
| R: ATCACTTCCTTGGCGGGTTC | |||
| IL-6 | NM_214399.1 | F: ACAAAGCCACCACCCCTAAC | 185 |
| R: CGTGGACGGCATCAATCTCA | |||
| IL-12 | NC_010458.4 | F: CAACCCTGTGCCTTAGCAGT | 113 |
| R: AGAGCCTGCATCAGCTCAGT | |||
| NF-κB | NM_001114281.1 | F: GGGGCGATGAGATCTTCCTG | 110 |
| R: CACGTCGGCTTGTGAAAAGG | |||
| ERK | XM_021088019.1 | F: CAGTCTCTGCCCTCCAAGAC | 139 |
| R: GGGTAGATCATCCAGCTCCA | |||
| JNK | XM_001929166.6 | F: TGGATGAAAGGGAACACACA | 104 |
| R: ATGATGACGATGGATGCTGA | |||
| P38 | XM_021091323.1 | F: AAGACGGGGTCCTCATCTCC | 230 |
| R: TCTCATCGTAGGGCTCTGCT |
| Item | Normal | CUG | NCUG | SEM | p-Value |
|---|---|---|---|---|---|
| BW, kg | 1.388 a ± 0.010 | 1.059 b ± 0.035 | 1.045 b ± 0.037 | 0.0203 | <0.001 |
| WW, kg | 7.530 a ± 0.526 | 7.457 a ± 0.368 | 5.592 b ± 0.912 | 0.1367 | <0.001 |
| ADG, kg | 0.236 a ± 0.017 | 0.246 a ± 0.014 | 0.175 b ± 0.035 | 0.0048 | <0.001 |
| ADG Per Week, kg | Normal | CUG | NCUG | SEM | p-Value |
|---|---|---|---|---|---|
| 1 d–7 d | 0.168 a ± 0.019 | 0.163 a ± 0.026 | 0.126 b ± 0.042 | 0.004 | <0.001 |
| 7 d–14 d | 0.204 a ± 0.064 | 0.241 a ± 0.046 | 0.161 b ± 0.069 | 0.007 | <0.001 |
| 14 d–21 d | 0.228 b ± 0.017 | 0.264 a ± 0.044 | 0.198 c ± 0.045 | 0.005 | <0.001 |
| 21 d–26 d | 0.343 a ± 0.031 | 0.346 a ± 0.035 | 0.231 b ± 0.077 | 0.009 | <0.001 |
| Organ Weight, kg | Normal | CUG | NCUG | SEM | p-Value |
|---|---|---|---|---|---|
| Heart | 0.093 a ± 0.011 | 0.097 a ± 0.006 | 0.057 b ± 0.006 | 0.007 | 0.02 |
| Liver | 0.447 a ± 0.025 | 0.397 b ± 0.030 | 0.280 c ± 0.017 | 0.020 | <0.001 |
| Kidney | 0.090 a ± 0.010 | 0.087 a ± 0.006 | 0.067 b ± 0.006 | 0.006 | 0.017 |
| Spleen | 0.047 a ± 0.047 | 0.033 ab ± 0.033 | 0.020 b ± 0.020 | 0.008 | 0.037 |
| Lung | 0.220 a ± 0.046 | 0.180 a ± 0.010 | 0.120 b | 0.022 | 0.011 |
| Pancreas | 0.010 | 0.013 ± 0.006 | 0.013 ± 0.006 | 0.004 | 0.63 |
| Organ Index, % | Normal | CUG | NCUG | SEM | p-Value |
|---|---|---|---|---|---|
| Heart | 1.15 ± 0.17 | 1.19 ± 0.050 | 1.18 ± 0.150 | 0.110 | 0.932 |
| Liver | 5.49 ab ± 0.436 | 4.86 b ± 0.303 | 5.83 a ± 0.212 | 0.270 | 0.031 |
| Kidney | 1.10 a ± 0.096 | 1.07 b ± 0.084 | 1.39 a ± 0.159 | 0.096 | 0.028 |
| Spleen | 0.58 ± 0.200 | 0.41 ± 0.066 | 0.42 ± 0.013 | 0.010 | 0.245 |
| Lung | 2.71 ± 0.610 | 2.21 ± 0.167 | 2.50 ± 0.081 | 0.300 | 0.322 |
| Pancreas | 0.12 ± 0.003 | 0.16 ± 0.069 | 0.28 ± 0.120 | 0.065 | 0.123 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Zhu, P.; Zheng, X.; Ma, Z.; Cui, C.; Wu, C.; Zeng, X.; Guan, W.; Chen, F. Altered Liver Metabolism, Mitochondrial Function, Oxidative Status, and Inflammatory Response in Intrauterine Growth Restriction Piglets with Different Growth Patterns before Weaning. Metabolites 2022, 12, 1053. https://doi.org/10.3390/metabo12111053
Wang J, Zhu P, Zheng X, Ma Z, Cui C, Wu C, Zeng X, Guan W, Chen F. Altered Liver Metabolism, Mitochondrial Function, Oxidative Status, and Inflammatory Response in Intrauterine Growth Restriction Piglets with Different Growth Patterns before Weaning. Metabolites. 2022; 12(11):1053. https://doi.org/10.3390/metabo12111053
Chicago/Turabian StyleWang, Jun, Pengwei Zhu, Xiaoyu Zheng, Ziwei Ma, Chang Cui, Caichi Wu, Xiangfang Zeng, Wutai Guan, and Fang Chen. 2022. "Altered Liver Metabolism, Mitochondrial Function, Oxidative Status, and Inflammatory Response in Intrauterine Growth Restriction Piglets with Different Growth Patterns before Weaning" Metabolites 12, no. 11: 1053. https://doi.org/10.3390/metabo12111053
APA StyleWang, J., Zhu, P., Zheng, X., Ma, Z., Cui, C., Wu, C., Zeng, X., Guan, W., & Chen, F. (2022). Altered Liver Metabolism, Mitochondrial Function, Oxidative Status, and Inflammatory Response in Intrauterine Growth Restriction Piglets with Different Growth Patterns before Weaning. Metabolites, 12(11), 1053. https://doi.org/10.3390/metabo12111053