Scopoletin Induced Metabolomic Profile Disturbances in Zebrafish Embryos
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Zebrafish Maintenance and Husbandry
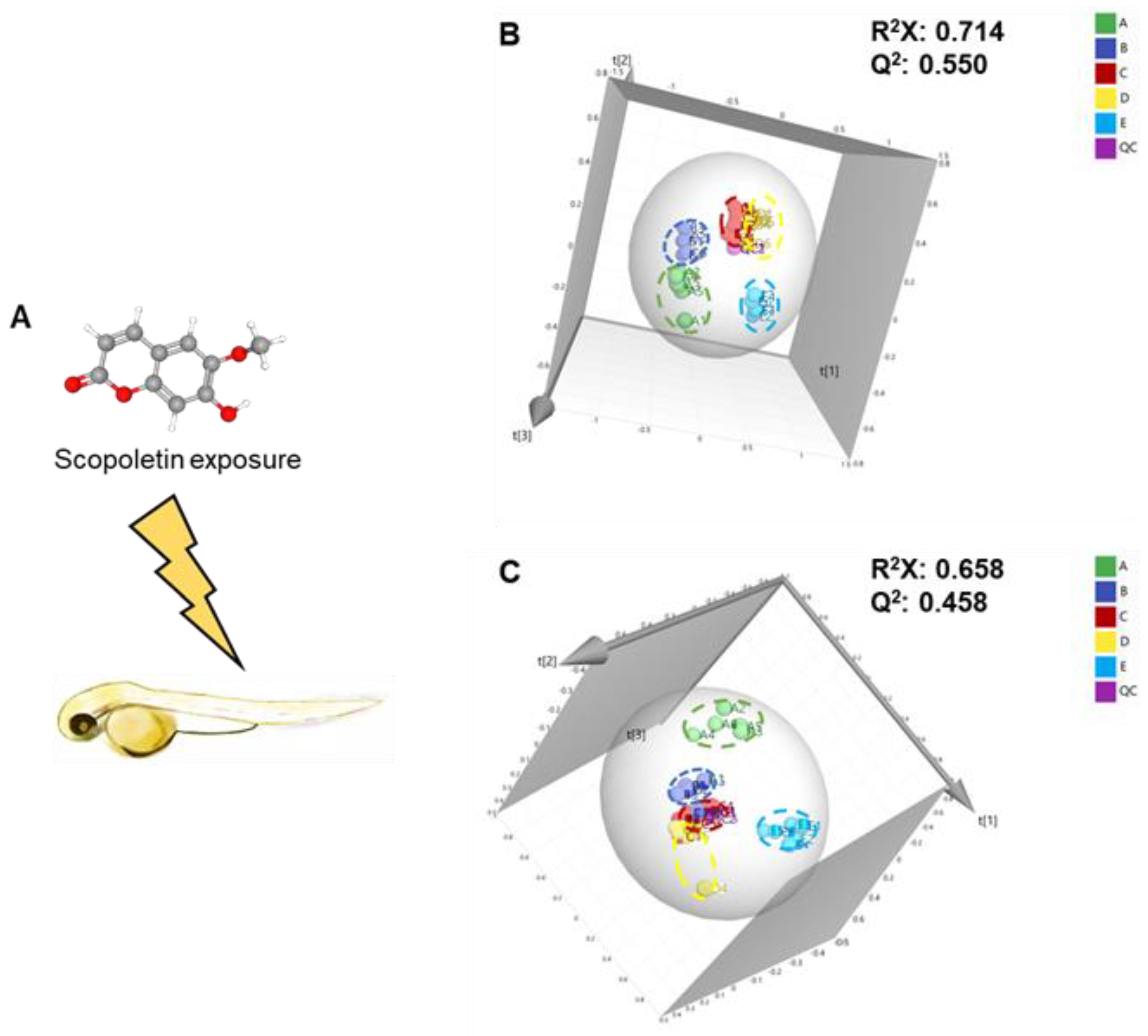
2.3. Scopoletin Exposure Setup
2.4. Metabolite Extraction
2.5. UHPLC-Q-Obitrap-HRMS Analysis
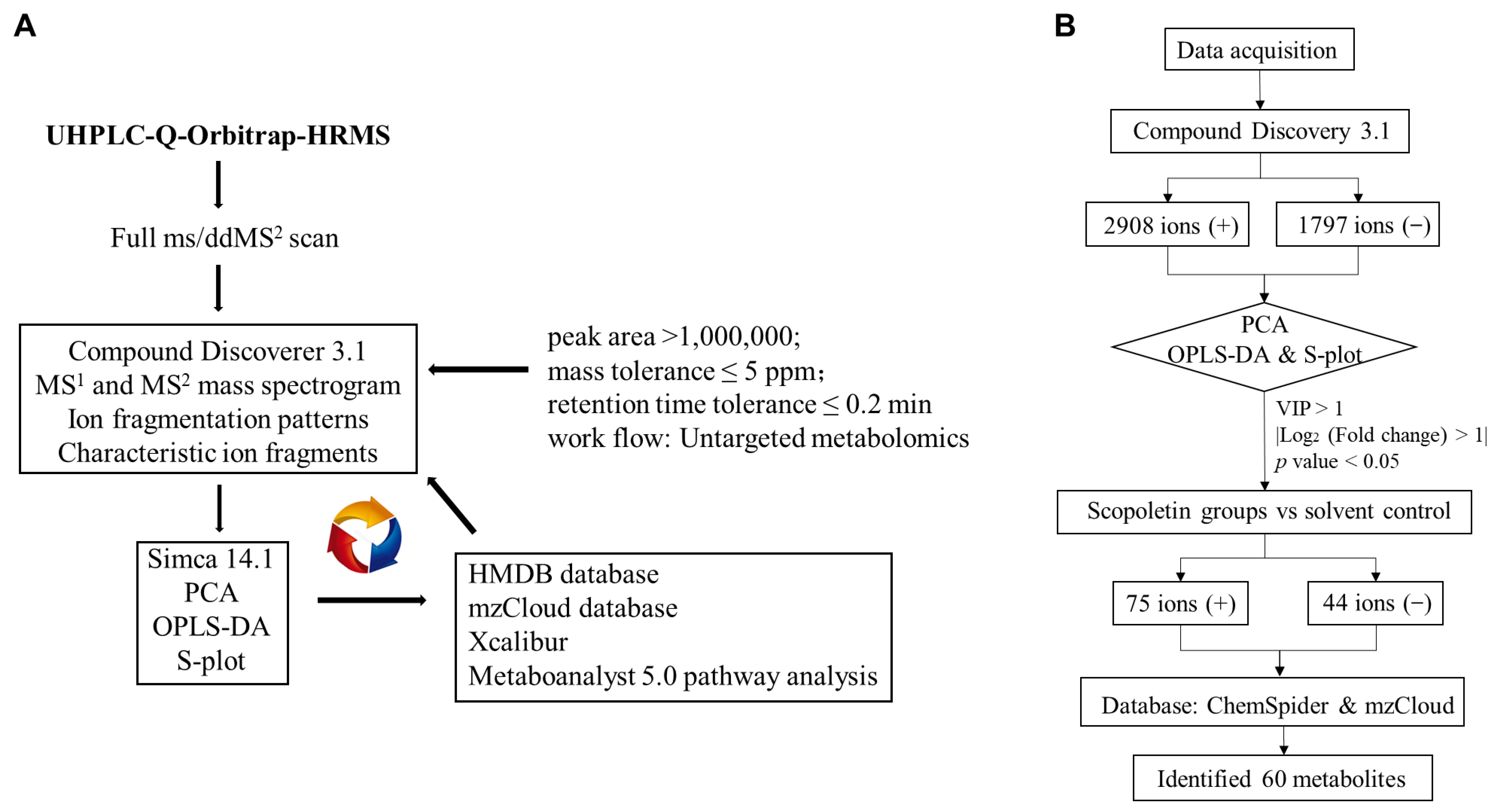
2.6. Data Processing and Pathway Analysis
2.7. Statistical Analysis
3. Results
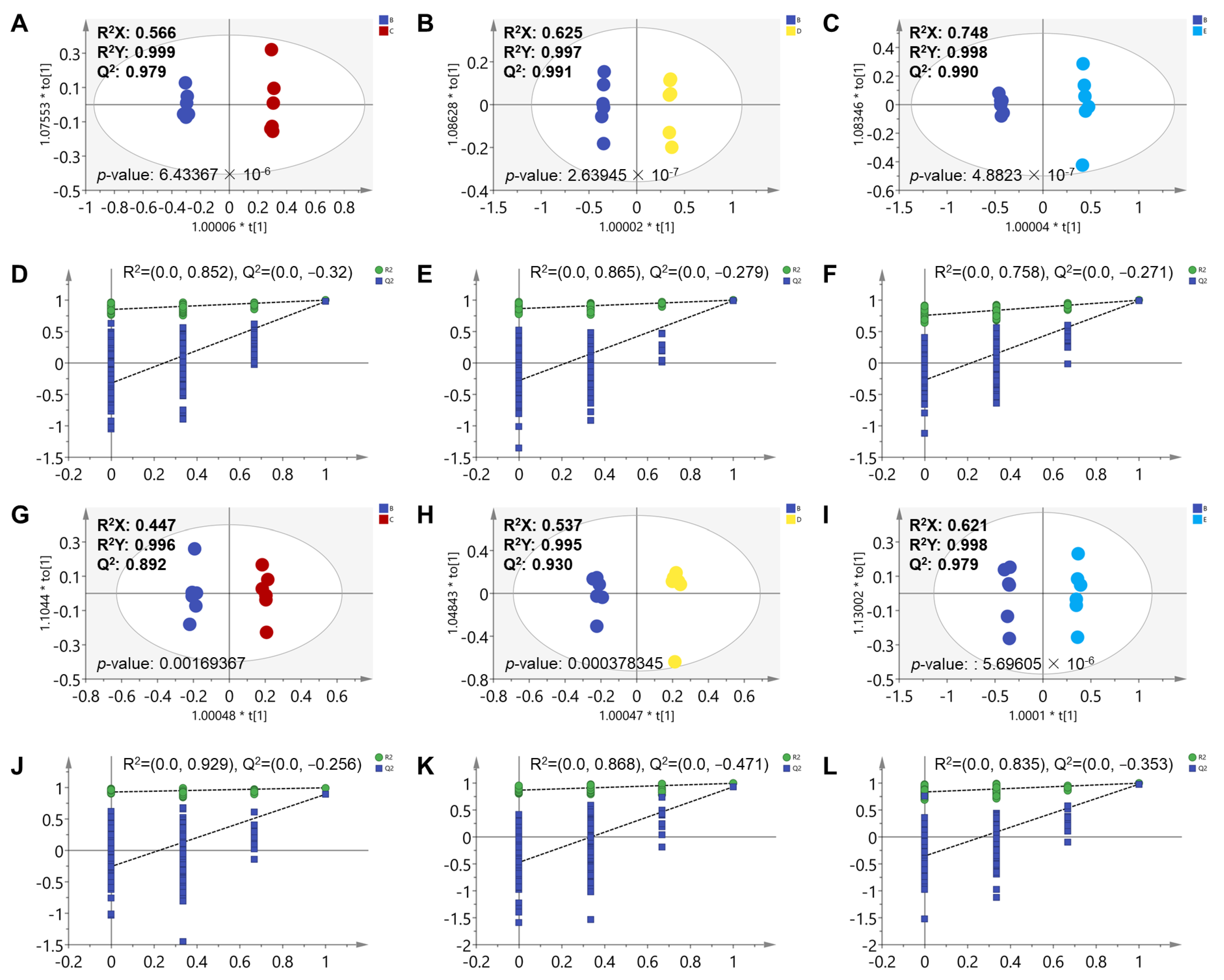
3.1. Metabolomic Alteration Induced by Scopoletin Exposure
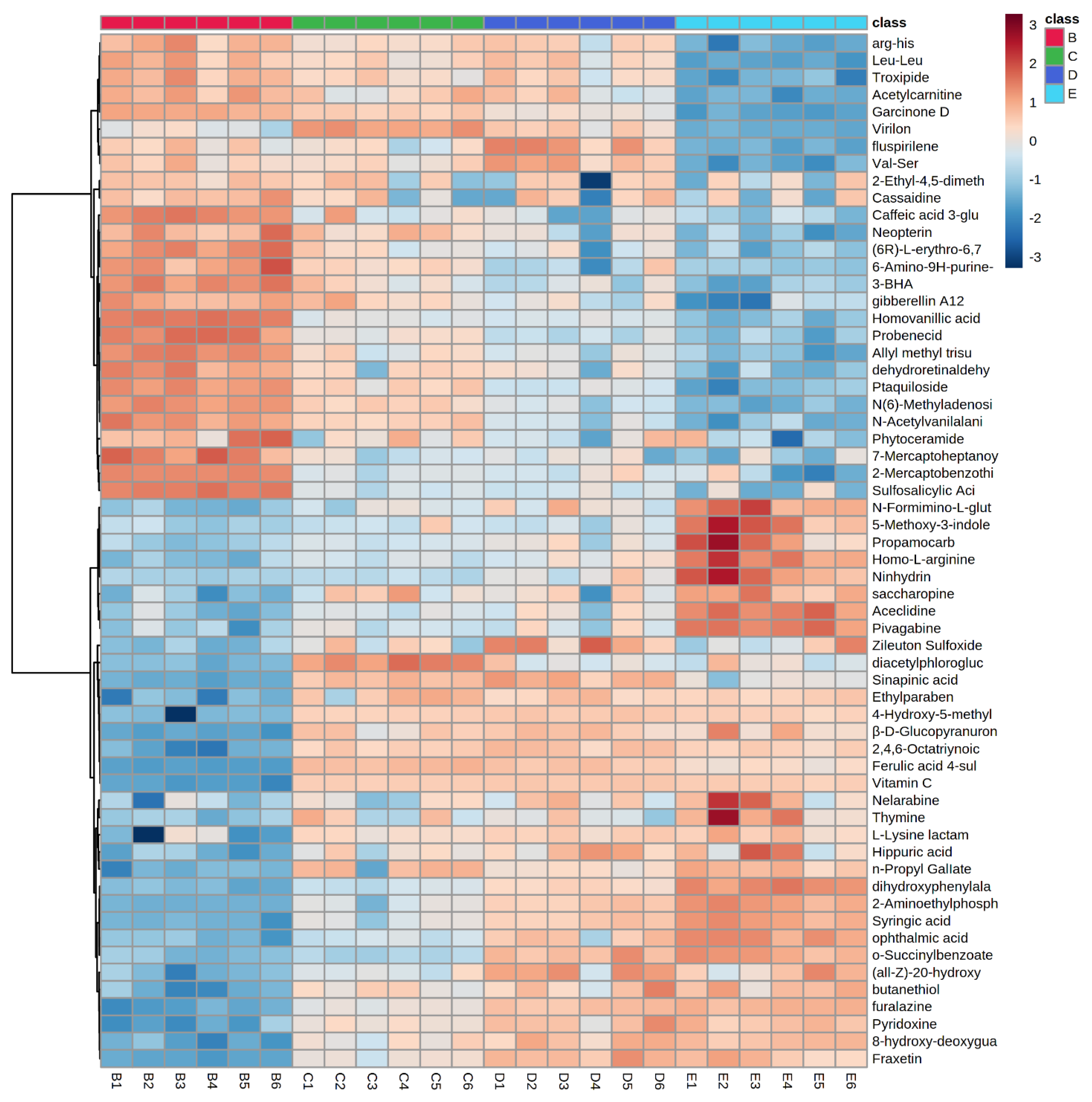
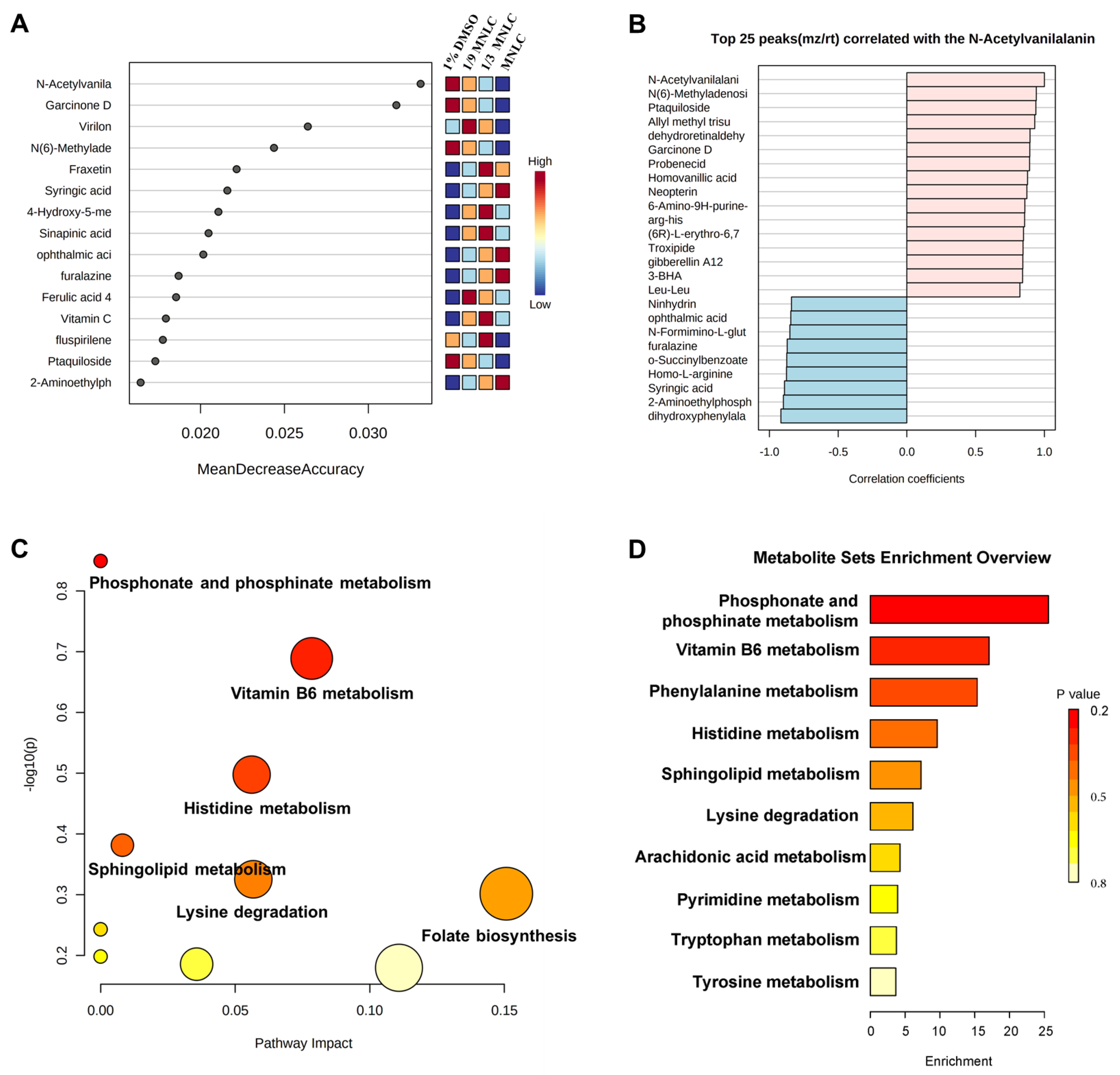
3.2. Analysis of Significantly Different Metabolites
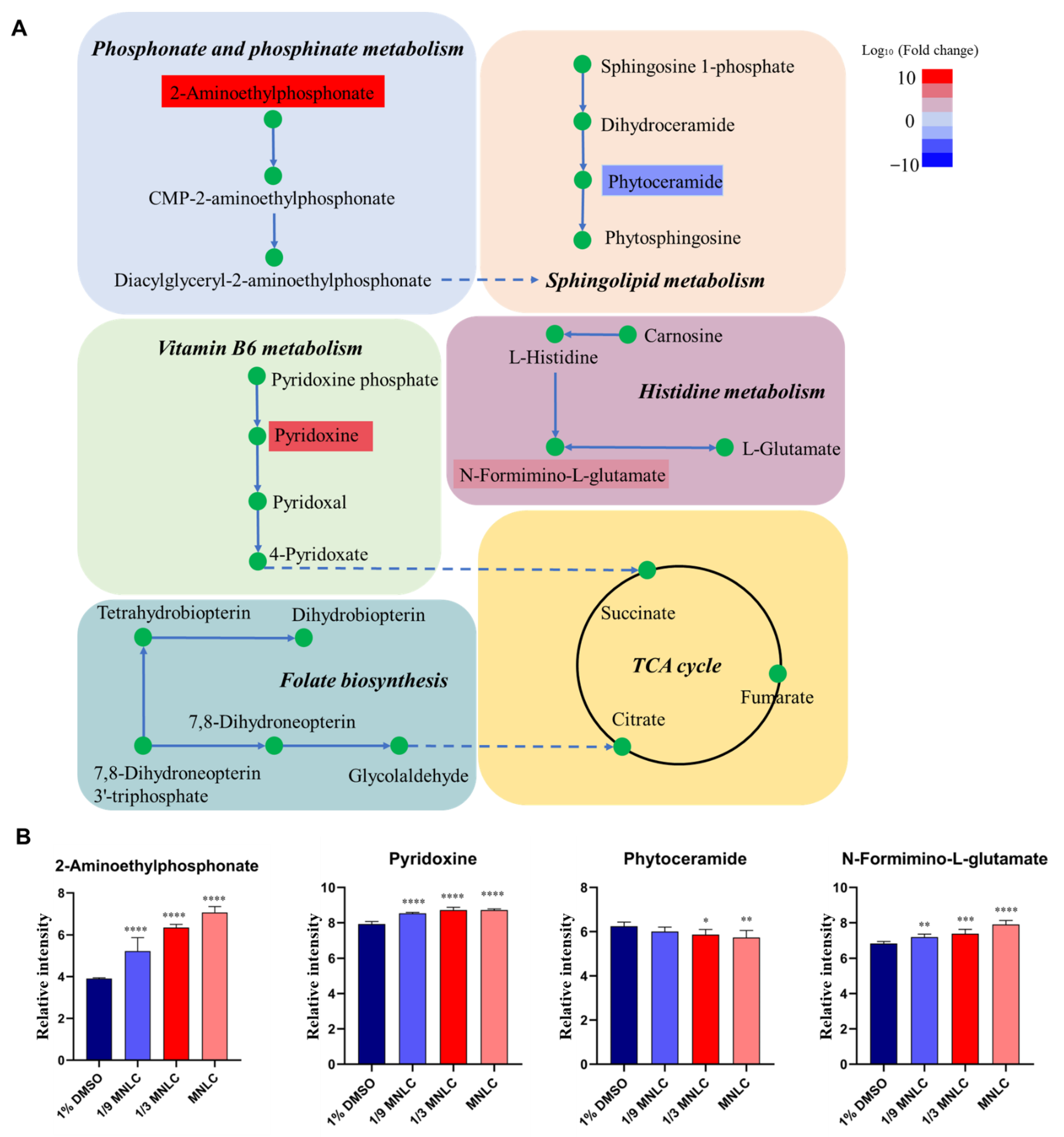
3.3. Metabolic Pathway and Function Analysis
3.4. Biological Networks of Differential Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seo, E.-J.; Saeed, M.; Law, B.; Wu, A.; Kadioglu, O.; Greten, H.; Efferth, T. Pharmacogenomics of Scopoletin in Tumor Cells. Molecules 2016, 21, 496. [Google Scholar] [CrossRef] [PubMed]
- Antika, L.D.; Tasfiyati, A.N.; Hikmat, H.; Septama, A.W. Scopoletin: A review of its source, biosynthesis, methods of extraction, and pharmacological activities. Z. Naturforsch. C J. Biosci. 2022, 77, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Zhang, L.; Fu, X.L.; Chen, K.; Qian, B.C. Effect of scopoletin on PC3 cell proliferation and apoptosis. Acta Pharmacol. Sin. 2001, 22, 929–933. [Google Scholar] [PubMed]
- Shi, Z.; Li, N.; Chen, C.; Wang, Y.; Lei, Z.; Chen, L.; Sun, J. Novel NO-releasing scopoletin derivatives induce cell death via mitochondrial apoptosis pathway and cell cycle arrest. Eur. J. Med. Chem. 2020, 200, 112386. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Dai, Y.; Gao, X.-H.; Lu, D.; Xia, Y.-F. Inhibition of vascular endothelial growth factor-induced angiogenesis by scopoletin through interrupting the autophosphorylation of VEGF receptor 2 and its downstream signaling pathways. Vascul. Pharmacol. 2010, 54, 18–28. [Google Scholar] [CrossRef]
- Graña, E.; Costas-Gil, A.; Longueira, S.; Celeiro, M.; Teijeira, M.; Reigosa, M.J.; Sánchez-Moreiras, A.M. Auxin-like effects of the natural coumarin scopoletin on Arabidopsis cell structure and morphology. J. Plant Physiol. 2017, 218, 45–55. [Google Scholar] [CrossRef]
- Gao, J.; Wang, F.; Cui, J.; Zhang, Q.; Wang, P.; Liu, D.; Zhou, Z. Assessment of toxicity and environmental behavior of chiral ethiprole and its metabolites using zebrafish model. J. Hazard. Mater. 2021, 414, 125492. [Google Scholar] [CrossRef]
- Wang, X.; Qin, Y.; Li, X.; Yan, B.; Martyniuk, C.J. Comprehensive Interrogation of Metabolic and Bioenergetic Responses of Early-Staged Zebrafish (Danio rerio) to a Commercial Copper Hydroxide Nanopesticide. Environ. Sci. Technol. 2021, 55, 13033–13044. [Google Scholar] [CrossRef]
- Weng, Y.; Huang, Z.; Wu, A.; Yu, Q.; Lu, H.; Lou, Z.; Lu, L.; Bao, Z.; Jin, Y. Embryonic toxicity of epoxiconazole exposure to the early life stage of zebrafish. Sci. Total Environ. 2021, 778, 146407. [Google Scholar] [CrossRef]
- Lee, H.; Sung, E.J.; Seo, S.; Min, E.K.; Lee, J.-Y.; Shim, I.; Kim, P.; Kim, T.-Y.; Lee, S.; Kim, K.-T. Integrated multi-omics analysis reveals the underlying molecular mechanism for developmental neurotoxicity of perfluorooctanesulfonic acid in zebrafish. Environ. Int. 2021, 157, 106802. [Google Scholar] [CrossRef]
- Merino, C.; Casado, M.; Piña, B.; Vinaixa, M.; Ramírez, N. Toxicity of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in early development: A wide-scope metabolomics assay in zebrafish embryos. J. Hazard. Mater. 2021, 429, 127746. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, L.; Huang, J.; Zhang, J.; Duan, J.; Guo, X.; Wu, S.; Sun, Z. Impact of PM2.5 Exposure on Plasma Metabolome in Healthy Adults during Air Pollution Waves: A Randomized, Crossover Trial. J. Hazard. Mater. 2022, 436, 129180. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wang, Y.-K.; Xiao, X.-R.; Zhao, Q.; Huang, J.-F.; Zhu, W.-F.; Li, F. Metabolic profiling of coumarins by the combination of UPLC-MS-based metabolomics and multiple mass defect filter. Xenobiotica 2020, 50, 1076–1089. [Google Scholar] [CrossRef]
- Chen, P.; Yang, J.; Wang, R.; Xiao, B.; Liu, Q.; Sun, B.; Wang, X.; Zhu, L. Graphene oxide enhanced the endocrine disrupting effects of bisphenol A in adult male zebrafish: Integrated deep learning and metabolomics studies. Sci. Total Environ. 2021, 809, 151103. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, X.-M.; Liu, H.-N.; Gonzalez, F.J.; Li, F. Metabolic map of osthole and its effect on lipids. Xenobiotica 2017, 48, 285–299. [Google Scholar] [CrossRef]
- Raldúa, D.; Casado, M.; Prats, E.; Faria, M.; Puig-Castellví, F.; Pérez, Y.; Alfonso, I.; Hsu, C.-Y.; Arick Ii, M.A.; Garcia-Reyero, N.; et al. Targeting redox metabolism: The perfect storm induced by acrylamide poisoning in the brain. Sci. Rep. 2020, 10, 312. [Google Scholar] [CrossRef]
- Lee, H.-K.; Kim, K.; Lee, J.; Lee, J.; Lee, J.; Kim, S.; Lee, S.-E.; Kim, J.-H. Targeted toxicometabolomics of endosulfan sulfate in adult zebrafish (Danio rerio) using GC-MS/MS in multiple reaction monitoring mode. J. Hazard. Mater. 2020, 389, 122056. [Google Scholar] [CrossRef]
- Basu, S.; Duren, W.; Evans, C.R.; Burant, C.F.; Michailidis, G.; Karnovsky, A. Sparse network modeling and metscape-based visualization methods for the analysis of large-scale metabolomics data. Bioinformatics 2017, 33, 1545–1553. [Google Scholar] [CrossRef]
- Martínez-Navarro, F.J.; Martínez-Morcillo, F.J.; López-Muñoz, A.; Pardo-Sánchez, I.; Martínez-Menchón, T.; Corbalán-Vélez, R.; Cayuela, M.L.; Pérez-Oliva, A.B.; García-Moreno, D.; Mulero, V. The vitamin B6-dependent enzymes PYGL and G6PD fuel NADPH oxidases to promote skin inflammation. Dev. Comp. Immunol. 2020, 108, 103666. [Google Scholar] [CrossRef]
- Cabrini, L.; Bergami, R.; Fiorentini, D.; Marchetti, M.; Landi, L.; Tolomelli, B. Vitamin B6 deficiency affects antioxidant defences in rat liver and heart. Biochem Mol Biol Int. 1998, 46, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-Y.; Flor, S.; Ruiz, P.; Dhakal, R.; Hu, X.; Teesch, L.M.; Ludewig, G.; Lehmler, H.-J. 3,3’-Dichlorobiphenyl Is Metabolized to a Complex Mixture of Oxidative Metabolites, Including Novel Methoxylated Metabolites, by HepG2 Cells. Environ. Sci. Technol. 2020, 54, 12345–12357. [Google Scholar] [CrossRef] [PubMed]
- Eifel, P.J.; Brown, D.M.; Lee, W.W.; Brown, J.M. Misonidazole neurotoxicity in mice decreased by administration with pyridoxine. Int. J. Radiat. Oncol. Biol. Phys. 1983, 9, 1513–1519. [Google Scholar] [CrossRef]
- Slováčková, J.; Slavík, J.; Kulich, P.; Večeřa, J.; Kováč, O.; Paculová, H.; Straková, N.; Fedr, R.; Silva, J.P.; Carvalho, F.; et al. Polychlorinated environmental toxicants affect sphingolipid metabolism during neurogenesis in vitro. Toxicology 2021, 463, 152986. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, Y. Plasma metabolic profiling analysis of neurotoxicity induced by oxaliplatin using metabonomics and multivariate data analysis. Toxicol. Res. 2018, 7, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Alaamery, M.; Albesher, N.; Aljawini, N.; Alsuwailm, M.; Massadeh, S.; Wheeler, M.A.; Chao, C.-C.; Quintana, F.J. Role of sphingolipid metabolism in neurodegeneration. J. Neurochem. 2020, 158, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulou, M.; Gordillo, R.; Koliaki, C.; Gancheva, S.; Jelenik, T.; De Filippo, E.; Herder, C.; Markgraf, D.; Jankowiak, F.; Esposito, I.; et al. Specific Hepatic Sphingolipids Relate to Insulin Resistance, Oxidative Stress, and Inflammation in Nonalcoholic Steatohepatitis. Diabetes Care 2018, 41, 1235–1243. [Google Scholar] [CrossRef]
- DeVos, L.; Chanson, A.; Liu, Z.; Ciappio, E.D.; Parnell, L.D.; Mason, J.B.; Tucker, K.L.; Crott, J.W. Associations between single nucleotide polymorphisms in folate uptake and metabolizing genes with blood folate, homocysteine, and DNA uracil concentrations. Am. J. Clin. Nutr. 2008, 88, 1149–1158. [Google Scholar] [CrossRef]
- Kang, S.-S.; Wong, P.W.K.; Norusis, M. Homocysteinemia due to folate deficiency. Metabolism 1987, 36, 458–462. [Google Scholar] [CrossRef]
- Yin, J.; Hong, X.; Ma, L.; Liu, R.; Bu, Y. Non-targeted metabolomic profiling of atrazine in Caenorhabditis elegans using UHPLC-QE Orbitrap/MS. Ecotoxicol. Environ. Saf. 2020, 206, 111170. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, W.; Chen, J.; Lin, Z.; Wang, N.; Wang, A.; Wang, B.; Wu, Y.; Xu, Z.; Wang, J. Scopoletin Induced Metabolomic Profile Disturbances in Zebrafish Embryos. Metabolites 2022, 12, 934. https://doi.org/10.3390/metabo12100934
Yao W, Chen J, Lin Z, Wang N, Wang A, Wang B, Wu Y, Xu Z, Wang J. Scopoletin Induced Metabolomic Profile Disturbances in Zebrafish Embryos. Metabolites. 2022; 12(10):934. https://doi.org/10.3390/metabo12100934
Chicago/Turabian StyleYao, Weixuan, Jingpei Chen, Zhanyu Lin, Nani Wang, Anli Wang, Binjie Wang, Yuanzhao Wu, Zhongshi Xu, and Jiye Wang. 2022. "Scopoletin Induced Metabolomic Profile Disturbances in Zebrafish Embryos" Metabolites 12, no. 10: 934. https://doi.org/10.3390/metabo12100934
APA StyleYao, W., Chen, J., Lin, Z., Wang, N., Wang, A., Wang, B., Wu, Y., Xu, Z., & Wang, J. (2022). Scopoletin Induced Metabolomic Profile Disturbances in Zebrafish Embryos. Metabolites, 12(10), 934. https://doi.org/10.3390/metabo12100934





