Transcriptomic-Metabolomic Profiling in Mouse Lung Tissues Reveals Sex- and Strain-Based Differences
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. High-Resolution Metabolomics
2.3. Transcriptomics by RNA-Sequencing
2.4. Transcriptome–Metabolome Wide Association Study (TMWAS)
3. Results
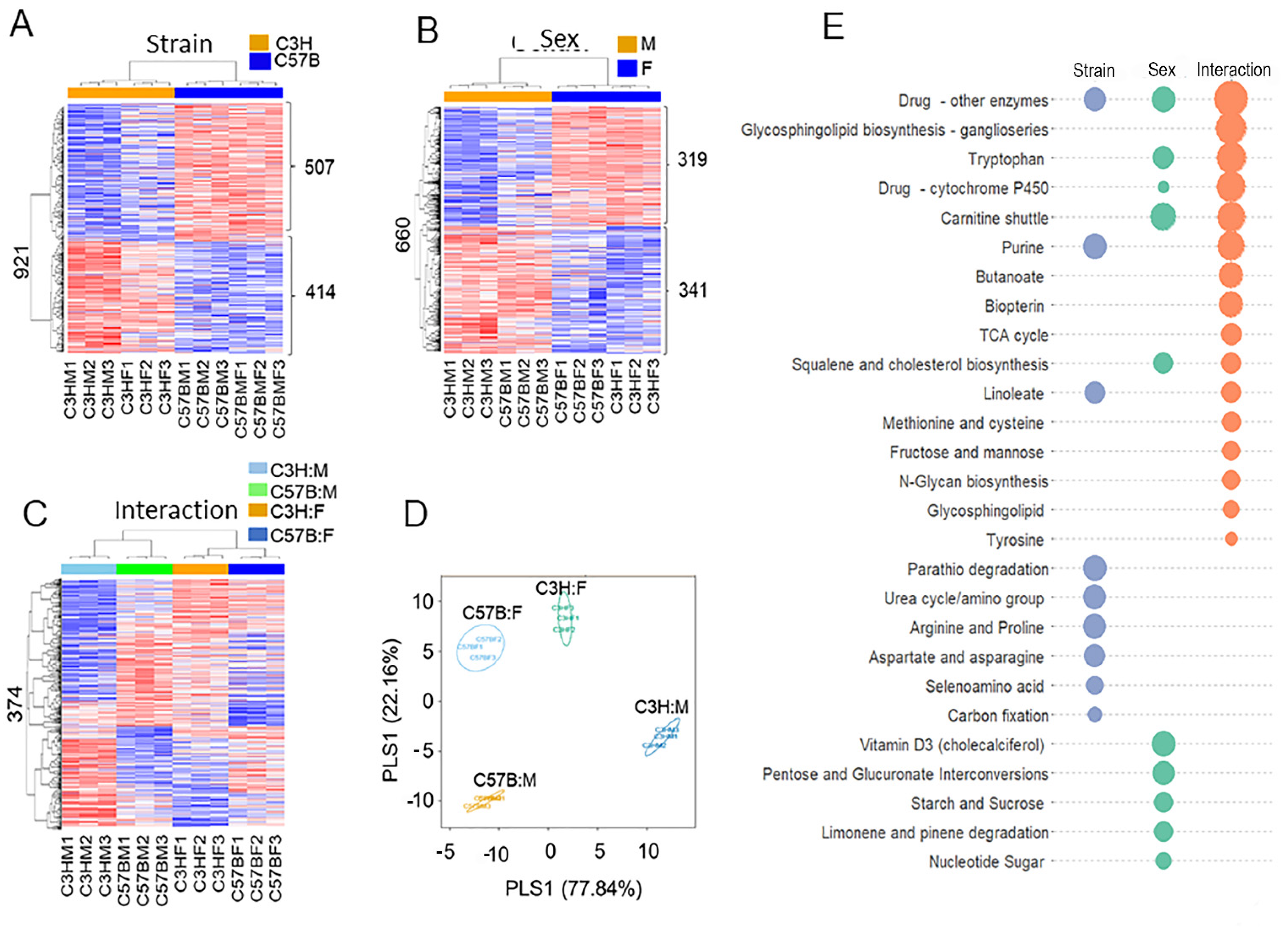
3.1. Differences in Metabolome
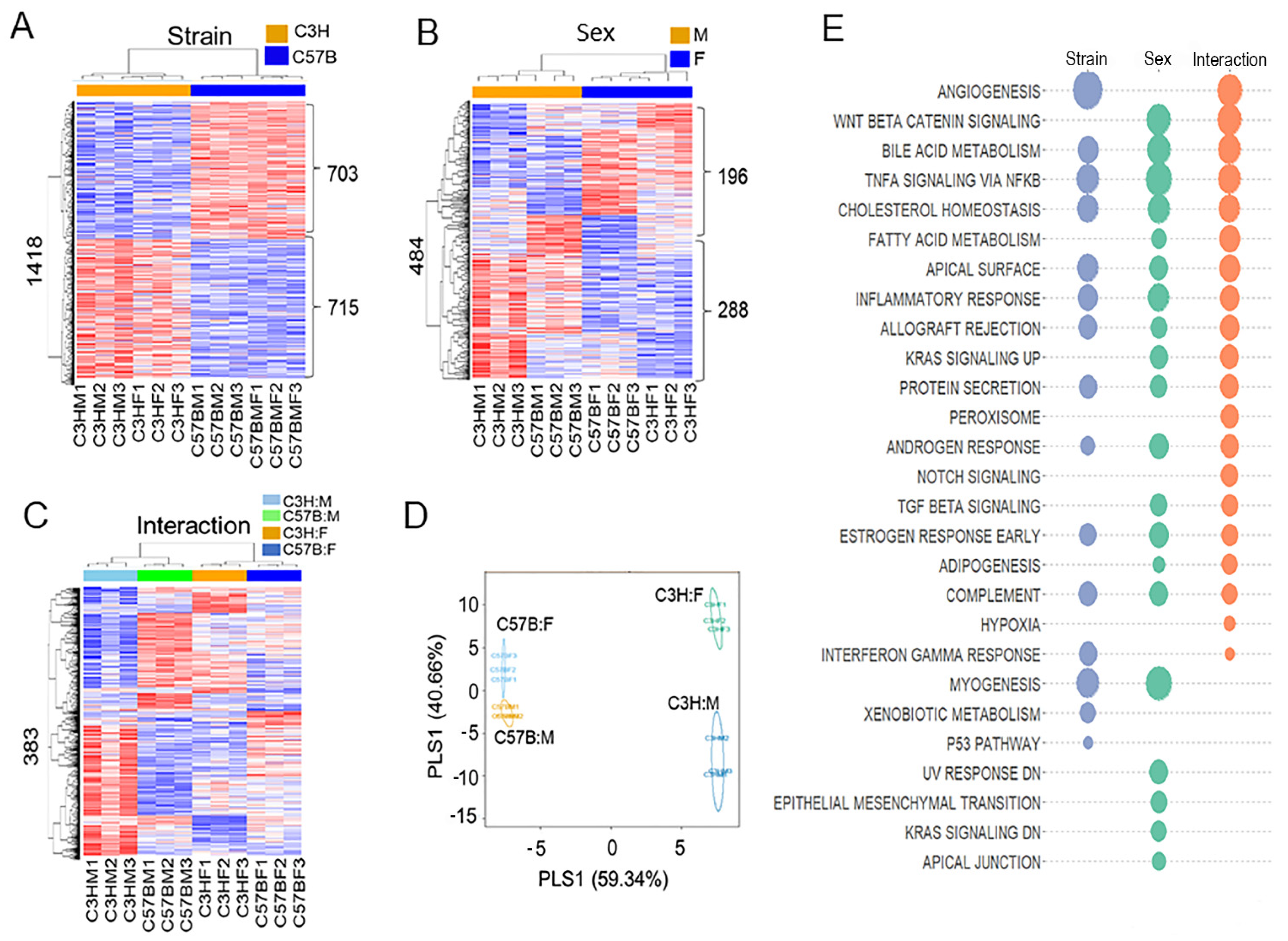
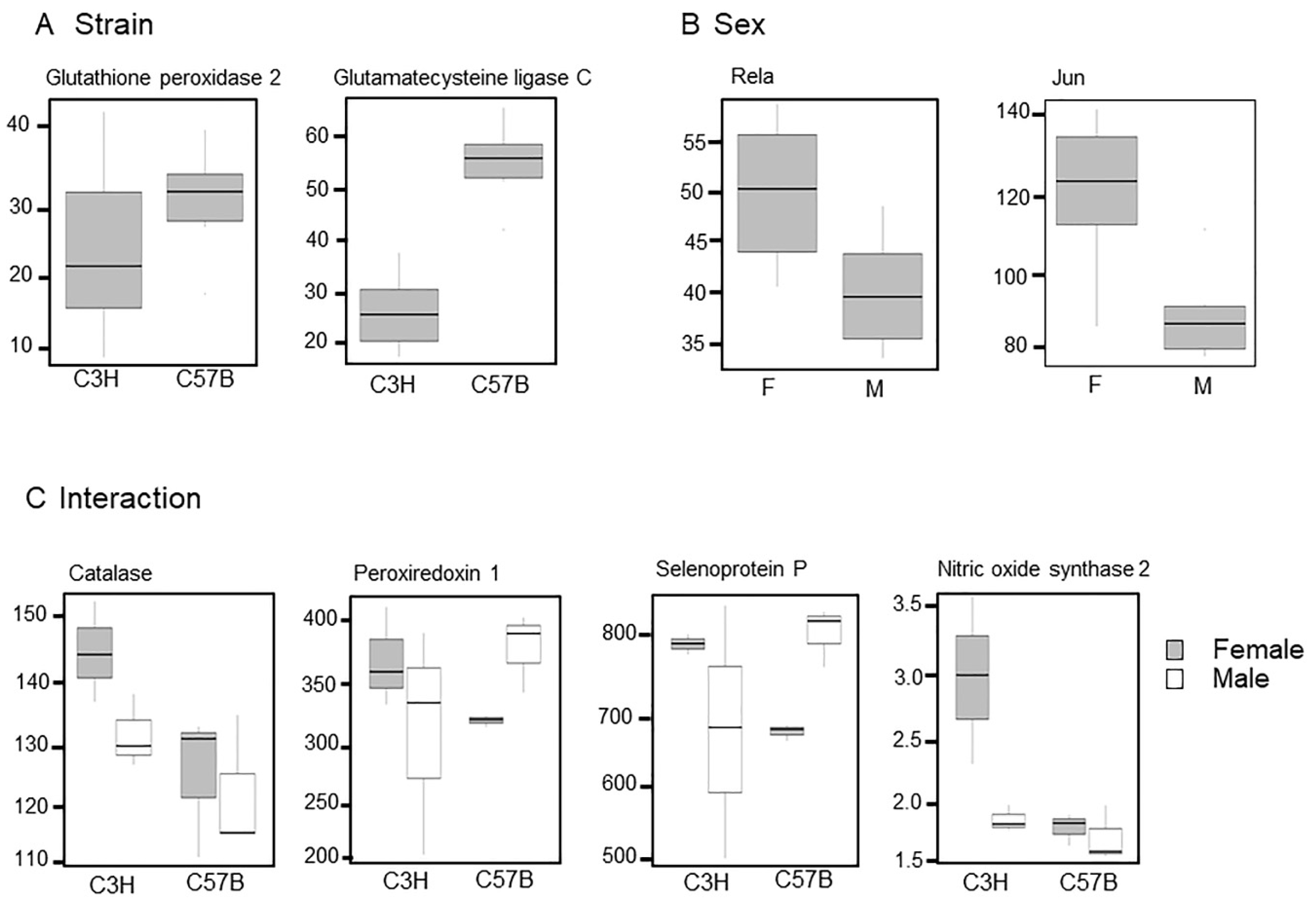
3.2. Differences in Transcriptome
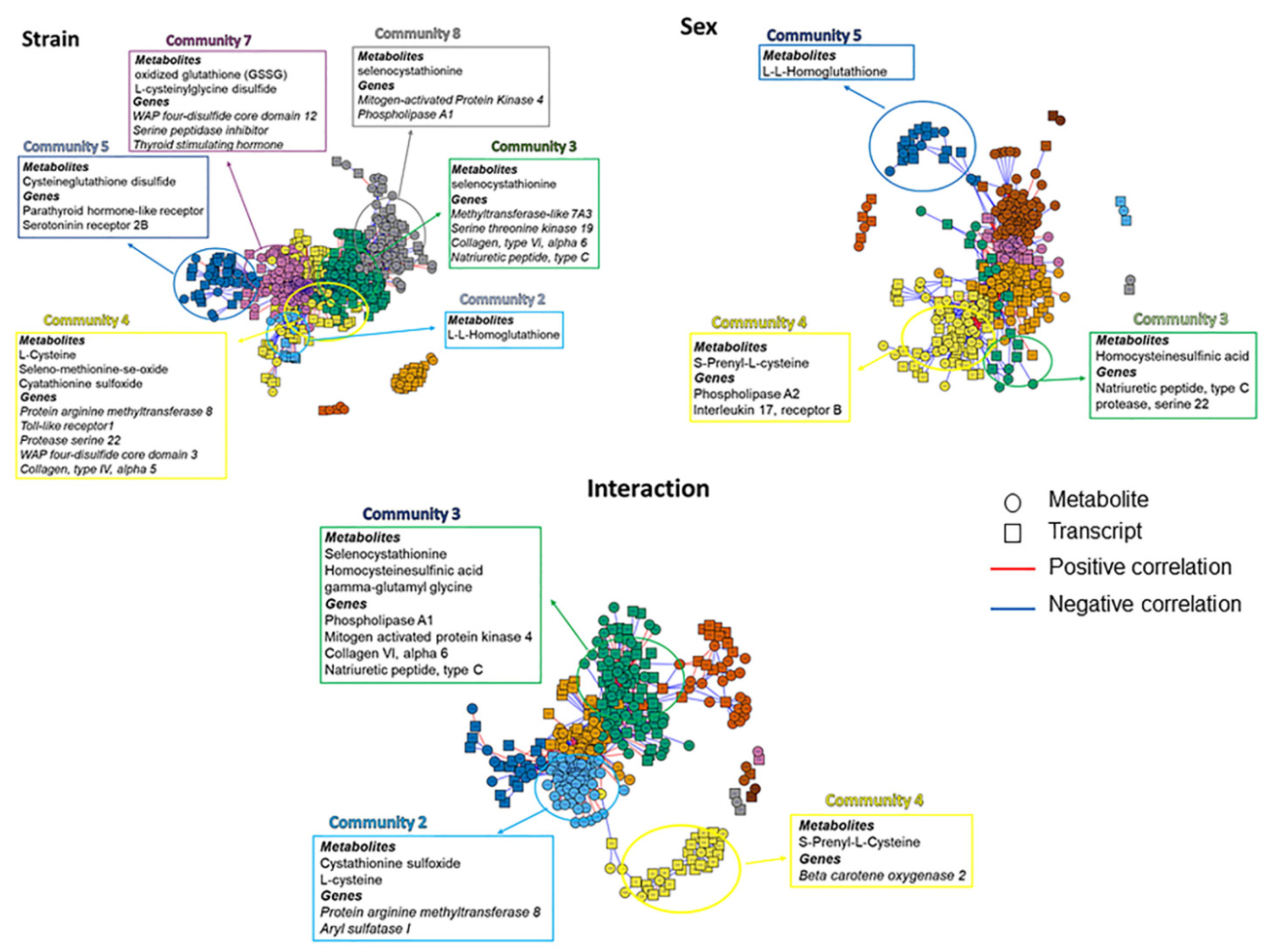
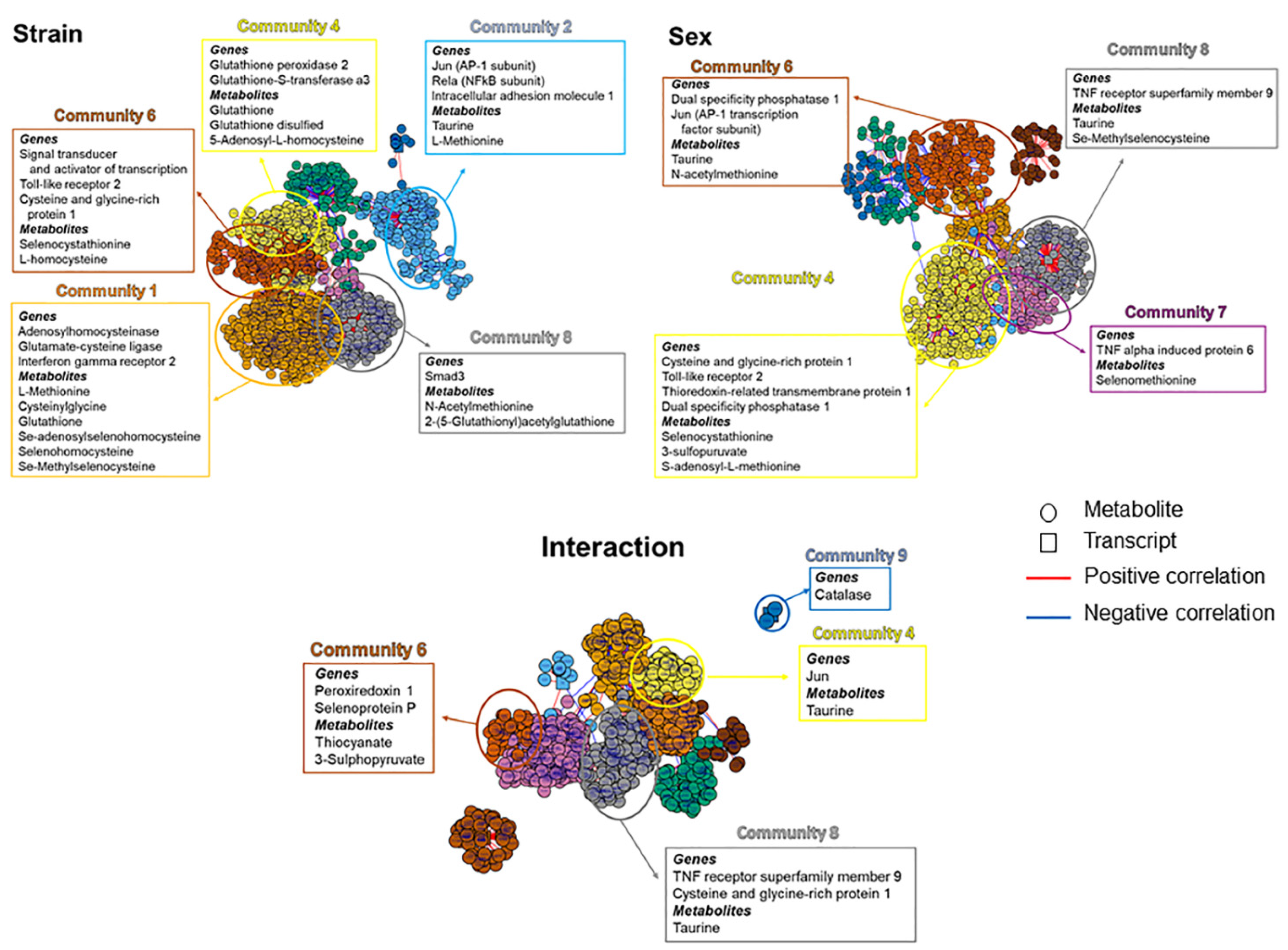
3.3. Integrated Transcriptome-Metabolome Wide Association Study
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coarfa, C.; Zhang, Y.; Maity, S.; Perera, D.N.; Jiang, W.; Wang, L.; Couroucli, X.; Moorthy, B.; Lingappan, K. Sexual dimorphism of the pulmonary transcriptome in neonatal hyperoxic lung injury: Identification of angiogenesis as a key pathway. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 313, L991–L1005. [Google Scholar] [CrossRef]
- Leary, S.; Das, P.; Ponnalagu, D.; Singh, H.; Bhandari, V. Genetic Strain and Sex Differences in a Hyperoxia-Induced Mouse Model of Varying Severity of Bronchopulmonary Dysplasia. Am. J. Pathol. 2019, 189, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.L.; Gladwell, W.; Verhein, K.C.; Cho, H.Y.; Wess, J.; Suzuki, O.; Wiltshire, T.; Kleeberger, S.R. Genome-wide association mapping of acute lung injury in neonatal inbred mice. FASEB J. 2014, 28, 2538–2550. [Google Scholar] [CrossRef] [PubMed]
- Tiono, J.; Surate Solaligue, D.E.; Mizikova, I.; Nardiello, C.; Vadasz, I.; Bottcher-Friebertshauser, E.; Ehrhardt, H.; Herold, S.; Seeger, W.; Morty, R.E. Mouse genetic background impacts susceptibility to hyperoxia-driven perturbations to lung maturation. Pediatr. Pulmonol. 2019, 54, 1060–1077. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, R.; Wall, S.B.; Dunigan, K.; Ren, C.; Jilling, T.; Rogers, L.K.; Tipple, T.E. Aurothioglucose does not improve alveolarization or elicit sustained Nrf2 activation in C57BL/6 models of bronchopulmonary dysplasia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018, 314, L736–L742. [Google Scholar] [CrossRef] [PubMed]
- Prows, D.R.; Gibbons, W.J., Jr.; Smith, J.J.; Pilipenko, V.; Martin, L.J. Age and Sex of Mice Markedly Affect Survival Times Associated with Hyperoxic Acute Lung Injury. PLoS ONE 2015, 10, e0130936. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Coarfa, C.; Dong, X.; Jiang, W.; Hayward-Piatkovskyi, B.; Gleghorn, J.P.; Lingappan, K. MicroRNA-30a as a candidate underlying sex-specific differences in neonatal hyperoxic lung injury: Implications for BPD. Am. J. Physiol. Lung Cell. Mol. Physiol. 2019, 316, L144–L156. [Google Scholar] [CrossRef]
- Lingappan, K.; Jiang, W.; Wang, L.; Moorthy, B. Sex-specific differences in neonatal hyperoxic lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 311, L481–L493. [Google Scholar] [CrossRef] [PubMed]
- Lingappan, K.; Jiang, W.; Wang, L.; Couroucli, X.I.; Barrios, R.; Moorthy, B. Sex-specific differences in hyperoxic lung injury in mice: Implications for acute and chronic lung disease in humans. Toxicol. Appl. Pharmacol. 2013, 272, 281–290. [Google Scholar] [CrossRef][Green Version]
- Ramirez, T.; Daneshian, M.; Kamp, H.; Bois, F.Y.; Clench, M.R.; Coen, M.; Donley, B.; Fischer, S.M.; Ekman, D.R.; Fabian, E.; et al. Metabolomics in toxicology and preclinical research. ALTEX 2013, 30, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wall, S.B.; Ren, C.; Velten, M.; Hill, C.L.; Locy, M.L.; Rogers, L.K.; Tipple, T.E. Thioredoxin Reductase Inhibition Attenuates Neonatal Hyperoxic Lung Injury and Enhances Nuclear Factor E2-Related Factor 2 Activation. Am. J. Respir. Cell Mol. Biol. 2016, 55, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.K.; Tipple, T.E.; Nelin, L.D.; Welty, S.E. Differential responses in the lungs of newborn mouse pups exposed to 85% or >95% oxygen. Pediatr. Res. 2009, 65, 33–38. [Google Scholar] [CrossRef]
- Tipple, T.E.; Welty, S.E.; Rogers, L.K.; Hansen, T.N.; Choi, Y.E.; Kehrer, J.P.; Smith, C.V. Thioredoxin-related mechanisms in hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 2007, 37, 405–413. [Google Scholar] [CrossRef]
- Britt, R.D., Jr.; Velten, M.; Locy, M.L.; Rogers, L.K.; Tipple, T.E. The thioredoxin reductase-1 inhibitor aurothioglucose attenuates lung injury and improves survival in a murine model of acute respiratory distress syndrome. Antioxid. Redox Signal. 2014, 20, 2681–2691. [Google Scholar] [CrossRef] [PubMed]
- Velten, M.; Britt, R.D., Jr.; Heyob, K.M.; Welty, S.E.; Eiberger, B.; Tipple, T.E.; Rogers, L.K. Prenatal inflammation exacerbates hyperoxia-induced functional and structural changes in adult mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 303, R279–R290. [Google Scholar] [CrossRef]
- Jendoubi, T. Approaches to Integrating Metabolomics and Multi-Omics Data: A Primer. Metabolites 2021, 11, 184. [Google Scholar] [CrossRef]
- Chandler, J.D.; Hu, X.; Ko, E.J.; Park, S.; Lee, Y.T.; Orr, M.; Fernandes, J.; Uppal, K.; Kang, S.M.; Jones, D.P.; et al. Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 311, R906–R916. [Google Scholar] [CrossRef]
- Go, Y.M.; Kim, C.W.; Walker, D.I.; Kang, D.W.; Kumar, S.; Orr, M.; Uppal, K.; Quyyumi, A.A.; Jo, H.; Jones, D.P. Disturbed flow induces systemic changes in metabolites in mouse plasma: A metabolomics study using ApoE−/− mice with partial carotid ligation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 308, R62–R72. [Google Scholar] [CrossRef]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef]
- Yu, T.; Park, Y.; Li, S.; Jones, D.P. Hybrid feature detection and information accumulation using high-resolution LC-MS metabolomics data. J. Proteome Res. 2013, 12, 1419–1427. [Google Scholar] [CrossRef]
- Uppal, K.; Soltow, Q.A.; Strobel, F.H.; Pittard, W.S.; Gernert, K.M.; Yu, T.; Jones, D.P. xMSanalyzer: Automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinform. 2013, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Royal Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Uppal, K.; Walker, D.I.; Jones, D.P. xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal. Chem. 2017, 89, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Baird, N.L.; Bowlin, J.L.; Cohrs, R.J.; Gilden, D.; Jones, K.L. Comparison of varicella-zoster virus RNA sequences in human neurons and fibroblasts. J. Virol. 2014, 88, 5877–5880. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010, 26, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Uppal, K.; Ma, C.; Go, Y.M.; Jones, D.P.; Wren, J. xMWAS: A data-driven integration and differential network analysis tool. Bioinformatics 2018, 34, 701–702. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Algesheimer, R.; Tessone, C.J. A Comparative Analysis of Community Detection Algorithms on Artificial Networks. Sci. Rep. 2016, 6, 30750. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Tabak, L.A. Policy: NIH plans to enhance reproducibility. Nature 2014, 505, 612–613. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, J.A.; Brown, L.L.; Murphy, S.J.; Grieder, F.; Silberberg, S.D. Accelerating Biomedical Discoveries through Rigor and Transparency. ILAR J. 2017, 58, 115–128. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, J.; Dunigan-Russell, K.; Zhong, H.; Lin, V.; Silverberg, M.; Moore, S.B.; Tran, V.; Jones, D.P.; Vitiello, P.F.; Rogers, L.K.; et al. Transcriptomic-Metabolomic Profiling in Mouse Lung Tissues Reveals Sex- and Strain-Based Differences. Metabolites 2022, 12, 932. https://doi.org/10.3390/metabo12100932
Fernandes J, Dunigan-Russell K, Zhong H, Lin V, Silverberg M, Moore SB, Tran V, Jones DP, Vitiello PF, Rogers LK, et al. Transcriptomic-Metabolomic Profiling in Mouse Lung Tissues Reveals Sex- and Strain-Based Differences. Metabolites. 2022; 12(10):932. https://doi.org/10.3390/metabo12100932
Chicago/Turabian StyleFernandes, Jolyn, Katelyn Dunigan-Russell, Hua Zhong, Vivian Lin, Mary Silverberg, Stephanie B. Moore, ViLinh Tran, Dean P. Jones, Peter F. Vitiello, Lynette K. Rogers, and et al. 2022. "Transcriptomic-Metabolomic Profiling in Mouse Lung Tissues Reveals Sex- and Strain-Based Differences" Metabolites 12, no. 10: 932. https://doi.org/10.3390/metabo12100932
APA StyleFernandes, J., Dunigan-Russell, K., Zhong, H., Lin, V., Silverberg, M., Moore, S. B., Tran, V., Jones, D. P., Vitiello, P. F., Rogers, L. K., & Tipple, T. E. (2022). Transcriptomic-Metabolomic Profiling in Mouse Lung Tissues Reveals Sex- and Strain-Based Differences. Metabolites, 12(10), 932. https://doi.org/10.3390/metabo12100932







