Bioinformatic Analysis of Kynurenine Pathway Enzymes and Their Relationship with Glioma Hallmarks
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Sample Selection
2.2. Selection of Gene Pools and Gene Expression Comparison
2.3. Statistical Analysis
3. Results
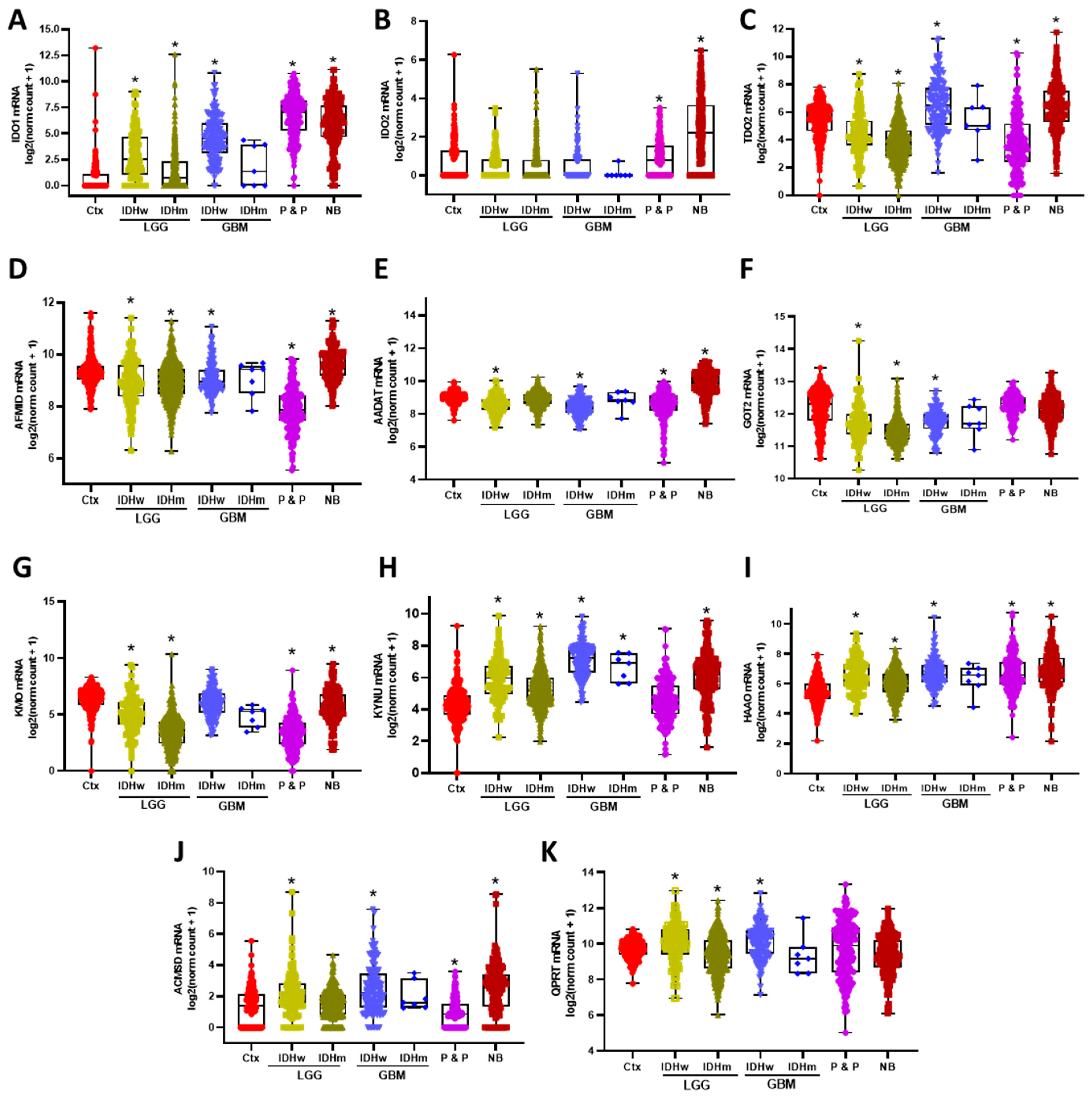
3.1. KP Enzymes Distribution on Brain Tumors
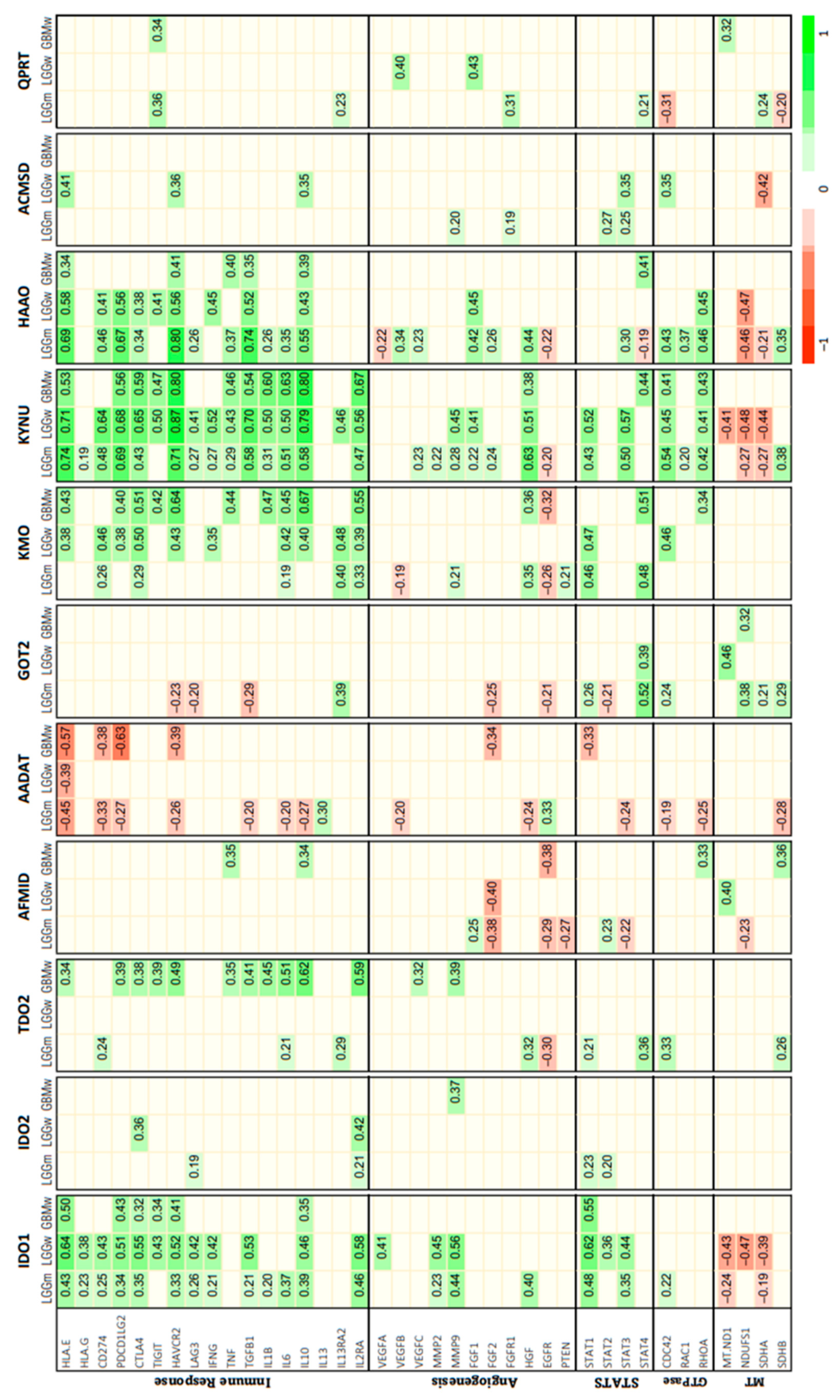
3.2. Correlation between KP Enzymes and Glioma Hallmarks
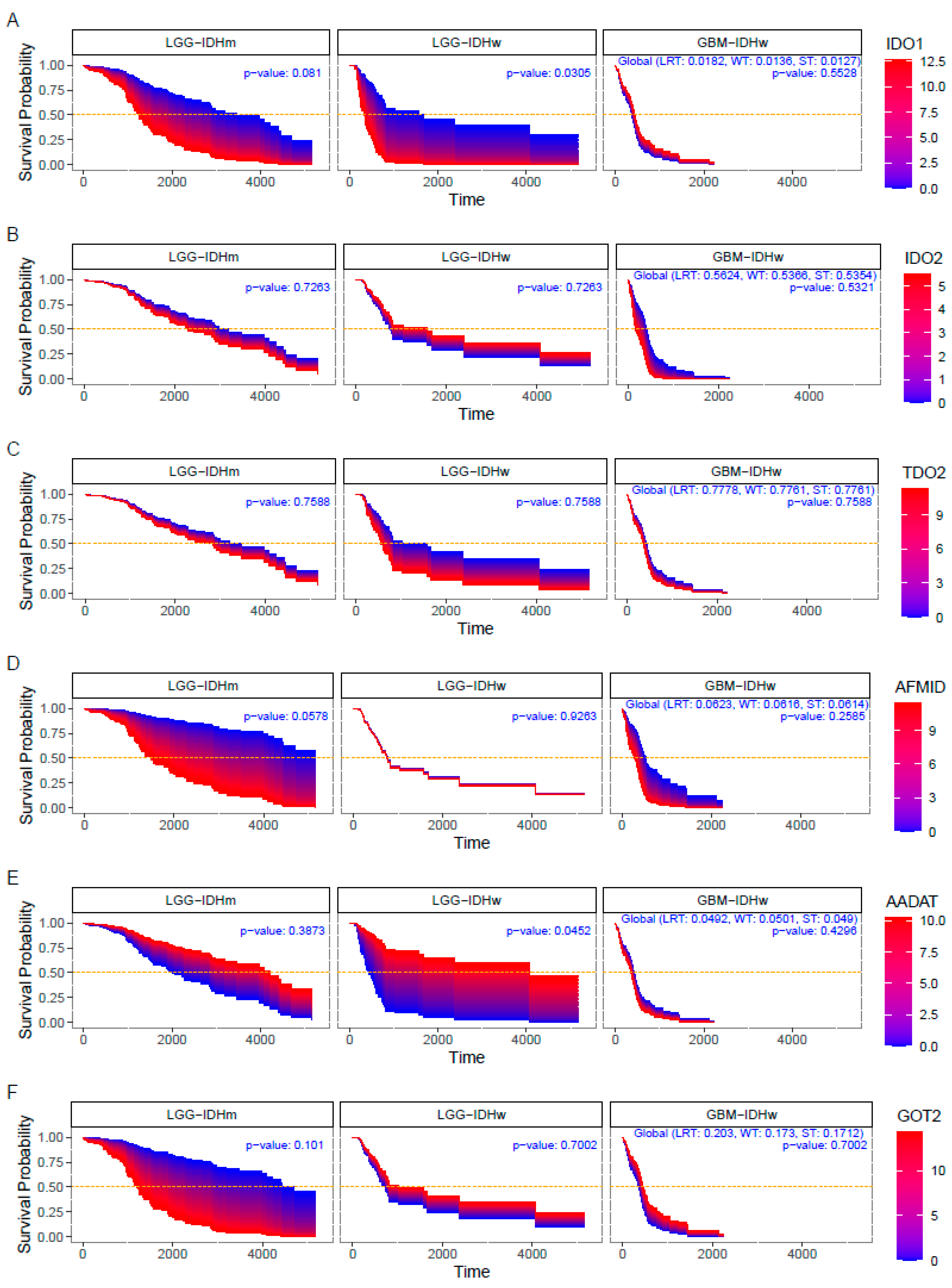
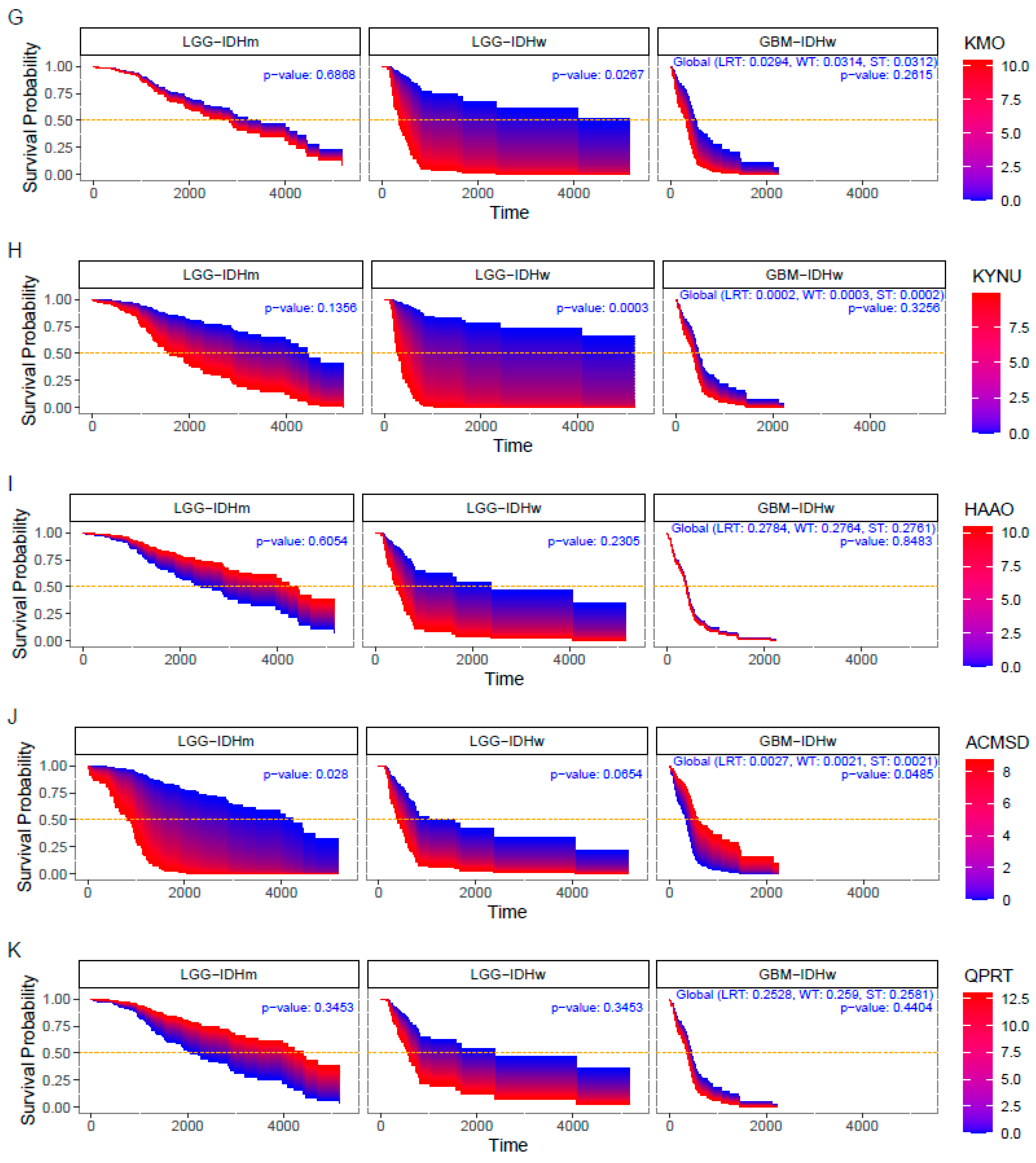
3.3. Impact of KP Enzymes on Survival Estimation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cahill, D.; Turcan, S. Origin of Gliomas. Semin. Neurol. 2018, 38, 5–10. [Google Scholar] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; A Cree, I.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Stupp, R.; Brada, M.; van de Bent, M.J.; Tonn, J.-C.; Pentheroudakis, G. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25 (Suppl. 3), iii93–iii101. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van de Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013-2017. Neuro. Oncol. 2020, 22 (Suppl. 2), iv1–iv96. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016–1036. [Google Scholar]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability--an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Yuan, Q.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. The definition of primary and secondary glioblastoma. Clin. Cancer Res. 2013, 19, 764–772. [Google Scholar] [CrossRef]
- Agnihotri, S.; Zadeh, G. Metabolic reprogramming in glioblastoma: The influence of cancer metabolism on epigenetics and unanswered questions. Neuro Oncol. 2016, 18, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Strickland, M.; Stoll, E.A. Metabolic Reprogramming in Glioma. Front. Cell Dev. Biol. 2017, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Shibao, S.; Minami, N.; Koike, N.; Niboyuki, F.; Yoshida, K.; Saya, H.; Sampetrean, O. Metabolic heterogeneity and plasticity of glioma stem cells in a mouse glioblastoma model. Neuro Oncol. 2018, 20, 343–354. [Google Scholar] [CrossRef]
- Vlashi, E.; Lagadec, C.; Vergnes, L.; Matsutani, T.; Masui, K.; Poulou, M.; Popescu, R.; Donna, L.D.; Evers, P.; Dekmezian, C. Metabolic state of glioma stem cells and nontumorigenic cells. Proc. Natl. Acad. Sci. USA 2011, 108, 16062–16067. [Google Scholar] [CrossRef]
- Ye, F.; Zhang, Y.; Liu, Y.; Yamada, K.; Tso, J.L.; Menjivar, J.C.; Tian, J.Y.; Yong, W.H.; Schaue, D.; Mischel, P.S. Protective properties of radio-chemoresistant glioblastoma stem cell clones are associated with metabolic adaptation to reduced glucose dependence. PLoS ONE 2013, 8, e80397. [Google Scholar] [CrossRef]
- Saga, I.; Shibao, S.; Okubo, J.; Osuka, S.; Kobayashi, Y.; Yamada, S.; Fujita, S.; Urakami, K.; Kusuhara, M.; Yoshida, K. Integrated analysis identifies different metabolic signatures for tumor-initiating cells in a murine glioblastoma model. Neuro Oncol. 2014, 16, 1048–1056. [Google Scholar] [CrossRef]
- Kim, J.; Han, J.; Jang, Y.; Kim, S.J.; Lee, M.J.; Ryu, M.J.; Kweon, G.R.; Heo, J.Y. High-capacity glycolytic and mitochondrial oxidative metabolisms mediate the growth ability of glioblastoma. Int. J. Oncol. 2015, 47, 1009–1016. [Google Scholar] [CrossRef]
- Waziri, A. Glioblastoma-derived mechanisms of systemic immunosuppression. Neurosurg. Clin. N. Am. 2010, 21, 31–42. [Google Scholar] [CrossRef]
- Broekman, M.L.; Mass, S.L.N.; Abels, E.R.; Mempel, T.R.; Krichevsky, A.M.; Breakefield, X.O. Multidimensional communication in the microenvirons of glioblastoma. Nat. Rev. Neurol. 2018, 14, 482–495. [Google Scholar] [CrossRef]
- Gieryng, A.; Pszczolkowska, D.; Walentynowicz, K.A.; Rajan, W.D.; Kaminska, B. Immune microenvironment of gliomas. Lab. Investig. 2017, 97, 498–518. [Google Scholar] [CrossRef] [PubMed]
- Perng, P.; Lim, M. Immunosuppressive Mechanisms of Malignant Gliomas: Parallels at Non-CNS Sites. Front. Oncol. 2015, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Badawy, A.A. Tryptophan availability for kynurenine pathway metabolism across the life span: Control mechanisms and focus on aging, exercise, diet and nutritional supplements. Neuropharmacology 2017, 112 Pt B, 248–263. [Google Scholar] [CrossRef]
- Bender, D.A.; Magboul, B.I.; Wynick, D. Probable mechanisms of regulation of the utilization of dietary tryptophan, nicotinamide and nicotinic acid as precursors of nicotinamide nucleotides in the rat. Br. J. Nutr. 1982, 48, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Gossmann, T.I.; Ziegler, M.; Puntervoll, P.; de Figueiredo, L.F.; Schuster, S.; Heiland, I. NAD(+) biosynthesis and salvage--a phylogenetic perspective. FEBS J. 2012, 279, 3355–3363. [Google Scholar] [CrossRef] [PubMed]
- Frumento, G.; Rotondo, R.; Tonetti, M.; Damonte, G.; Benatti, U.; Ferrara, G.B. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J. Exp. Med. 2002, 196, 459–468. [Google Scholar] [CrossRef]
- Belladonna, M.L.; Puccetti, P.; Orabona, C.; Fallarino, F.; Vacca, C.; Volpi, C.; Gizzi, S.; Pallotta, M.T.; Fioretti, M.C.; Grohmann, U. Immunosuppression via tryptophan catabolism: The role of kynurenine pathway enzymes. Transplantation 2007, 84 (Suppl. 1), S17–S20. [Google Scholar] [CrossRef]
- Tanaka, M.; Tóth, F.; Polyák, H.; Szabó, A.; Mándi, Y.; Vécsei, L. Immune Influencers in Action: Metabolites and Enzymes of the Tryptophan-Kynurenine Metabolic Pathway. Biomedicines 2021, 9, 734. [Google Scholar]
- Kudo, T.; Prentzell, M.T.; Mohapatra, S.R.; Sahm, F.; Zhao, Z.; Grummt, I.; Wick, W.; Opitz, C.A.; Platten, M.; Green, E.W. Constitutive Expression of the Immunosuppressive Tryptophan Dioxygenase TDO2 in Glioblastoma Is Driven by the Transcription Factor C/EBPbeta. Front. Immunol. 2020, 11, 657. [Google Scholar] [CrossRef]
- Mohapatra, S.R.; Sadik, A.; Tykocinski, L.O.; Dietze, J.; Poschet, G.; Heiland, I.; Opitz, C.A. Hypoxia Inducible Factor 1alpha Inhibits the Expression of Immunosuppressive Tryptophan-2,3-Dioxygenase in Glioblastoma. Front. Immunol. 2019, 10, 2762. [Google Scholar] [CrossRef]
- Wainwright, D.A.; Balyasnikova, I.V.; Chang, A.L.; Ahmed, A.U.; Moon, K.S.; Auffinger, B.; Tobias, A.L.; Han, Y.; Lesniak, M.S. IDO expression in brain tumors increases the recruitment of regulatory T cells and negatively impacts survival. Clin. Cancer Res. 2012, 18, 6110–6121. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.T.; Tseng, L.M.; Chen, J.L.; Chu, P.Y.; Lee, C.H.; Huang, C.T.; Wang, W.L.; Lau, K.Y.; Tseng, M.F.; Chang, Y.Y. Kynurenine 3-monooxygenase upregulates pluripotent genes through beta-catenin and promotes triple-negative breast cancer progression. EBioMedicine 2020, 54, 102717. [Google Scholar] [CrossRef]
- Liu, C.Y.; Huang, T.T.; Chen, J.l.; Chu, P.Y.; Lee, C.H.; Lee, H.C.; Lee, Y.H.; Chang, Y.Y.; Yang, S.H.; Jiang, J.K. Significance of Kynurenine 3-Monooxygenase Expression in Colorectal Cancer. Front. Oncol. 2021, 11, 620361. [Google Scholar] [CrossRef] [PubMed]
- Liu, I.L.; Chung, T.F.; Huang, W.H.; Hsu, C.H.; Liu, C.C.; Chiu, Y.Y.; Huang, K.C.; Liao, A.T.C.; Chen, S.L. Kynurenine 3-monooxygenase (KMO), and signal transducer and activator of transcription 3 (STAT3) expression is involved in tumour proliferation and predicts poor survival in canine melanoma. Vet. Comp. Oncol. 2021, 19, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Vazquez Cervantes, G.I.; Pineda, B.; Ramírez Ortega, D.; Salazar, A.; González Esquivel, D.F.; Rembao, D.; Zavala Vega, S.; Gómez Manzo, S.; Pérez de la Cruz, G.; Pérez de la Cruz, V. Kynurenine Monooxygenase Expression and Activity in Human Astrocytomas. Cells 2021, 10, 2028. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Yunhai, L.; Rodgers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. 2020. Available online: https://www.r-project.org (accessed on 1 September 2022).
- Therneau, T.M. A Package for Survival Analysis in R. R package version 3.4-0. 2022. Available online: https://CRAN.R-project.org/package=survival (accessed on 1 September 2022).
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biom J. 2008, 50, 346–363. [Google Scholar] [CrossRef]
- Revelle, W. psych: Procedures for Psychological, Psychometric, and Personality Research. R Package Version 1.9.12, Northwestern University. 2019. Available online: https://CRAN.R-project.org/package=psych (accessed on 1 September 2022).
- Gonzalez Esquivel, D.; Ramirez Ortega, D.; Pineda, B.; Castro, N.; Rios, C.; Pérez de la Cruz, V. Kynurenine pathway metabolites and enzymes involved in redox reactions. Neuropharmacology 2017, 112 Pt B, 331–345. [Google Scholar] [CrossRef]
- Ramirez Ortega, D.; Ovalle Rodríguez, P.; Pineda, B.; González-Esquivel, D.F.; Ramos Chávez, L.A.; Vazquez-Cervantes, G.I.; Roldán-Roldán, G.; Pérez de la Cruz, G.; Díaz Ruiz, A.; Méndez Armenta, M.; et al. Kynurenine Pathway as a New Target of Cognitive Impairment Induced by Lead Toxicity During the Lactation. Sci. Rep. 2020, 10, 3184. [Google Scholar] [CrossRef]
- Ramos-Chavez, L.A.; Roldán-Roldán, G.; García-Juárez, B.; González-Esquivel, D.; Pérez de la Cruz, V.; Pineda, B.; Ramírez- Ortega, D.; García Muñoz, I.; Jímenez Herrera, B.; Ríos, C.; et al. Low Serum Tryptophan Levels as an Indicator of Global Cognitive Performance in Nondemented Women over 50 Years of Age. Oxid. Med. Cell Longev. 2018, 2018, 8604718. [Google Scholar] [CrossRef]
- Prendergast, G.C. Cancer: Why tumours eat tryptophan. Nature 2011, 478, 192–194. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Braidy, N.; Bessede, A.; Brew, B.J.; Grant, R.; Teo, C.; Guillemin, G.J. The kynurenine pathway in brain tumor pathogenesis. Cancer Res. 2012, 72, 5649–5657. [Google Scholar] [CrossRef]
- Du, L.; Xing, Z.; Tao, B.; Li, T.; Yang, D.; Li, W.; Zheng, Y.; Kuang, C.; Yang, Q. Both IDO1 and TDO contribute to the malignancy of gliomas via the Kyn-AhR-AQP4 signaling pathway. Signal. Transduct. Target Ther. 2020, 5, 10. [Google Scholar] [CrossRef]
- Wainwright, D.A.; Chang, A.L.; Dey, M.; Balyasnikova, I.V.; Kim, C.-K.; Tobias, A.; Cheng, Y.; Kim, J.W.; Qiao, J.; Zhang, L.; et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin. Cancer Res. 2014, 20, 5290–5301. [Google Scholar] [CrossRef]
- Hanihara, M.; Oh-Oka, K.; Mitsuka, K.; Nakao, A.; Kinouchi, H. Synergistic antitumor effect with indoleamine 2,3-dioxygenase inhibition and temozolomide in a murine glioma model. J. Neurosurg. 2016, 124, 1594–1601. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezi, M.; Rice, T.; Kosel, M.T.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, K.; Wakimoto, H.; Lafrate, A.J.; Tanaka, S.; Loebel, F.; Lelic, N.; Wiederschain, D.; Bedel, O.; Deng, G.; Zhang, B. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell 2015, 28, 773–784. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH mutation in glioma: Molecular mechanisms and potential therapeutic targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef] [PubMed]
- Butturini, E.; de Prati, A.C.; Mariotto, S. Redox Regulation of STAT1 and STAT3 Signaling. Int. J. Mol. Sci. 2020, 21, 7034. [Google Scholar] [CrossRef] [PubMed]
- Meissl, K.; Simonovic, N.; Amenitsch, L.; Witalisz-Siepracka, A.; Klein, K.; Lassnig, C.; Puga, A.; Vogl, C.; Poelzl, A.; Bosmann, M.; et al. STAT1 Isoforms Differentially Regulate NK Cell Maturation and Anti-tumor Activity. Front. Immunol. 2020, 11, 2189. [Google Scholar] [CrossRef]
- Goder, A.; Ginter, T.; Heinzel, T.; Stroh, S.; Fahrer, J.; Henke, A.; Kramer, O.H. STAT1 N-terminal domain discriminatively controls type I and type II IFN signaling. Cytokine 2021, 144, 155552. [Google Scholar] [CrossRef]
- Yu, C.P.; Pan, Z.Z.; Luo, D.Y. TDO as a therapeutic target in brain diseases. Metab. Brain Dis. 2016, 31, 737–747. [Google Scholar] [CrossRef]
- Walczak, K.; Wnorowski, A.; Turski, W.A.; Plech, T. Kynurenic acid and cancer: Facts and controversies. Cell. Mol. Life Sci. 2020, 77, 1531–1550. [Google Scholar] [CrossRef] [PubMed]
- Venkateswaran, N.; Lafita-Navarro, M.C.; Hao, Y.; Kilgore, J.A.; Perez-Castro, L.; Braverman, J.; Borenstein-Auerbach, N.; Kim, M.; Lesner, N.P.; Mishra, P. MYC promotes tryptophan uptake and metabolism by the kynurenine pathway in colon cancer. Genes Dev. 2019, 33, 1236–1251. [Google Scholar] [CrossRef] [PubMed]
- Mezrich, J.D.; Fechner, J.H.; Zhang, X.; Jhonson, B.P.; Burlingham, W.J.; Bradfield, C.A. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 2010, 185, 3190–3198. [Google Scholar] [CrossRef] [PubMed]
- Kesarwani, P.; Kant, S.; Prabhu, A.; Chinnaiyan, P. The interplay between metabolic remodeling and immune regulation in glioblastoma. Neuro Oncol. 2017, 19, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- Heng, B.; Bilgin, A.A.; Lovejoy, D.B.; Tan, V.X.; Milioli, H.H.; Gluch, L.; Bustamante, S.; Sabaretnam, T.; Moscato, P.; Lim, C.K.; et al. Differential kynurenine pathway metabolism in highly metastatic aggressive breast cancer subtypes: Beyond IDO1-induced immunosuppression. Breast Cancer Res. 2020, 22, 113. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, X.; Lai, J.; Yi, W.; Yang, J.; Du, T.; Long, X.; Zhang, Y.; Xiao, Y. A novel role of kynureninase in the growth control of breast cancer cells and its relationships with breast cancer. J. Cell. Mol. Med. 2019, 23, 6700–6707. [Google Scholar] [CrossRef]
- Fahrmann, J.F.; Tanaka, I.; Irajizad, E.; Mao, X.; Dennison, J.B.; Murage, E.; Casabar, J.; Mayo, J.; Peng, Q.; Celiktas, M.; et al. Mutational Activation of the NRF2 Pathway Upregulates Kynureninase Resulting in Tumor Immunosuppression and Poor Outcome in Lung Adenocarcinoma. Cancers 2022, 14, 2543. [Google Scholar] [CrossRef]
- Ci, C.; Wu, C.; Lyu, D.; Chang, X.; He, C.; Liu, W.; Chen, L. Downregulation of kynureninase restrains cutaneous squamous cell carcinoma proliferation and represses the PI3K/AKT pathway. Clin. Exp. Dermatol. 2020, 45, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Sahm, F.; Oezen, I.; Optiz, C.A.; Radlwimmer, B.; von Deimling, A.; Ahrendt, T.; Adams, S.; Bode, H.B.; Guillemin, G.G.; Wick, W.; et al. The endogenous tryptophan metabolite and NAD+ precursor quinolinic acid confers resistance of gliomas to oxidative stress. Cancer Res. 2013, 73, 3225–3234. [Google Scholar] [CrossRef]
- Tang, M.; Xie, Q.; Gimple, R.C.; Zhong, Z.; Tam, T.; Tian, J.; Kidwell, R.L.; Wu, Q.; Praguer, B.C.; Qiu, Z.; et al. Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res. 2020, 30, 833–853. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Wang, W.; Li, H.; Jiao, Y.; Huo, R.; Zihan, Y.; Wang, J.; Wang, S.; Wang, J.; Chen, D. Single-Cell Atlas Reveals Complexity of the Immunosuppressive Microenvironment of Initial and Recurrent Glioblastoma. Front. Immunol. 2020, 11, 835. [Google Scholar] [CrossRef] [PubMed]
- Feger, G.; Angelov, B.; Angelova, A. Prediction of Amphiphilic Cell-Penetrating Peptide Building Blocks from Protein-Derived Amino Acid Sequences for Engineering of Drug Delivery Nanoassemblies. J. Phys. Chem. B 2020, 124, 4069–4078. [Google Scholar] [CrossRef] [PubMed]
| Grouping | Name | Abbreviation |
|---|---|---|
| Neoplasias | Glioblastoma | GBM |
| Low-grade gliomas | LGG | |
| Low-grade glioma with IDH-1 mutation | LLG IDHm | |
| Low-grade glioma with IDH-1 wildtype | LLG IDHw | |
| Glioblastoma multiforme with IDH-1 wildtype | GBM IDHw | |
| Glioblastoma multiforme with IDH-1 mutation | GBM IDHm | |
| Kynurenine pathway | Tryptophan dioxygenase | TDO |
| Indoleamine dioxygenase | IDO | |
| Arylformamidase | AFMID | |
| Glutamic-oxaloacetic transaminase | GOT2 | |
| Aminoadipate aminotransferase | AADAT | |
| Kynureninase | KYNU | |
| Kynurenine monooxygenase | KMO | |
| Quinolinic acid phosphoribosyl transferase | QPRT | |
| 3-HANA dioxygenase | HAAO | |
| Aminocarboxymuconate semialdehyde decarboxylase | ACMSD | |
| Immune response | Human leukocyte antigen E | HLA-E |
| Human leukocyte antigen G | HLA-G | |
| Programmed cell death 1 | PD-1/CD274 | |
| Programmed cell death 1 ligand 2 | PDCD1LG2 | |
| Cytotoxic T lymphocyte antigen 4 | CTLA4 | |
| T cell immunoreceptor with Ig and ITIM domains | TIGIT | |
| Hepatitis A virus cellular receptor 2 | HAVCR2 | |
| Lymphocyte activating 3 | LAG3 | |
| Interferon gamma | IFNG | |
| Tumor necrosis factor | TNF | |
| Transforming growth factor beta 1 | TGFB1 | |
| Interleukin 1 beta | IL1B | |
| Interleukin 6 | IL6 | |
| Interleukin 10 | IL10 | |
| Interleukin 13 | IL13 | |
| Interleukin 13 receptor subunit alpha 2 | IL13RA2 | |
| Interleukin 2 receptor subunit alpha | IL2RA | |
| Angiogenesis | Vascular endothelial growth factor | VEGF |
| Vascular endothelial growth factor A | VEGFA | |
| Vascular endothelial growth factor B | VEGFB | |
| Vascular endothelial growth factor C | VEGFC | |
| Matrix metalloproteinase 2 | MMP2 | |
| Matrix metalloproteinase 9 | MMP9 | |
| Fibroblast growth factor 1 | FGF1 | |
| Fibroblast growth factor 2 | FGF2 | |
| Fibroblast growth factor receptor 1 | FGFR1 | |
| Hepatocyte growth factor | HGF | |
| Epidermal growth factor receptor | EGFR | |
| Phosphatase and tensin homolog | PTEN | |
| Signal transducer and activator of transcription (STAT) | Signal transducer and activator of transcription 1 | STAT1 |
| Signal transducer and activator of transcription 2 | STAT2 | |
| Signal transducer and activator of transcription 3 | STAT3 | |
| Signal transducer and activator of transcription 4 | STAT4 | |
| Rho GTPases | Cell division control protein 42 homolog | CDC42 |
| Rac family small GTPase 1 | RAC1 | |
| Ras homolog family member A | RHOA | |
| Electron transport chain: complexes I and II | Mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 1 | MT-ND1 |
| NADH:ubiquinone oxidoreductase core subunit S1 | NDUFS1 | |
| Succinate dehydrogenase complex flavoprotein subunit A | SDHA | |
| Succinate dehydrogenase complex iron sulfur subunit B | SDHB |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez Cervantes, G.I.; Navarro Cossio, J.Á.; Pérez de la Cruz, G.; Salazar, A.; Pérez de la Cruz, V.; Pineda, B. Bioinformatic Analysis of Kynurenine Pathway Enzymes and Their Relationship with Glioma Hallmarks. Metabolites 2022, 12, 1054. https://doi.org/10.3390/metabo12111054
Vázquez Cervantes GI, Navarro Cossio JÁ, Pérez de la Cruz G, Salazar A, Pérez de la Cruz V, Pineda B. Bioinformatic Analysis of Kynurenine Pathway Enzymes and Their Relationship with Glioma Hallmarks. Metabolites. 2022; 12(11):1054. https://doi.org/10.3390/metabo12111054
Chicago/Turabian StyleVázquez Cervantes, Gustavo Ignacio, Javier Ángel Navarro Cossio, Gonzalo Pérez de la Cruz, Aleli Salazar, Verónica Pérez de la Cruz, and Benjamin Pineda. 2022. "Bioinformatic Analysis of Kynurenine Pathway Enzymes and Their Relationship with Glioma Hallmarks" Metabolites 12, no. 11: 1054. https://doi.org/10.3390/metabo12111054
APA StyleVázquez Cervantes, G. I., Navarro Cossio, J. Á., Pérez de la Cruz, G., Salazar, A., Pérez de la Cruz, V., & Pineda, B. (2022). Bioinformatic Analysis of Kynurenine Pathway Enzymes and Their Relationship with Glioma Hallmarks. Metabolites, 12(11), 1054. https://doi.org/10.3390/metabo12111054







