Protein O-GlcNAcylation in Metabolic Modulation of Skeletal Muscle: A Bright but Long Way to Go
Abstract
1. Introduction
2. O-GlcNAcylation Is an Essential Metabolic Modulator in Skeletal Muscle Physiology
2.1. O-GlcNAcylation Is an Extremely Sensitive Sensor for the Nutrition Availability in Skeletal Muscle via the HBP
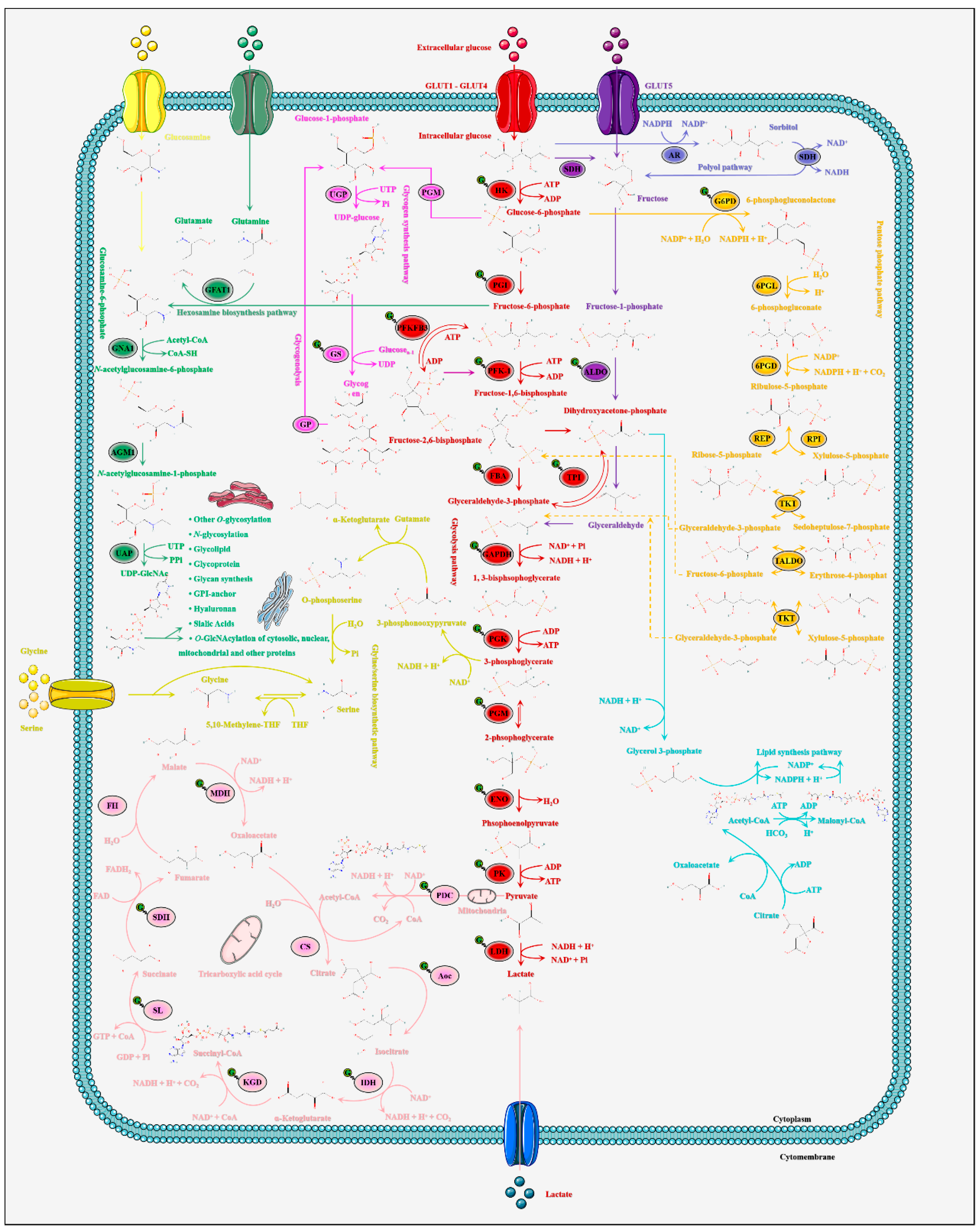
2.2. O-GlcNAcylation Is the Pivotal Modulator of Glucose Metabolic Homeostasis in Skeletal Muscle
2.3. O-GlcNAcylation Is the Precise Spatiotemporal Regulator of Insulin Signal Transduction in Skeletal Muscle
2.4. O-GlcNAcylation Is the Pivotal Maintainer of the TCA Cycle and Mitochondrial Homeostasis in Skeletal Muscle
3. The Fine Characteristics of O-GlcNAcylation, Skeletal Muscle Fibre Types and Exercises
4. O-GlcNAcylation, Autophagy and Skeletal Muscle Pathology
5. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Conibear, A.C. Deciphering protein post-translational modifications using chemical biology tools. Nat. Rev. Chem. 2020, 4, 674–695. [Google Scholar] [CrossRef]
- Altelaar, A.F.; Munoz, J.; Heck, A.J. Next-generation proteomics: Towards an integrative view of proteome dynamics. Nat. Rev. Genet. 2013, 14, 35–48. [Google Scholar] [CrossRef]
- Aebersold, R.; Agar, J.N.; Amster, I.J.; Baker, M.S.; Bertozzi, C.R.; Boja, E.S.; Costello, C.E.; Cravatt, B.F.; Fenselau, C.; Garcia, B.A.; et al. How many human proteoforms are there? Nat. Chem. Biol. 2018, 14, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef] [PubMed]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Saha, A.; Bello, D.; Fernández-Tejada, A. Advances in chemical probing of protein O-GlcNAc glycosylation: Structural role and molecular mechanisms. Chem. Soc. Rev. 2021, 50, 10451–10485. [Google Scholar] [CrossRef]
- Chatham, J.C.; Zhang, J.; Wende, A.R. Role of O-Linked N-Acetylglucosamine Protein Modification in Cellular (Patho)Physiology. Physiol. Rev. 2021, 101, 427–493. [Google Scholar] [CrossRef]
- Joiner, C.M.; Levine, Z.G.; Aonbangkhen, C.; Woo, C.M.; Walker, S. Aspartate Residues Far from the Active Site Drive O-GlcNAc Transferase Substrate Selection. J. Am. Chem. Soc. 2019, 141, 12974–12978. [Google Scholar] [CrossRef]
- Li, B.; Li, H.; Hu, C.W.; Jiang, J. Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase. Nat. Commun. 2017, 8, 666. [Google Scholar] [CrossRef]
- Yang, X.; Qian, K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452–465. [Google Scholar] [CrossRef]
- Kohler, J.J. Carb cutting works better with a partner. Nat. Struct. Mol. Biol. 2017, 24, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Pathak, S.; Alonso, J.; Schimpl, M.; Rafie, K.; Blair, D.E.; Borodkin, V.S.; Albarbarawi, O.; van Aalten, D.M.F. The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat. Struct. Mol. Biol. 2015, 22, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Levine, Z.G.; Walker, S. The Biochemistry of O-GlcNAc Transferase: Which Functions Make It Essential in Mammalian Cells? Annu. Rev. Biochem. 2016, 85, 631–657. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.B.; Singh, J.P.; Li, M.D.; Wu, J.; Yang, X. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol. Metab. TEM 2013, 24, 301–309. [Google Scholar] [CrossRef]
- Fehl, C.; Hanover, J.A. Tools, tactics and objectives to interrogate cellular roles of O-GlcNAc in disease. Nat. Chem. Biol. 2022, 18, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yao, R.Z.; Lian, S.; Liu, P.; Hu, Y.J.; Shi, H.Z.; Lv, H.M.; Yang, Y.Y.; Xu, B.; Li, S.Z. O-GlcNAcylation: The “stress and nutrition receptor” in cell stress response. Cell Stress Chaperones 2021, 26, 297–309. [Google Scholar] [CrossRef]
- Han, C.; Gu, Y.; Shan, H.; Mi, W.; Sun, J.; Shi, M.; Zhang, X.; Lu, X.; Han, F.; Gong, Q.; et al. O-GlcNAcylation of SIRT1 enhances its deacetylase activity and promotes cytoprotection under stress. Nat. Commun. 2017, 8, 1491. [Google Scholar] [CrossRef]
- Wulff-Fuentes, E.; Berendt, R.R.; Massman, L.; Danner, L.; Malard, F.; Vora, J.; Kahsay, R.; Olivier-Van Stichelen, S. The human O-GlcNAcome database and meta-analysis. Sci Data 2021, 8, 25. [Google Scholar] [CrossRef]
- Xu, S.; Tong, M.; Suttapitugsakul, S.; Wu, R. Spatial and temporal proteomics reveals the distinct distributions and dynamics of O-GlcNAcylated proteins. Cell Rep. 2022, 39, 110946. [Google Scholar] [CrossRef]
- Muha, V.; Authier, F.; Szoke-Kovacs, Z.; Johnson, S.; Gallagher, J.; McNeilly, A.; McCrimmon, R.J.; Teboul, L.; van Aalten, D.M.F. Loss of O-GlcNAcase catalytic activity leads to defects in mouse embryogenesis. J. Biol. Chem. 2021, 296, 100439. [Google Scholar] [CrossRef]
- Moulton, M.J.; Humphreys, G.B.; Kim, A.; Letsou, A. O-GlcNAcylation Dampens Dpp/BMP Signaling to Ensure Proper Drosophila Embryonic Development. Dev. Cell 2020, 53, 330–343.e333. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qian, C.; Cao, X. Post-Translational Modification Control of Innate Immunity. Immunity 2016, 45, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.H.; Weng, C.L.; Lin, K.I. O-GlcNAcylation and its role in the immune system. J. Biomed. Sci. 2020, 27, 57. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gong, W.; Wang, H.; Li, T.; Attri, K.S.; Lewis, R.E.; Kalil, A.C.; Bhinderwala, F.; Powers, R.; Yin, G.; et al. O-GlcNAc Transferase Suppresses Inflammation and Necroptosis by Targeting Receptor-Interacting Serine/Threonine-Protein Kinase 3. Immunity 2019, 50, 576–590.e576. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.B.; Ma, Y.; Torres, S.; Zhang, B.; Feriod, C.; Heck, R.M.; Qian, K.; Fu, M.; Li, X.; Nathanson, M.H.; et al. Calcium-dependent O-GlcNAc signaling drives liver autophagy in adaptation to starvation. Genes Dev. 2017, 31, 1655–1665. [Google Scholar] [CrossRef]
- Guo, B.; Liang, Q.; Li, L.; Hu, Z.; Wu, F.; Zhang, P.; Ma, Y.; Zhao, B.; Kovács, A.L.; Zhang, Z.; et al. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 2014, 16, 1215–1226. [Google Scholar] [CrossRef]
- Zhang, B.; Li, M.D.; Yin, R.; Liu, Y.; Yang, Y.; Mitchell-Richards, K.A.; Nam, J.H.; Li, R.; Wang, L.; Iwakiri, Y.; et al. O-GlcNAc transferase suppresses necroptosis and liver fibrosis. JCI Insight 2019, 4, e127709. [Google Scholar] [CrossRef]
- Hao, Y.; Fan, X.; Shi, Y.; Zhang, C.; Sun, D.E.; Qin, K.; Qin, W.; Zhou, W.; Chen, X. Next-generation unnatural monosaccharides reveal that ESRRB O-GlcNAcylation regulates pluripotency of mouse embryonic stem cells. Nat. Commun. 2019, 10, 4065. [Google Scholar] [CrossRef]
- Jang, H.; Kim, T.W.; Yoon, S.; Choi, S.Y.; Kang, T.W.; Kim, S.Y.; Kwon, Y.W.; Cho, E.J.; Youn, H.D. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell 2012, 11, 62–74. [Google Scholar] [CrossRef]
- Hart, G.W. Nutrient regulation of signaling and transcription. J. Biol. Chem. 2019, 294, 2211–2231. [Google Scholar] [CrossRef]
- Li, X.; Zhu, Q.; Shi, X.; Cheng, Y.; Li, X.; Xu, H.; Duan, X.; Hsieh-Wilson, L.C.; Chu, J.; Pelletier, J.; et al. O-GlcNAcylation of core components of the translation initiation machinery regulates protein synthesis. Proc. Natl. Acad. Sci. USA 2019, 116, 7857–7866. [Google Scholar] [CrossRef] [PubMed]
- Wells, L.; Vosseller, K.; Hart, G.W. Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science 2001, 291, 2376–2378. [Google Scholar] [CrossRef] [PubMed]
- Trapannone, R.; Mariappa, D.; Ferenbach, A.T.; van Aalten, D.M. Nucleocytoplasmic human O-GlcNAc transferase is sufficient for O-GlcNAcylation of mitochondrial proteins. Biochem. J. 2016, 473, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Forma, E.; Jóźwiak, P.; Bryś, M.; Krześlak, A. The potential role of O-GlcNAc modification in cancer epigenetics. Cell. Mol. Biol. Lett. 2014, 19, 438–460. [Google Scholar] [CrossRef] [PubMed]
- Na, H.J.; Akan, I.; Abramowitz, L.K.; Hanover, J.A. Nutrient-Driven O-GlcNAcylation Controls DNA Damage Repair Signaling and Stem/Progenitor Cell Homeostasis. Cell Rep. 2020, 31, 107632. [Google Scholar] [CrossRef]
- Nie, H.; Ju, H.; Fan, J.; Shi, X.; Cheng, Y.; Cang, X.; Zheng, Z.; Duan, X.; Yi, W. O-GlcNAcylation of PGK1 coordinates glycolysis and TCA cycle to promote tumor growth. Nat. Commun. 2020, 11, 36. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, X.; Wu, J.L.; Fu, L.; Liu, K.; Liu, D.; Chen, G.G.; Lai, P.B.; Wong, N.; Yu, J. O-GlcNAc transferase promotes fatty liver-associated liver cancer through inducing palmitic acid and activating endoplasmic reticulum stress. J. Hepatol. 2017, 67, 310–320. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Q.; Zhang, N.; Zhang, K.; Dou, T.; Cao, Y.; Liu, Y.; Li, K.; Hao, X.; Xie, X.; et al. Proteomic profiling and genome-wide mapping of O-GlcNAc chromatin-associated proteins reveal an O-GlcNAc-regulated genotoxic stress response. Nat. Commun. 2020, 11, 5898. [Google Scholar] [CrossRef]
- Schwein, P.A.; Woo, C.M. The O-GlcNAc Modification on Kinases. ACS Chem. Biol. 2020, 15, 602–617. [Google Scholar] [CrossRef]
- Hart, G.W.; Slawson, C.; Ramirez-Correa, G.; Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011, 80, 825–858. [Google Scholar] [CrossRef]
- Butkinaree, C.; Park, K.; Hart, G.W. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 2010, 1800, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.E.; Suh, P.G.; Kim, J.I. O-GlcNAcylation in health and neurodegenerative diseases. Exp. Mol. Med. 2021, 53, 1674–1682. [Google Scholar] [CrossRef]
- Aulak, K.S.; Barnes, J.W.; Tian, L.; Mellor, N.E.; Haque, M.M.; Willard, B.; Li, L.; Comhair, S.C.; Stuehr, D.J.; Dweik, R.A. Specific O-GlcNAc modification at Ser-615 modulates eNOS function. Redox Biol. 2020, 36, 101625. [Google Scholar] [CrossRef] [PubMed]
- Umapathi, P.; Mesubi, O.O.; Banerjee, P.S.; Abrol, N.; Wang, Q.; Luczak, E.D.; Wu, Y.; Granger, J.M.; Wei, A.C.; Gaido, O.E.R.; et al. Excessive O-GlcNAcylation Causes Heart Failure and Sudden Death. Circulation 2021, 143, 1687–1703. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.S.; Ma, J.; Hart, G.W. Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc. Natl. Acad. Sci. USA 2015, 112, 6050–6055. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fu, M.; Li, M.D.; Zhang, K.; Zhang, B.; Wang, S.; Liu, Y.; Ni, W.; Ong, Q.; Mi, J.; et al. O-GlcNAc transferase inhibits visceral fat lipolysis and promotes diet-induced obesity. Nat. Commun. 2020, 11, 181. [Google Scholar] [CrossRef]
- Petrus, P.; Lecoutre, S.; Dollet, L.; Wiel, C.; Sulen, A.; Gao, H.; Tavira, B.; Laurencikiene, J.; Rooyackers, O.; Checa, A.; et al. Glutamine Links Obesity to Inflammation in Human White Adipose Tissue. Cell Metab. 2020, 31, 375–390.e311. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, Y.J.; Fan, W.X.; Quan, X.; Xu, B.; Li, S.Z. O-GlcNAcylation: The Underestimated Emerging Regulators of Skeletal Muscle Physiology. Cells 2022, 11, 1789. [Google Scholar] [CrossRef]
- Ciraku, L.; Bacigalupa, Z.A.; Ju, J.; Moeller, R.A.; Le Minh, G.; Lee, R.H.; Smith, M.D.; Ferrer, C.M.; Trefely, S.; Izzo, L.T.; et al. O-GlcNAc transferase regulates glioblastoma acetate metabolism via regulation of CDK5-dependent ACSS2 phosphorylation. Oncogene 2022, 41, 2122–2136. [Google Scholar] [CrossRef]
- Lu, S.; Liao, Z.; Lu, X.; Katschinski, D.M.; Mercola, M.; Chen, J.; Brown, J.H.; Molkentin, J.D.; Bossuyt, J.; Bers, D.M. Hyperglycemia Acutely Increases Cytosolic Reactive Oxygen Species via O-linked GlcNAcylation and CaMKII Activation in Mouse Ventricular Myocytes. Circ. Res. 2020, 126, e80–e96. [Google Scholar] [CrossRef]
- Pedowitz, N.J.; Batt, A.R.; Darabedian, N.; Pratt, M.R. MYPT1 O-GlcNAc modification regulates sphingosine-1-phosphate mediated contraction. Nat. Chem. Biol. 2021, 17, 169–177. [Google Scholar] [CrossRef]
- Akimoto, Y.; Yan, K.; Miura, Y.; Tsumoto, H.; Toda, T.; Fukutomi, T.; Sugahara, D.; Kudo, A.; Arai, T.; Chiba, Y.; et al. O-GlcNAcylation and phosphorylation of β-actin Ser(199) in diabetic nephropathy. Am. J. Physiol. Ren. Physiol. 2019, 317, F1359–F1374. [Google Scholar] [CrossRef] [PubMed]
- Leung, M.C.; Hitchen, P.G.; Ward, D.G.; Messer, A.E.; Marston, S.B. Z-band alternatively spliced PDZ motif protein (ZASP) is the major O-linked β-N-acetylglucosamine-substituted protein in human heart myofibrils. J. Biol. Chem. 2013, 288, 4891–4898. [Google Scholar] [CrossRef] [PubMed]
- Hédou, J.; Bastide, B.; Page, A.; Michalski, J.C.; Morelle, W. Mapping of O-linked beta-N-acetylglucosamine modification sites in key contractile proteins of rat skeletal muscle. Proteomics 2009, 9, 2139–2148. [Google Scholar] [CrossRef]
- Basu, H.; Pekkurnaz, G.; Falk, J.; Wei, W.; Chin, M.; Steen, J.; Schwarz, T.L. FHL2 anchors mitochondria to actin and adapts mitochondrial dynamics to glucose supply. J. Cell Biol. 2021, 220, e201912077. [Google Scholar] [CrossRef]
- Ramirez-Correa, G.A.; Jin, W.; Wang, Z.; Zhong, X.; Gao, W.D.; Dias, W.B.; Vecoli, C.; Hart, G.W.; Murphy, A.M. O-linked GlcNAc modification of cardiac myofilament proteins: A novel regulator of myocardial contractile function. Circ. Res. 2008, 103, 1354–1358. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Liu, Y.; Yang, Y.; Lv, H.; Lian, S.; Xu, B.; Li, S. OGT upregulates myogenic IL-6 by mediating O-GlcNAcylation of p65 in mouse skeletal muscle under cold exposure. J. Cell. Physiol. 2022, 237, 1341–1352. [Google Scholar] [CrossRef]
- Huang, P.; Ho, S.R.; Wang, K.; Roessler, B.C.; Zhang, F.; Hu, Y.; Bowe, D.B.; Kudlow, J.E.; Paterson, A.J. Muscle-specific overexpression of NCOATGK, splice variant of O-GlcNAcase, induces skeletal muscle atrophy. Am. J. Physiol. Cell Physiol. 2011, 300, C456–C465. [Google Scholar] [CrossRef]
- Stevens, L.; Bastide, B.; Hedou, J.; Cieniewski-Bernard, C.; Montel, V.; Cochon, L.; Dupont, E.; Mounier, Y. Potential regulation of human muscle plasticity by MLC2 post-translational modifications during bed rest and countermeasures. Arch. Biochem. Biophys. 2013, 540, 125–132. [Google Scholar] [CrossRef]
- Myslicki, J.P.; Belke, D.D.; Shearer, J. Role of O-GlcNAcylation in nutritional sensing, insulin resistance and in mediating the benefits of exercise. Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2014, 39, 1205–1213. [Google Scholar] [CrossRef]
- Lipina, C.; Hundal, H.S. Lipid modulation of skeletal muscle mass and function. J. Cachexia Sarcopenia Muscle 2017, 8, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Shamim, B.; Hawley, J.A.; Camera, D.M. Protein Availability and Satellite Cell Dynamics in Skeletal Muscle. Sports Med. 2018, 48, 1329–1343. [Google Scholar] [CrossRef] [PubMed]
- McGee, S.L.; Hargreaves, M. AMPK-mediated regulation of transcription in skeletal muscle. Clin. Sci. 2010, 118, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Egan, B.; Zierath, J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013, 17, 162–184. [Google Scholar] [CrossRef] [PubMed]
- Mengeste, A.M.; Rustan, A.C.; Lund, J. Skeletal muscle energy metabolism in obesity. Obesity 2021, 29, 1582–1595. [Google Scholar] [CrossRef] [PubMed]
- Henselmans, M.; Bjørnsen, T.; Hedderman, R.; Vårvik, F.T. The Effect of Carbohydrate Intake on Strength and Resistance Training Performance: A Systematic Review. Nutrients 2022, 14, 856. [Google Scholar] [CrossRef] [PubMed]
- Yang, J. Enhanced skeletal muscle for effective glucose homeostasis. Prog. Mol. Biol. Transl. Sci. 2014, 121, 133–163. [Google Scholar] [CrossRef]
- Giudice, J.; Taylor, J.M. Muscle as a paracrine and endocrine organ. Curr. Opin. Pharmacol. 2017, 34, 49–55. [Google Scholar] [CrossRef]
- Pedersen, B.K.; Febbraio, M.A. Muscles, exercise and obesity: Skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 2012, 8, 457–465. [Google Scholar] [CrossRef]
- Singh, J.P.; Qian, K.; Lee, J.S.; Zhou, J.; Han, X.; Zhang, B.; Ong, Q.; Ni, W.; Jiang, M.; Ruan, H.B.; et al. O-GlcNAcase targets pyruvate kinase M2 to regulate tumor growth. Oncogene 2020, 39, 560–573. [Google Scholar] [CrossRef]
- Yi, W.; Clark, P.M.; Mason, D.E.; Keenan, M.C.; Hill, C.; Goddard, W.A., 3rd; Peters, E.C.; Driggers, E.M.; Hsieh-Wilson, L.C. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012, 337, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Munk, A.; Nielsen, T.S.; Daughtry, M.R.; Larsson, L.; Li, S.; Høyer, K.F.; Geisler, H.W.; Sulek, K.; Kjøbsted, R.; et al. Skeletal muscle O-GlcNAc transferase is important for muscle energy homeostasis and whole-body insulin sensitivity. Mol. Metab. 2018, 11, 160–177. [Google Scholar] [CrossRef] [PubMed]
- Durning, S.P.; Flanagan-Steet, H.; Prasad, N.; Wells, L. O-Linked β-N-acetylglucosamine (O-GlcNAc) Acts as a Glucose Sensor to Epigenetically Regulate the Insulin Gene in Pancreatic Beta Cells. J. Biol. Chem. 2016, 291, 2107–2118. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, B.; Hu, Y.; Liu, P.; Lian, S.; Lv, H.; Yang, Y.; Ji, H.; Yang, H.; Liu, J.; et al. O-GlcNAc/Akt pathway regulates glucose metabolism and reduces apoptosis in liver of piglets with acute cold stress. Cryobiology 2021, 100, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Pandey, A.; Hart, G.W. Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol. Cell. Proteom. MCP 2007, 6, 1365–1379. [Google Scholar] [CrossRef]
- Hanover, J.A.; Krause, M.W.; Love, D.C. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta 2010, 1800, 80–95. [Google Scholar] [CrossRef]
- Filhoulaud, G.; Guillemain, G.; Scharfmann, R. The hexosamine biosynthesis pathway is essential for pancreatic beta cell development. J. Biol. Chem. 2009, 284, 24583–24594. [Google Scholar] [CrossRef]
- Richter, E.A.; Hargreaves, M. Exercise, GLUT4, and skeletal muscle glucose uptake. Physiol. Rev. 2013, 93, 993–1017. [Google Scholar] [CrossRef]
- Love, D.C.; Hanover, J.A. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci. STKE Signal Transduct. Knowl. Environ. 2005, 2005, re13. [Google Scholar] [CrossRef]
- Ruegenberg, S.; Horn, M.; Pichlo, C.; Allmeroth, K.; Baumann, U.; Denzel, M.S. Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis. Nat. Commun. 2020, 11, 687. [Google Scholar] [CrossRef]
- Campbell, S.; Mesaros, C.; Izzo, L.; Affronti, H.; Noji, M.; Schaffer, B.E.; Tsang, T.; Sun, K.; Trefely, S.; Kruijning, S.; et al. Glutamine deprivation triggers NAGK-dependent hexosamine salvage. eLife 2021, 10, e62644. [Google Scholar] [CrossRef]
- Horn, M.; Denzel, S.I.; Srinivasan, B.; Allmeroth, K.; Schiffer, I.; Karthikaisamy, V.; Miethe, S.; Breuer, P.; Antebi, A.; Denzel, M.S. Hexosamine Pathway Activation Improves Protein Homeostasis through the Integrated Stress Response. iScience 2020, 23, 100887. [Google Scholar] [CrossRef] [PubMed]
- Lam, C.; Low, J.Y.; Tran, P.T.; Wang, H. The hexosamine biosynthetic pathway and cancer: Current knowledge and future therapeutic strategies. Cancer Lett. 2021, 503, 11–18. [Google Scholar] [CrossRef]
- Zachara, N.E.; Hart, G.W. O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 2004, 1673, 13–28. [Google Scholar] [CrossRef]
- Zhao, L.; Shah, J.A.; Cai, Y.; Jin, J. ‘O-GlcNAc Code’ Mediated Biological Functions of Downstream Proteins. Molecules 2018, 23, 1967. [Google Scholar] [CrossRef]
- Chatham, J.C.; Nöt, L.G.; Fülöp, N.; Marchase, R.B. Hexosamine biosynthesis and protein O-glycosylation: The first line of defense against stress, ischemia, and trauma. Shock 2008, 29, 431–440. [Google Scholar] [CrossRef]
- Groves, J.A.; Lee, A.; Yildirir, G.; Zachara, N.E. Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis. Cell Stress Chaperones 2013, 18, 535–558. [Google Scholar] [CrossRef]
- Lambert, M.; Bastide, B.; Cieniewski-Bernard, C. Involvement of O-GlcNAcylation in the Skeletal Muscle Physiology and Physiopathology: Focus on Muscle Metabolism. Front. Endocrinol. 2018, 9, 578. [Google Scholar] [CrossRef]
- Wang, X.; Feng, Z.; Wang, X.; Yang, L.; Han, S.; Cao, K.; Xu, J.; Zhao, L.; Zhang, Y.; Liu, J. O-GlcNAcase deficiency suppresses skeletal myogenesis and insulin sensitivity in mice through the modulation of mitochondrial homeostasis. Diabetologia 2016, 59, 1287–1296. [Google Scholar] [CrossRef]
- Guo, X.; Sun, W.; Luo, G.; Wu, L.; Xu, G.; Hou, D.; Hou, Y.; Guo, X.; Mu, X.; Qin, L.; et al. Panax notoginseng saponins alleviate skeletal muscle insulin resistance by regulating the IRS1-PI3K-AKT signaling pathway and GLUT4 expression. FEBS Open Bio 2019, 9, 1008–1019. [Google Scholar] [CrossRef]
- Hawkins, M.; Angelov, I.; Liu, R.; Barzilai, N.; Rossetti, L. The tissue concentration of UDP-N-acetylglucosamine modulates the stimulatory effect of insulin on skeletal muscle glucose uptake. J. Biol. Chem. 1997, 272, 4889–4895. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, C.M.; Lynch, T.P.; Sodi, V.L.; Falcone, J.N.; Schwab, L.P.; Peacock, D.L.; Vocadlo, D.J.; Seagroves, T.N.; Reginato, M.J. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol. Cell 2014, 54, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Baldini, S.F.; Steenackers, A.; Stichelen, S.O.-V.; Mir, A.M.; Mortuaire, M.; Lefebvre, T.; Guinez, C. Glucokinase expression is regulated by glucose through O-GlcNAc glycosylation. Biochem. Biophys. Res. Commun. 2016, 478, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Duan, X.; Mao, W.; Li, X.; Li, Z.; Li, Q.; Zheng, Z.; Xu, H.; Chen, M.; Wang, P.G.; et al. O-GlcNAcylation of G6PD promotes the pentose phosphate pathway and tumor growth. Nat. Commun. 2015, 6, 8468. [Google Scholar] [CrossRef]
- Lei, Y.; Chen, T.; Li, Y.; Shang, M.; Zhang, Y.; Jin, Y.; Yu, Q.; Guo, F.; Wang, T. O-GlcNAcylation of PFKFB3 is required for tumor cell proliferation under hypoxia. Oncogenesis 2020, 9, 21. [Google Scholar] [CrossRef]
- Lim, K.; Yoon, B.H.; Ha, C.H. O-Linked N-acetylglucosaminylation of Sp1 interferes with Sp1 activation of glycolytic genes. Biochem. Biophys. Res. Commun. 2015, 468, 349–353. [Google Scholar] [CrossRef]
- Bacigalupa, Z.A.; Bhadiadra, C.H.; Reginato, M.J. O-GlcNAcylation: Key regulator of glycolytic pathways. J. Bioenerg. Biomembr. 2018, 50, 189–198. [Google Scholar] [CrossRef]
- Nachit, M.; Leclercq, I.A. Emerging awareness on the importance of skeletal muscle in liver diseases: Time to dig deeper into mechanisms! Clin. Sci. 2019, 133, 465–481. [Google Scholar] [CrossRef]
- Kazemi, Z.; Chang, H.; Haserodt, S.; McKen, C.; Zachara, N.E. O-linked beta-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3beta-dependent manner. J. Biol. Chem. 2010, 285, 39096–39107. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.J.; Lund, K.C.; Taylor, R.P.; McClain, D.A. Insulin resistance of glycogen synthase mediated by o-linked N-acetylglucosamine. J. Biol. Chem. 2003, 278, 10022–10027. [Google Scholar] [CrossRef]
- Zumbaugh, M.D.; Yen, C.N.; Bodmer, J.S.; Shi, H.; Gerrard, D.E. Skeletal Muscle O-GlcNAc Transferase Action on Global Metabolism is Partially Mediated Through Interleukin-15. Front. Physiol. 2021, 12, 682052. [Google Scholar] [CrossRef] [PubMed]
- Laczy, B.; Fülöp, N.; Onay-Besikci, A.; Des Rosiers, C.; Chatham, J.C. Acute regulation of cardiac metabolism by the hexosamine biosynthesis pathway and protein O-GlcNAcylation. PLoS ONE 2011, 6, e18417. [Google Scholar] [CrossRef] [PubMed]
- Guinez, C.; Filhoulaud, G.; Rayah-Benhamed, F.; Marmier, S.; Dubuquoy, C.; Dentin, R.; Moldes, M.; Burnol, A.F.; Yang, X.; Lefebvre, T.; et al. O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes 2011, 60, 1399–1413. [Google Scholar] [CrossRef] [PubMed]
- Sodi, V.L.; Bacigalupa, Z.A.; Ferrer, C.M.; Lee, J.V.; Gocal, W.A.; Mukhopadhyay, D.; Wellen, K.E.; Ivan, M.; Reginato, M.J. Nutrient sensor O-GlcNAc transferase controls cancer lipid metabolism via SREBP-1 regulation. Oncogene 2018, 37, 924–934. [Google Scholar] [CrossRef]
- Benhamed, F.; Filhoulaud, G.; Caron, S.; Lefebvre, P.; Staels, B.; Postic, C. O-GlcNAcylation Links ChREBP and FXR to Glucose-Sensing. Front. Endocrinol. 2014, 5, 230. [Google Scholar] [CrossRef]
- Anthonisen, E.H.; Berven, L.; Holm, S.; Nygård, M.; Nebb, H.I.; Grønning-Wang, L.M. Nuclear receptor liver X receptor is O-GlcNAc-modified in response to glucose. J. Biol. Chem. 2010, 285, 1607–1615. [Google Scholar] [CrossRef]
- Wong, Y.K.; Wang, J.; Lim, T.K.; Lin, Q.; Yap, C.T.; Shen, H.M. O-GlcNAcylation promotes fatty acid synthase activity under nutritional stress as a pro-survival mechanism in cancer cells. Proteomics 2022, 22, e2100175. [Google Scholar] [CrossRef]
- Deshmukh, A.S. Insulin-stimulated glucose uptake in healthy and insulin-resistant skeletal muscle. Horm. Mol. Biol. Clin. Investig. 2016, 26, 13–24. [Google Scholar] [CrossRef] [PubMed]
- McLarty, J.L.; Marsh, S.A.; Chatham, J.C. Post-translational protein modification by O-linked N-acetyl-glucosamine: Its role in mediating the adverse effects of diabetes on the heart. Life Sci 2013, 92, 621–627. [Google Scholar] [CrossRef]
- Wells, L.; Vosseller, K.; Hart, G.W. A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell. Mol. Life Sci. CMLS 2003, 60, 222–228. [Google Scholar] [CrossRef]
- Virkamäki, A.; Daniels, M.C.; Hämäläinen, S.; Utriainen, T.; McClain, D.; Yki-Järvinen, H. Activation of the hexosamine pathway by glucosamine in vivo induces insulin resistance in multiple insulin sensitive tissues. Endocrinology 1997, 138, 2501–2507. [Google Scholar] [CrossRef]
- Arias, E.B.; Kim, J.; Cartee, G.D. Prolonged incubation in PUGNAc results in increased protein O-Linked glycosylation and insulin resistance in rat skeletal muscle. Diabetes 2004, 53, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Walgren, J.L.; Vincent, T.S.; Schey, K.L.; Buse, M.G. High glucose and insulin promote O-GlcNAc modification of proteins, including alpha-tubulin. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E424–E434. [Google Scholar] [CrossRef] [PubMed]
- Yki-Järvinen, H.; Virkamäki, A.; Daniels, M.C.; McClain, D.; Gottschalk, W.K. Insulin and glucosamine infusions increase O-linked N-acetyl-glucosamine in skeletal muscle proteins in vivo. Metab. Clin. Exp. 1998, 47, 449–455. [Google Scholar] [CrossRef]
- Ansari, S.A.; Emerald, B.S. The Role of Insulin Resistance and Protein O-GlcNAcylation in Neurodegeneration. Front. Neurosci. 2019, 13, 473. [Google Scholar] [CrossRef] [PubMed]
- Copeland, R.J.; Bullen, J.W.; Hart, G.W. Cross-talk between GlcNAcylation and phosphorylation: Roles in insulin resistance and glucose toxicity. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E17–E28. [Google Scholar] [CrossRef]
- Rojas, F.A.; Hirata, A.E.; Saad, M.J. Regulation of insulin receptor substrate-2 tyrosine phosphorylation in animal models of insulin resistance. Endocrine 2003, 21, 115–122. [Google Scholar] [CrossRef]
- Copps, K.D.; White, M.F. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia 2012, 55, 2565–2582. [Google Scholar] [CrossRef]
- Yudushkin, I. Getting the Akt Together: Guiding Intracellular Akt Activity by PI3K. Biomolecules 2019, 9, 67. [Google Scholar] [CrossRef]
- Ebner, M.; Lučić, I.; Leonard, T.A.; Yudushkin, I. PI(3,4,5)P(3) Engagement Restricts Akt Activity to Cellular Membranes. Mol. Cell 2017, 65, 416–431.e416. [Google Scholar] [CrossRef]
- Wick, K.L.; Liu, F. A new molecular target of insulin action: Regulating the pivotal PDK1. Curr. Drug Targets Immune Endocr. Metab. Disord. 2001, 1, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Yudushkin, I. Control of Akt activity and substrate phosphorylation in cells. IUBMB Life 2020, 72, 1115–1125. [Google Scholar] [CrossRef] [PubMed]
- Goldbraikh, D.; Neufeld, D.; Eid-Mutlak, Y.; Lasry, I.; Gilda, J.E.; Parnis, A.; Cohen, S. USP1 deubiquitinates Akt to inhibit PI3K-Akt-FoxO signaling in muscle during prolonged starvation. EMBO Rep. 2020, 21, e48791. [Google Scholar] [CrossRef]
- Xie, Y.; Shi, X.; Sheng, K.; Han, G.; Li, W.; Zhao, Q.; Jiang, B.; Feng, J.; Li, J.; Gu, Y. PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (Review). Mol. Med. Rep. 2019, 19, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Camaforte, N.A.P.; Saldanha, L.L.; Vareda, P.M.P.; Rezende-Neto, J.M.; Senger, M.R.; Delgado, A.Q.; Morgan, H.J.N.; Violato, N.M.; Pieroni, L.G.; Dokkedal, A.L.; et al. Hypoglycaemic activity of Bauhinia holophylla through GSK3-β inhibition and glycogenesis activation. Pharm. Biol. 2019, 57, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ongusaha, P.P.; Miles, P.D.; Havstad, J.C.; Zhang, F.; So, W.V.; Kudlow, J.E.; Michell, R.H.; Olefsky, J.M.; Field, S.J.; et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 2008, 451, 964–969. [Google Scholar] [CrossRef]
- Whelan, S.A.; Lane, M.D.; Hart, G.W. Regulation of the O-linked beta-N-acetylglucosamine transferase by insulin signaling. J. Biol. Chem. 2008, 283, 21411–21417. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.A.; Dias, W.B.; Thiruneelakantapillai, L.; Lane, M.D.; Hart, G.W. Regulation of insulin receptor substrate 1 (IRS-1)/AKT kinase-mediated insulin signaling by O-Linked beta-N-acetylglucosamine in 3T3-L1 adipocytes. J. Biol. Chem. 2010, 285, 5204–5211. [Google Scholar] [CrossRef]
- Issad, T.; Masson, E.; Pagesy, P. O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes Metab. 2010, 36, 423–435. [Google Scholar] [CrossRef]
- Vosseller, K.; Wells, L.; Lane, M.D.; Hart, G.W. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. USA 2002, 99, 5313–5318. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Dodd, G.T.; Tiganis, T. Protein Tyrosine Phosphatases in Hypothalamic Insulin and Leptin Signaling. Trends Pharmacol. Sci. 2015, 36, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tang, Z.; Shen, A.; Tao, T.; Wan, C.; Zhu, X.; Huang, J.; Zhang, W.; Xia, N.; Wang, S.; et al. The Role of PTP1B O-GlcNAcylation in Hepatic Insulin Resistance. Int. J. Mol. Sci. 2015, 16, 22856–22869. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, X.; Luan, H.H.; Zhang, B.; Zhang, K.; Nam, J.H.; Li, Z.; Fu, M.; Munk, A.; Zhang, D.; et al. OGT suppresses S6K1-mediated macrophage inflammation and metabolic disturbance. Proc. Natl. Acad. Sci. USA 2020, 117, 16616–16625. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, J.; Bai, Z.; Sink, S.; Zhao, C.; Lorenzo, F.R.; McClain, D.A. Iron down-regulates leptin by suppressing protein O-GlcNAc modification in adipocytes, resulting in decreased levels of O-glycosylated CREB. J. Biol. Chem. 2019, 294, 5487–5495. [Google Scholar] [CrossRef]
- Rexach, J.E.; Clark, P.M.; Mason, D.E.; Neve, R.L.; Peters, E.C.; Hsieh-Wilson, L.C. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat. Chem. Biol. 2012, 8, 253–261. [Google Scholar] [CrossRef]
- Dentin, R.; Hedrick, S.; Xie, J.; Yates, J., 3rd; Montminy, M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science 2008, 319, 1402–1405. [Google Scholar] [CrossRef]
- Romanello, V.; Sandri, M. The connection between the dynamic remodeling of the mitochondrial network and the regulation of muscle mass. Cell. Mol. Life Sci. CMLS 2021, 78, 1305–1328. [Google Scholar] [CrossRef]
- Parry, H.A.; Roberts, M.D.; Kavazis, A.N. Human Skeletal Muscle Mitochondrial Adaptations Following Resistance Exercise Training. Int. J. Sports Med. 2020, 41, 349–359. [Google Scholar] [CrossRef]
- Banerjee, P.S.; Lagerlöf, O.; Hart, G.W. Roles of O-GlcNAc in chronic diseases of aging. Mol. Asp. Med. 2016, 51, 1–15. [Google Scholar] [CrossRef]
- Wang, Z.; Qin, J.; Zhao, J.; Li, J.; Li, D.; Popp, M.; Popp, F.; Alakus, H.; Kong, B.; Dong, Q.; et al. Inflammatory IFIT3 renders chemotherapy resistance by regulating post-translational modification of VDAC2 in pancreatic cancer. Theranostics 2020, 10, 7178–7192. [Google Scholar] [CrossRef]
- Hanover, J.A.; Yu, S.; Lubas, W.B.; Shin, S.H.; Ragano-Caracciola, M.; Kochran, J.; Love, D.C. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 2003, 409, 287–297. [Google Scholar] [CrossRef]
- Hu, Y.; Suarez, J.; Fricovsky, E.; Wang, H.; Scott, B.T.; Trauger, S.A.; Han, W.; Hu, Y.; Oyeleye, M.O.; Dillmann, W.H. Increased enzymatic O-GlcNAcylation of mitochondrial proteins impairs mitochondrial function in cardiac myocytes exposed to high glucose. J. Biol. Chem. 2009, 284, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Cao, J.; Huang, J.; Yao, J.; Yan, G.; Xu, H.; Yang, P. Discovery and confirmation of O-GlcNAcylated proteins in rat liver mitochondria by combination of mass spectrometry and immunological methods. PLoS ONE 2013, 8, e76399. [Google Scholar] [CrossRef]
- Zumbaugh, M.D.; Johnson, S.E.; Shi, T.H.; Gerrard, D.E. Molecular and biochemical regulation of skeletal muscle metabolism. J. Anim. Sci. 2022, 100, skac035. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Banerjee, P.; Whelan, S.A.; Liu, T.; Wei, A.C.; Ramirez-Correa, G.; McComb, M.E.; Costello, C.E.; O’Rourke, B.; Murphy, A.; et al. Comparative Proteomics Reveals Dysregulated Mitochondrial O-GlcNAcylation in Diabetic Hearts. J. Proteome Res. 2016, 15, 2254–2264. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, T.; Wei, A.C.; Banerjee, P.; O’Rourke, B.; Hart, G.W. O-GlcNAcomic Profiling Identifies Widespread O-Linked β-N-Acetylglucosamine Modification (O-GlcNAcylation) in Oxidative Phosphorylation System Regulating Cardiac Mitochondrial Function. J. Biol. Chem. 2015, 290, 29141–29153. [Google Scholar] [CrossRef]
- Wu, J.; Liu, J.; Lapenta, K.; Desrouleaux, R.; Li, M.D.; Yang, X. Regulation of the urea cycle by CPS1 O-GlcNAcylation in response to dietary restriction and aging. J. Mol. Cell Biol. 2022, 14, mjac016. [Google Scholar] [CrossRef]
- Zhao, L.; Feng, Z.; Yang, X.; Liu, J. The regulatory roles of O-GlcNAcylation in mitochondrial homeostasis and metabolic syndrome. Free Radic. Res. 2016, 50, 1080–1088. [Google Scholar] [CrossRef]
- Ohashi, N.; Morino, K.; Ida, S.; Sekine, O.; Lemecha, M.; Kume, S.; Park, S.Y.; Choi, C.S.; Ugi, S.; Maegawa, H. Pivotal Role of O-GlcNAc Modification in Cold-Induced Thermogenesis by Brown Adipose Tissue Through Mitochondrial Biogenesis. Diabetes 2017, 66, 2351–2362. [Google Scholar] [CrossRef]
- Pekkurnaz, G.; Trinidad, J.C.; Wang, X.; Kong, D.; Schwarz, T.L. Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell 2014, 158, 54–68. [Google Scholar] [CrossRef]
- Gawlowski, T.; Suarez, J.; Scott, B.; Torres-Gonzalez, M.; Wang, H.; Schwappacher, R.; Han, X.; Yates, J.R., 3rd; Hoshijima, M.; Dillmann, W. Modulation of dynamin-related protein 1 (DRP1) function by increased O-linked-β-N-acetylglucosamine modification (O-GlcNAc) in cardiac myocytes. J. Biol. Chem. 2012, 287, 30024–30034. [Google Scholar] [CrossRef] [PubMed]
- Akinbiyi, E.O.; Abramowitz, L.K.; Bauer, B.L.; Stoll, M.S.K.; Hoppel, C.L.; Hsiao, C.P.; Hanover, J.A.; Mears, J.A. Blocked O-GlcNAc cycling alters mitochondrial morphology, function, and mass. Sci. Rep. 2021, 11, 22106. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.Y.; Wei, X.X.; Zhi, X.L.; Wang, X.H.; Meng, D. Drp1-dependent mitochondrial fission in cardiovascular disease. Acta Pharmacol. Sin. 2021, 42, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Champattanachai, V.; Marchase, R.B.; Chatham, J.C. Glucosamine protects neonatal cardiomyocytes from ischemia-reperfusion injury via increased protein O-GlcNAc and increased mitochondrial Bcl-2. Am. J. Physiol. Cell Physiol. 2008, 294, C1509–C1520. [Google Scholar] [CrossRef] [PubMed]
- Marsh, S.A.; Powell, P.C.; Dell’italia, L.J.; Chatham, J.C. Cardiac O-GlcNAcylation blunts autophagic signaling in the diabetic heart. Life Sci. 2013, 92, 648–656. [Google Scholar] [CrossRef]
- Cividini, F.; Scott, B.T.; Dai, A.; Han, W.; Suarez, J.; Diaz-Juarez, J.; Diemer, T.; Casteel, D.E.; Dillmann, W.H. O-GlcNAcylation of 8-Oxoguanine DNA Glycosylase (Ogg1) Impairs Oxidative Mitochondrial DNA Lesion Repair in Diabetic Hearts. J. Biol. Chem. 2016, 291, 26515–26528. [Google Scholar] [CrossRef]
- Jones, S.P.; Zachara, N.E.; Ngoh, G.A.; Hill, B.G.; Teshima, Y.; Bhatnagar, A.; Hart, G.W.; Marbán, E. Cardioprotection by N-acetylglucosamine linkage to cellular proteins. Circulation 2008, 117, 1172–1182. [Google Scholar] [CrossRef]
- Dinić, S.; Arambašić, J.; Mihailović, M.; Uskoković, A.; Grdović, N.; Marković, J.; Karadžić, B.; Poznanović, G.; Vidaković, M. Decreased O-GlcNAcylation of the key proteins in kinase and redox signalling pathways is a novel mechanism of the beneficial effect of α-lipoic acid in diabetic liver. Br. J. Nutr. 2013, 110, 401–412. [Google Scholar] [CrossRef]
- Bottinelli, R.; Reggiani, C. Human skeletal muscle fibres: Molecular and functional diversity. Prog. Biophys. Mol. Biol. 2000, 73, 195–262. [Google Scholar] [CrossRef]
- Bottinelli, R. Functional heterogeneity of mammalian single muscle fibres: Do myosin isoforms tell the whole story? Pflug. Arch. Eur. J. Physiol. 2001, 443, 6–17. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef]
- Schiaffino, S. Muscle fiber type diversity revealed by anti-myosin heavy chain antibodies. FEBS J. 2018, 285, 3688–3694. [Google Scholar] [CrossRef] [PubMed]
- Westerblad, H.; Bruton, J.D.; Katz, A. Skeletal muscle: Energy metabolism, fiber types, fatigue and adaptability. Exp. Cell Res. 2010, 316, 3093–3099. [Google Scholar] [CrossRef] [PubMed]
- Frontera, W.R.; Ochala, J. Skeletal muscle: A brief review of structure and function. Calcif. Tissue Int. 2015, 96, 183–195. [Google Scholar] [CrossRef]
- Cieniewski-Bernard, C.; Lambert, M.; Dupont, E.; Montel, V.; Stevens, L.; Bastide, B. O-GlcNAcylation, contractile protein modifications and calcium affinity in skeletal muscle. Front. Physiol. 2014, 5, 421. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.; Richard, E.; Duban-Deweer, S.; Krzewinski, F.; Deracinois, B.; Dupont, E.; Bastide, B.; Cieniewski-Bernard, C. O-GlcNAcylation is a key modulator of skeletal muscle sarcomeric morphometry associated to modulation of protein-protein interactions. Biochim. Biophys. Acta 2016, 1860, 2017–2030. [Google Scholar] [CrossRef]
- Cieniewski-Bernard, C.; Montel, V.; Stevens, L.; Bastide, B. O-GlcNAcylation, an original modulator of contractile activity in striated muscle. J. Muscle Res. Cell Motil. 2009, 30, 281–287. [Google Scholar] [CrossRef]
- Hortemo, K.H.; Lunde, P.K.; Anonsen, J.H.; Kvaløy, H.; Munkvik, M.; Rehn, T.A.; Sjaastad, I.; Lunde, I.G.; Aronsen, J.M.; Sejersted, O.M. Exercise training increases protein O-GlcNAcylation in rat skeletal muscle. Physiol. Rep. 2016, 4, e12896. [Google Scholar] [CrossRef] [PubMed]
- Cieniewski-Bernard, C.; Mounier, Y.; Michalski, J.C.; Bastide, B. O-GlcNAc level variations are associated with the development of skeletal muscle atrophy. J. Appl. Physiol. 2006, 100, 1499–1505. [Google Scholar] [CrossRef]
- Cieniewski-Bernard, C.; Dupont, E.; Richard, E.; Bastide, B. Phospho-GlcNAc modulation of slow MLC2 during soleus atrophy through a multienzymatic and sarcomeric complex. Pflug. Arch. 2014, 466, 2139–2151. [Google Scholar] [CrossRef]
- Fülöp, N.; Feng, W.; Xing, D.; He, K.; Nőt, L.G.; Brocks, C.A.; Marchase, R.B.; Miller, A.P.; Chatham, J.C. Aging leads to increased levels of protein O-linked N-acetylglucosamine in heart, aorta, brain and skeletal muscle in Brown-Norway rats. Biogerontology 2008, 9, 139. [Google Scholar] [CrossRef]
- Toivonen, M.H.; Pöllänen, E.; Ahtiainen, M.; Suominen, H.; Taaffe, D.R.; Cheng, S.; Takala, T.; Kujala, U.M.; Tammi, M.I.; Sipilä, S.; et al. OGT and OGA expression in postmenopausal skeletal muscle associates with hormone replacement therapy and muscle cross-sectional area. Exp. Gerontol. 2013, 48, 1501–1504. [Google Scholar] [CrossRef] [PubMed]
- Hortemo, K.H.; Aronsen, J.M.; Lunde, I.G.; Sjaastad, I.; Lunde, P.K.; Sejersted, O.M. Exhausting treadmill running causes dephosphorylation of sMLC2 and reduced level of myofilament MLCK2 in slow twitch rat soleus muscle. Physiol. Rep. 2015, 3, e12285. [Google Scholar] [CrossRef] [PubMed]
- Peternelj, T.T.; Marsh, S.A.; Strobel, N.A.; Matsumoto, A.; Briskey, D.; Dalbo, V.J.; Tucker, P.S.; Coombes, J.S. Glutathione depletion and acute exercise increase O-GlcNAc protein modification in rat skeletal muscle. Mol. Cell. Biochem. 2015, 400, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.A.; Robinson, K.A.; Koning, J.S.; Buse, M.G. Effects of exercise and feeding on the hexosamine biosynthetic pathway in rat skeletal muscle. Am. J. Physiol. 1997, 272, E848–E855. [Google Scholar] [CrossRef]
- Murata, K.; Morino, K.; Ida, S.; Ohashi, N.; Lemecha, M.; Park, S.Y.; Ishikado, A.; Kume, S.; Choi, C.S.; Sekine, O.; et al. Lack of O-GlcNAcylation enhances exercise-dependent glucose utilization potentially through AMP-activated protein kinase activation in skeletal muscle. Biochem. Biophys. Res. Commun. 2018, 495, 2098–2104. [Google Scholar] [CrossRef] [PubMed]
- Gransee, H.M.; Mantilla, C.B.; Sieck, G.C. Respiratory muscle plasticity. Compr. Physiol. 2012, 2, 1441–1462. [Google Scholar] [CrossRef] [PubMed]
- Konopka, A.R.; Harber, M.P. Skeletal muscle hypertrophy after aerobic exercise training. Exerc. Sport Sci. Rev. 2014, 42, 53–61. [Google Scholar] [CrossRef]
- Qaisar, R.; Bhaskaran, S.; Van Remmen, H. Muscle fiber type diversification during exercise and regeneration. Free Radic. Biol. Med. 2016, 98, 56–67. [Google Scholar] [CrossRef]
- Blaauw, B.; Schiaffino, S.; Reggiani, C. Mechanisms modulating skeletal muscle phenotype. Compr. Physiol. 2013, 3, 1645–1687. [Google Scholar] [CrossRef]
- Teixeira, E.; Duarte, J.A. Skeletal Muscle Loading Changes its Regenerative Capacity. Sports Med. 2016, 46, 783–792. [Google Scholar] [CrossRef]
- Davies, K.J.; Quintanilha, A.T.; Brooks, G.A.; Packer, L. Free radicals and tissue damage produced by exercise. Biochem. Biophys. Res. Commun. 1982, 107, 1198–1205. [Google Scholar] [CrossRef]
- Pette, D.; Staron, R.S. Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 2000, 50, 500–509. [Google Scholar] [CrossRef]
- Kim, Y.H.; Song, M.; Oh, Y.S.; Heo, K.; Choi, J.W.; Park, J.M.; Kim, S.H.; Lim, S.; Kwon, H.M.; Ryu, S.H.; et al. Inhibition of phospholipase C-beta1-mediated signaling by O-GlcNAc modification. J. Cell. Physiol. 2006, 207, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Slawson, C.; Housley, M.P.; Hart, G.W. O-GlcNAc cycling: How a single sugar post-translational modification is changing the way we think about signaling networks. J. Cell. Biochem. 2006, 97, 71–83. [Google Scholar] [CrossRef]
- Peternelj, T.T.; Marsh, S.A.; Morais, C.; Small, D.M.; Dalbo, V.J.; Tucker, P.S.; Coombes, J.S. O-GlcNAc protein modification in C2C12 myoblasts exposed to oxidative stress indicates parallels with endogenous antioxidant defense. Biochem. Cell Biol. Biochim. Biol. Cell. 2015, 93, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Lima, V.V.; Spitler, K.; Choi, H.; Webb, R.C.; Tostes, R.C. O-GlcNAcylation and oxidation of proteins: Is signalling in the cardiovascular system becoming sweeter? Clin. Sci. 2012, 123, 473–486. [Google Scholar] [CrossRef]
- Chen, P.H.; Chi, J.T.; Boyce, M. Functional crosstalk among oxidative stress and O-GlcNAc signaling pathways. Glycobiology 2018, 28, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Booth, F.W.; Laye, M.J.; Spangenburg, E.E. Gold standards for scientists who are conducting animal-based exercise studies. J. Appl. Physiol. 2010, 108, 219–221. [Google Scholar] [CrossRef]
- Sun, C.; Shang, J.; Yao, Y.; Yin, X.; Liu, M.; Liu, H.; Zhou, Y. O-GlcNAcylation: A bridge between glucose and cell differentiation. J. Cell. Mol. Med. 2016, 20, 769–781. [Google Scholar] [CrossRef]
- Sun, C.; Lan, W.; Li, B.; Zuo, R.; Xing, H.; Liu, M.; Li, J.; Yao, Y.; Wu, J.; Tang, Y.; et al. Glucose regulates tissue-specific chondro-osteogenic differentiation of human cartilage endplate stem cells via O-GlcNAcylation of Sox9 and Runx2. Stem Cell Res. Ther. 2019, 10, 357. [Google Scholar] [CrossRef]
- Gu, H.; Song, M.; Boonanantanasarn, K.; Baek, K.; Woo, K.M.; Ryoo, H.M.; Baek, J.H. Conditions Inducing Excessive O-GlcNAcylation Inhibit BMP2-Induced Osteogenic Differentiation of C2C12 Cells. Int. J. Mol. Sci. 2018, 19, 202. [Google Scholar] [CrossRef]
- Ogawa, M.; Mizofuchi, H.; Kobayashi, Y.; Tsuzuki, G.; Yamamoto, M.; Wada, S.; Kamemura, K. Terminal differentiation program of skeletal myogenesis is negatively regulated by O-GlcNAc glycosylation. Biochim. Biophys. Acta 2012, 1820, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Seo, H.G.; Son, S.; Choi, H.; Kim, B.G.; Kweon, T.H.; Kim, S.; Pai, J.; Shin, I.; Yang, W.H.; et al. O-GlcNAcylation of Mef2c regulates myoblast differentiation. Biochem. Biophys. Res. Commun. 2020, 529, 692–698. [Google Scholar] [CrossRef]
- Ogawa, M.; Sakakibara, Y.; Kamemura, K. Requirement of decreased O-GlcNAc glycosylation of Mef2D for its recruitment to the myogenin promoter. Biochem. Biophys. Res. Commun. 2013, 433, 558–562. [Google Scholar] [CrossRef]
- Anderson, J.E. Key concepts in muscle regeneration: Muscle “cellular ecology” integrates a gestalt of cellular cross-talk, motility, and activity to remodel structure and restore function. Eur. J. Appl. Physiol. 2022, 122, 273–300. [Google Scholar] [CrossRef] [PubMed]
- Byon, C.H.; Kim, S.W. Regulatory Effects of O-GlcNAcylation in Vascular Smooth Muscle Cells on Diabetic Vasculopathy. J. Lipid Atheroscler. 2020, 9, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Dang, K.; Jiang, S.; Gao, Y.; Qian, A. The role of protein glycosylation in muscle diseases. Mol. Biol. Rep. 2022, 49, 8037–8049. [Google Scholar] [CrossRef]
- Nakamura, S.; Nakano, S.; Nishii, M.; Kaneko, S.; Kusaka, H. Localization of O-GlcNAc-modified proteins in neuromuscular diseases. Med. Mol. Morphol. 2012, 45, 86–90. [Google Scholar] [CrossRef]
- Shan, X.; Vocadlo, D.J.; Krieger, C. Reduced protein O-glycosylation in the nervous system of the mutant SOD1 transgenic mouse model of amyotrophic lateral sclerosis. Neurosci. Lett. 2012, 516, 296–301. [Google Scholar] [CrossRef]
- Lüdemann, N.; Clement, A.; Hans, V.H.; Leschik, J.; Behl, C.; Brandt, R. O-glycosylation of the tail domain of neurofilament protein M in human neurons and in spinal cord tissue of a rat model of amyotrophic lateral sclerosis (ALS). J. Biol. Chem. 2005, 280, 31648–31658. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.H.; Hu, J.; Wu, J.; Huynh, D.T.; Smith, T.J.; Pan, S.; Bisnett, B.J.; Smith, A.B.; Lu, A.; Condon, B.M.; et al. Gigaxonin glycosylation regulates intermediate filament turnover and may impact giant axonal neuropathy etiology or treatment. JCI Insight 2020, 5, e127751. [Google Scholar] [CrossRef] [PubMed]
- Malicdan, M.C.; Noguchi, S.; Nonaka, I.; Hayashi, Y.K.; Nishino, I. A Gne knockout mouse expressing human GNE D176V mutation develops features similar to distal myopathy with rimmed vacuoles or hereditary inclusion body myopathy. Hum. Mol. Genet. 2007, 16, 2669–2682. [Google Scholar] [CrossRef]
- Bennmann, D.; Weidemann, W.; Thate, A.; Kreuzmann, D.; Horstkorte, R. Aberrant O-GlcNAcylation disrupts GNE enzyme activity in GNE myopathy. FEBS J. 2016, 283, 2285–2294. [Google Scholar] [CrossRef]
- Massaccesi, L.; Goi, G.; Tringali, C.; Barassi, A.; Venerando, B.; Papini, N. Dexamethasone-Induced Skeletal Muscle Atrophy Increases O-GlcNAcylation in C2C12 Cells. J. Cell. Biochem. 2016, 117, 1833–1842. [Google Scholar] [CrossRef]
- Akan, I.; Olivier-Van Stichelen, S.; Bond, M.R.; Hanover, J.A. Nutrient-driven O-GlcNAc in proteostasis and neurodegeneration. J. Neurochem. 2018, 144, 7–34. [Google Scholar] [CrossRef]
- Martin-Rincon, M.; Morales-Alamo, D.; Calbet, J.A.L. Exercise-mediated modulation of autophagy in skeletal muscle. Scand. J. Med. Sci. Sports 2018, 28, 772–781. [Google Scholar] [CrossRef]
- Rodney, G.G.; Pal, R.; Abo-Zahrah, R. Redox regulation of autophagy in skeletal muscle. Free Radic. Biol. Med. 2016, 98, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Demontis, F. Skeletal muscle autophagy and its role in sarcopenia and organismal aging. Curr. Opin. Pharmacol. 2017, 34, 1–6. [Google Scholar] [CrossRef]
- Xie, Y.; Kang, R.; Sun, X.; Zhong, M.; Huang, J.; Klionsky, D.J.; Tang, D. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 2015, 11, 28–45. [Google Scholar] [CrossRef]
- Fan, J.; Yang, X.; Li, J.; Shu, Z.; Dai, J.; Liu, X.; Li, B.; Jia, S.; Kou, X.; Yang, Y.; et al. Spermidine coupled with exercise rescues skeletal muscle atrophy from D-gal-induced aging rats through enhanced autophagy and reduced apoptosis via AMPK-FOXO3a signal pathway. Oncotarget 2017, 8, 17475–17490. [Google Scholar] [CrossRef] [PubMed]
- Mammucari, C.; Milan, G.; Romanello, V.; Masiero, E.; Rudolf, R.; Del Piccolo, P.; Burden, S.J.; Di Lisi, R.; Sandri, C.; Zhao, J.; et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007, 6, 458–471. [Google Scholar] [CrossRef]
- Herrenbruck, A.R.; Bollinger, L.M. Role of skeletal muscle autophagy in high-fat-diet-induced obesity and exercise. Nutr. Rev. 2020, 78, 56–64. [Google Scholar] [CrossRef]
- Neel, B.A.; Lin, Y.; Pessin, J.E. Skeletal muscle autophagy: A new metabolic regulator. Trends Endocrinol. Metab. TEM 2013, 24, 635–643. [Google Scholar] [CrossRef]
- Wang, X.; Lin, Y.; Liu, S.; Zhu, Y.; Lu, K.; Broering, R.; Lu, M. O-GlcNAcylation modulates HBV replication through regulating cellular autophagy at multiple levels. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 14473–14489. [Google Scholar] [CrossRef]
- Shen, Y.; Czaja, M.J. A Novel Mechanism of Starvation-Stimulated Hepatic Autophagy: Calcium-Induced O-GlcNAc-Dependent Signaling. Hepatology 2019, 69, 446–448. [Google Scholar] [CrossRef]
- Kim, K.; Yoo, H.C.; Kim, B.G.; Kim, S.; Sung, Y.; Yoon, I.; Yu, Y.C.; Park, S.J.; Kim, J.H.; Myung, K.; et al. O-GlcNAc modification of leucyl-tRNA synthetase 1 integrates leucine and glucose availability to regulate mTORC1 and the metabolic fate of leucine. Nat. Commun. 2022, 13, 2904. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Hanover, J.A. Nutrient-driven O-GlcNAc cycling influences autophagic flux and neurodegenerative proteotoxicity. Autophagy 2013, 9, 604–606. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kumar, A.; Singh, P.K.; Parihar, R.; Dwivedi, V.; Lakhotia, S.C.; Ganesh, S. Decreased O-linked GlcNAcylation protects from cytotoxicity mediated by huntingtin exon1 protein fragment. J. Biol. Chem. 2014, 289, 13543–13553. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Scruggs, S.B.; Gilda, J.E.; Ping, P.; Gomes, A.V. Regulation of cardiac proteasomes by ubiquitination, SUMOylation, and beyond. J. Mol. Cell. Cardiol. 2014, 71, 32–42. [Google Scholar] [CrossRef]
- Zhang, F.; Hu, Y.; Huang, P.; Toleman, C.A.; Paterson, A.J.; Kudlow, J.E. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J. Biol. Chem. 2007, 282, 22460–22471. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, S.; Zhang, H.; Xu, J. Identification of nitric oxide as an endogenous inhibitor of 26S proteasomes in vascular endothelial cells. PLoS ONE 2014, 9, e98486. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Cao, M.; Ruan, X.; Jiang, L.; Lee, S.; Lemanek, A.; Ghassemian, M.; Pizzo, D.P.; Wan, Y.; Qiao, Y.; et al. Cancer-cell-secreted miR-122 suppresses O-GlcNAcylation to promote skeletal muscle proteolysis. Nat. Cell Biol. 2022, 24, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Lazarus, B.D.; Forsythe, M.E.; Love, D.C.; Krause, M.W.; Hanover, J.A. O-GlcNAc cycling mutants modulate proteotoxicity in Caenorhabditis elegans models of human neurodegenerative diseases. Proc. Natl. Acad. Sci. USA 2012, 109, 17669–17674. [Google Scholar] [CrossRef]
- Love, D.C.; Ghosh, S.; Mondoux, M.A.; Fukushige, T.; Wang, P.; Wilson, M.A.; Iser, W.B.; Wolkow, C.A.; Krause, M.W.; Hanover, J.A. Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 7413–7418. [Google Scholar] [CrossRef]
- Park, S.; Lee, Y.; Pak, J.W.; Kim, H.; Choi, H.; Kim, J.W.; Roth, J.; Cho, J.W. O-GlcNAc modification is essential for the regulation of autophagy in Drosophila melanogaster. Cell. Mol. Life Sci. CMLS 2015, 72, 3173–3183. [Google Scholar] [CrossRef]
- Huang, L.; Yuan, P.; Yu, P.; Kong, Q.; Xu, Z.; Yan, X.; Shen, Y.; Yang, J.; Wan, R.; Hong, K.; et al. O-GlcNAc-modified SNAP29 inhibits autophagy-mediated degradation via the disturbed SNAP29-STX17-VAMP8 complex and exacerbates myocardial injury in type I diabetic rats. Int. J. Mol. Med. 2018, 42, 3278–3290. [Google Scholar] [CrossRef]
- Dodson, M.; Liu, P.; Jiang, T.; Ambrose, A.J.; Luo, G.; de la Vega, M.R.; Cholanians, A.B.; Wong, P.K.; Chapman, E.; Zhang, D.D. Increased O-GlcNAcylation of SNAP29 Drives Arsenic-Induced Autophagic Dysfunction. Mol. Cell. Biol. 2018, 38, e00595-17. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Lak, B.; Li, J.; Jokitalo, E.; Wang, Y. GRASP55 Senses Glucose Deprivation through O-GlcNAcylation to Promote Autophagosome-Lysosome Fusion. Dev. Cell 2018, 45, 245–261.e246. [Google Scholar] [CrossRef]
- Shi, Y.; Yan, S.; Shao, G.C.; Wang, J.; Jian, Y.P.; Liu, B.; Yuan, Y.; Qin, K.; Nai, S.; Huang, X.; et al. O-GlcNAcylation stabilizes the autophagy-initiating kinase ULK1 by inhibiting chaperone-mediated autophagy upon HPV infection. J. Biol. Chem. 2022, 298, 102341. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Hu, Y.; Li, S. Protein O-GlcNAcylation in Metabolic Modulation of Skeletal Muscle: A Bright but Long Way to Go. Metabolites 2022, 12, 888. https://doi.org/10.3390/metabo12100888
Liu Y, Hu Y, Li S. Protein O-GlcNAcylation in Metabolic Modulation of Skeletal Muscle: A Bright but Long Way to Go. Metabolites. 2022; 12(10):888. https://doi.org/10.3390/metabo12100888
Chicago/Turabian StyleLiu, Yang, Yajie Hu, and Shize Li. 2022. "Protein O-GlcNAcylation in Metabolic Modulation of Skeletal Muscle: A Bright but Long Way to Go" Metabolites 12, no. 10: 888. https://doi.org/10.3390/metabo12100888
APA StyleLiu, Y., Hu, Y., & Li, S. (2022). Protein O-GlcNAcylation in Metabolic Modulation of Skeletal Muscle: A Bright but Long Way to Go. Metabolites, 12(10), 888. https://doi.org/10.3390/metabo12100888




