The Pharmacometabodynamics of Gefitinib after Intravenous Administration to Mice: A Preliminary UPLC–IM–MS Study
Abstract
1. Introduction
2. Results and Discussion
2.1. Plasma Pharmacokinetics of Gefitinib and Metabolites
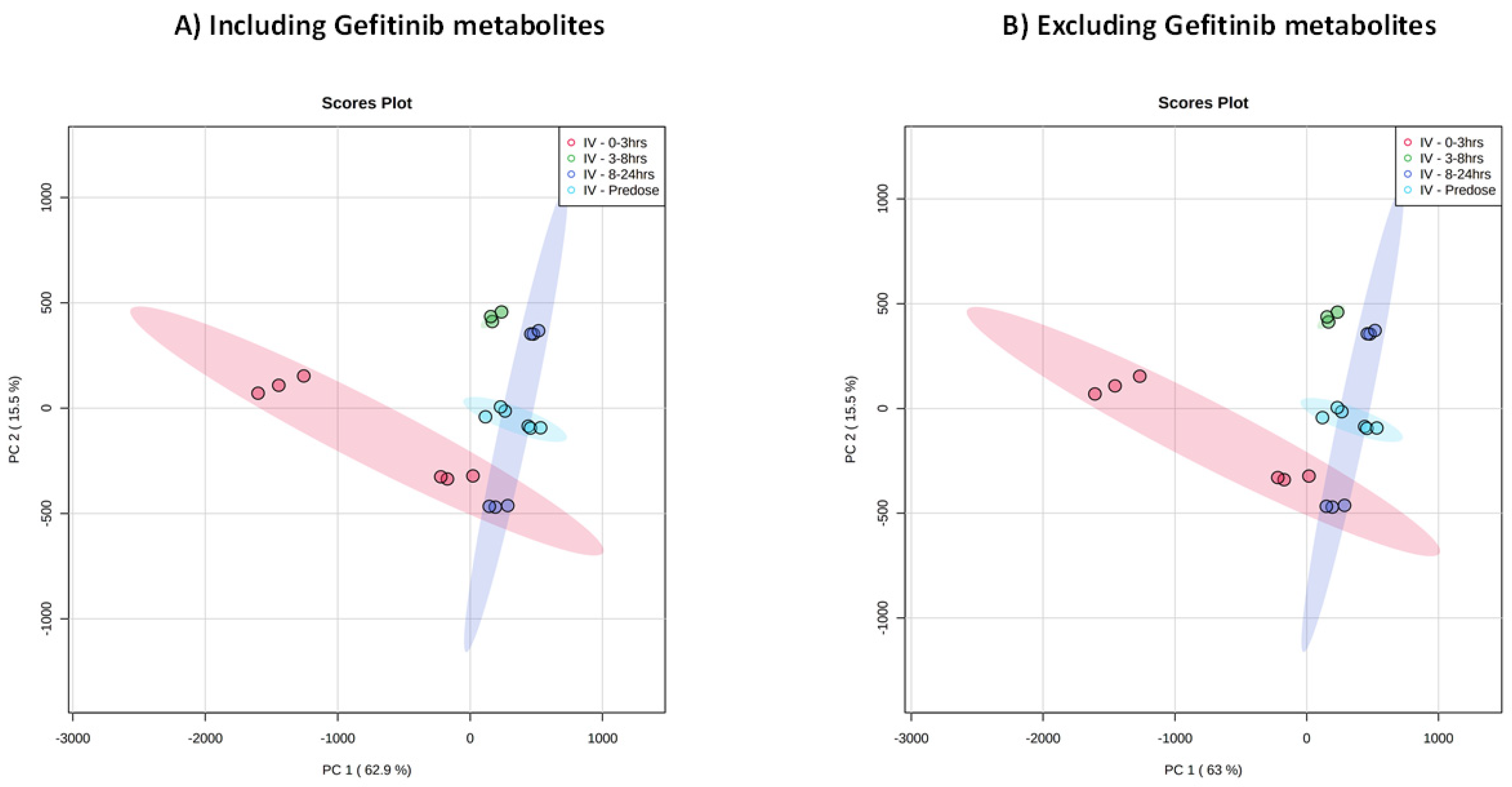
2.2. Untargeted Analysis of Urine Including Gefitinib and Metabolites
2.3. Untargeted Metabolic Phenotyping
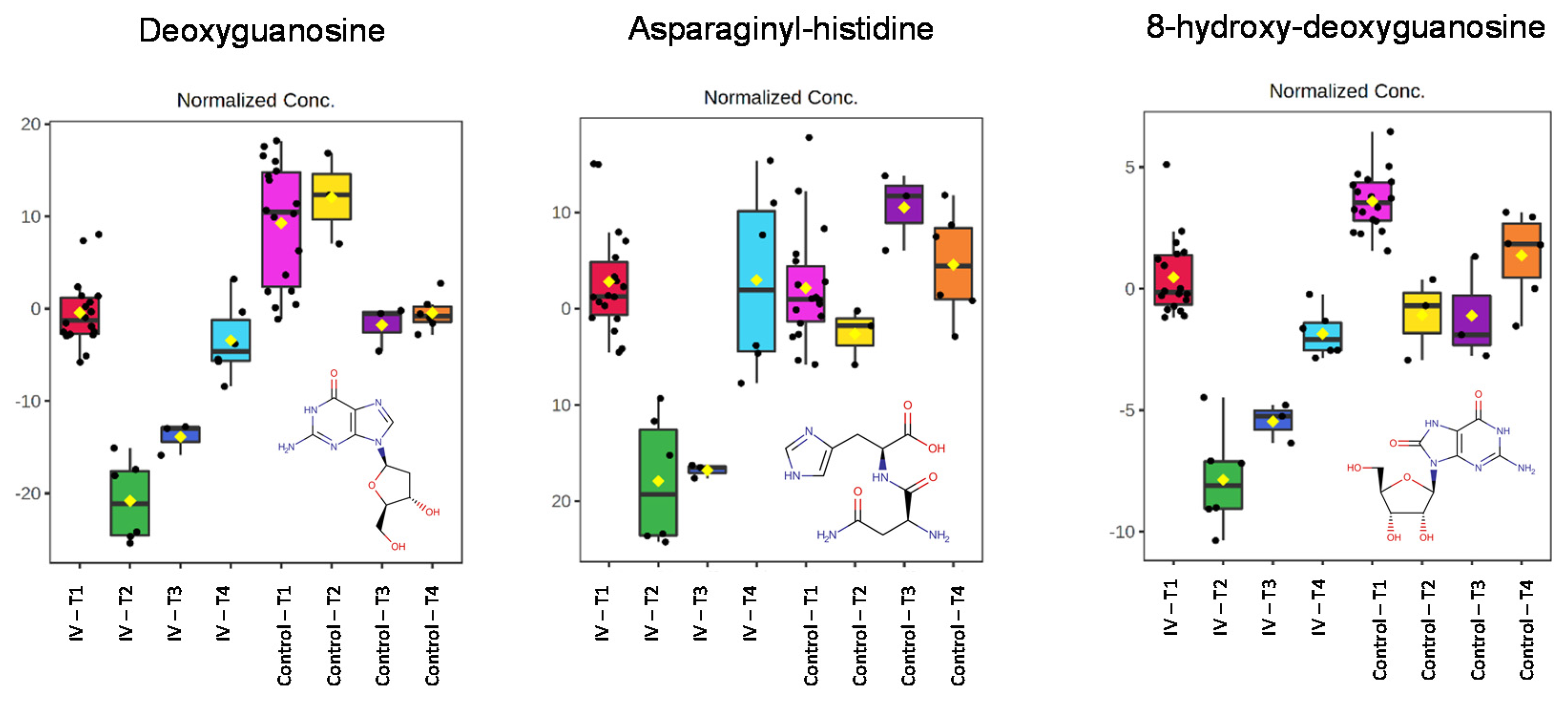
2.4. Endogenous Metabolite Identification
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Study Conduct
4.3. Metabolite Profiling
4.4. Reversed-Phase LC/IM/MS
4.5. Data Analysis for Metabolite Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clayton, T.; Lindon, J.; Cloarec, O.; Antti, H.; Charuel, C.; Hanton, G.; Provost, J.; Le Net, J.; Baker, D.; Walley, R.; et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 2006, 440, 1073–1077. [Google Scholar] [CrossRef]
- Clayton, T.A.; Baker, D.; Lindon, J.C.; Everett, J.R.; Nicholson, J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14728–14733. [Google Scholar] [CrossRef]
- Everett, J.R. NMR-based pharmacometabonomics: A new paradigm for personalised or precision medicine. Prog. Nucl. Magn. Reson. Spectros. 2017, 102–103, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Everett, J.R. Pharmacometabonomics: The Prediction of Drug Effects Using Metabolic Profiling. In Concepts and Principles of Pharmacology. Handbook of Experimental Pharmacology; Barrett, J., Page, C., Michel, M., Eds.; Springer: Berlin, Germany, 2019; Volume 260, pp. 263–299. [Google Scholar]
- Beger, R.D.; Schmidt, M.A.; Kaddurah-Daouk, R. Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine. Metabolites 2020, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.C.; Ekins, S.; Williams, A.J.; Tropsha, A. A bibliometric review of drug repurposing. Drug Discov. Today 2018, 3, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.A.; Williams, G.A.; Sridhara, R.G.; Chen, G.; Pazdur, R. United States Food and Drug Administration Drug Approval summary: Gefitinib (ZD1839; Iressa) tablets. Clin. Cancer Res. 2004, 10, 1212–1218. [Google Scholar] [CrossRef]
- Maemondo, M.; Inoue, A.; Kobayashi, K.; Sugawara, S.; Oizumi, S.; Isobe, H.; Gemma, A.; Harada, M.; Yoshizawa, H.; Kinoshita, I.; et al. North-East Japan Study Group. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010, 362, 2380–2388. [Google Scholar] [CrossRef]
- Dhillon, S. Gefitinib: A review of its use in adults with advanced non-small cell lung cancer. Target Oncol. 2015, 10, 153–170. [Google Scholar] [CrossRef]
- McKillop, D.; Partridge, E.A.; Hutchison, M.; Rhead, S.A.; Parry, A.C.; Bardsley, J.; Woodman, M.; Swaisland, H.C. Pharmacokinetics of gefitinib, an epidermal growth factor receptor tyrosine kinase inhibitor, in rat and dog. Xenobiotica 2004, 34, 901–915. [Google Scholar] [CrossRef]
- McKillop, D.; Hutchison, M.; Partridge, E.A.; Bushby, N.; Cooper, C.M.F.; Clarkson-Jones, J.A.; Herron, W.; Swaisland, H.C. Metabolic disposition of gefitinib, an epidermal growth factor receptor tyrosine kinase inhibitor, in rat, dog and man. Xenobiotica 2004, 34, 917–934. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Zhao, C.; He, X.-R.; Jiang, S.-T.; Han, S.-Y.; Xu, G.-B.; Li, P.-P. Simultaneous determination of gefitinib and its major metabolites in mouse plasma by HPLC-MS/MS and its application to a pharmacokinetics study. J. Chromatogr. B 2016, 1011, 215–222. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, R.; Xu Chen, X.; Lee, S.B.; Pan, J.; Xiong, D.; Hu, J.; Miller, M.S.; Szabo, E.; Lubet, R.A.; et al. Effect of weekly or daily dosing regimen of Gefitinib in mouse models of lung cancer. Oncotarget 2017, 42, 72447–72456. [Google Scholar] [CrossRef]
- Molloy, B.J.; King, A.; Mullin, L.; Gethings, L.A.; Riley, R.; Plumb, R.; Wilson, I.D. Rapid determination of the pharmacokinetics and metabolic fate of gefitinib in the mouse using a combination of UPLC/MS/MS, UPLC/QToF/MS, and ion mobility (IM)-enabled UPLC/QToF/MS. Xenobiotica 2021, 51, 434–446. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, Y.; Feng, T.; Cao, L.; Wu, W.; Qi, K. Comprehensive identification, fragmentation pattern, and metabolic pathways of gefitinib metabolites via UHPLC-Q-TOF-MS/MS: In vivo study of rat plasma, urine, bile, and faeces. Xenobiotica 2021, 51, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.K.; Stafford, L.E.; Swaisland, H.C.; Payne, R. A sensitive assay for ZD1839 (Iressa) in human plasma by liquid–liquid extraction and high performance liquid chromatography with mass spectrometric detection: Validation and use in Phase I clinical trials. J. Pharm. Biomed. Anal. 2002, 29, 221–228. [Google Scholar] [CrossRef]
- Wang, L.Z.; Lim, M.Y.; Chin, T.M.; Thuya, W.L.; Nye, P.L.; Wong, A.; Chan, S.-Y.; Goh, B.-C.; Ho, P.C. Rapid determination of gefitinib and its main metabolite, O-desmethyl gefitinib in human plasma using liquid chromatography-tandem mass spectrometry. J. Chromatogr. B 2011, 879, 2155–2161. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Chen, X.; Wang, F.; Xin, S.; Feng, W.; Zhu, X.; Zhuang, W.; Zhouab, S.; Huang, M.; Wang, X.; et al. Development and validation of a sensitive LC-MS/MS method for determination of gefitinib and its major metabolites in human plasma and its application in non-small cell lung cancer patients. J. Pharm. Biomed. Anal. 2019, 172, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, J.; Zhou, S.; Yu, L.; Han, F.; Ling, R.; Ling, J. Tentative identification of gefitinib metabolites in non-small-cell lung cancer patient plasma using ultra-performance liquid chromatography coupled with triple quadrupole time-of-flight mass spectrometry. PLoS ONE 2020, 15, e0236523. [Google Scholar] [CrossRef]
- McKillop, D.; McCormick, A.D.; Miles, G.S.; Phillips, P.J.; Pickup, K.J.; Bushby, N.; Hutchison, M. In vitro metabolism of gefitinib in human liver microsomes. Xenobiotica 2004, 34, 983–1000. [Google Scholar] [CrossRef]
- Mckillop, D.; McCormick, A.D.; Millar, A.; Miles, G.S.; Phillips, P.J.; Hutchison, M. Cytochrome P450-dependent metabolism of gefitinib. Xenobiotica 2005, 35, 39–50. [Google Scholar] [CrossRef]
- Li, J.; Zhao, M.; He, P.; Hidalgo, M.; Bake, S.D. Differential metabolism of gefitinib and erlotinib by human cytochrome P450 enzymes. Clin. Cancer Res. 2007, 13, 3731–3737. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lu, Y.; Guan, X.; Dong, B.; Chavan, H.; Wang, J.; Zhang, Y.; Krishnamurthy, P.; Li, F. Metabolomics reveals the formation of aldehydes and iminium in gefitinib metabolism. Biochem. Pharmacol. 2015, 97, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Connelly, J.; Lindon, J.C.; Holmes, E. Metabonomics: A platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 2002, 1, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.G. Metabonomics in Toxicology: A Review. Toxicol. Sci. 2005, 85, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Bales, J.R.; Sadler, P.J.; Nicholson, J.K.; Timbrell, J.A. Urinary excretion of acetaminophen and its metabolites as studied by proton NMR spectroscopy. Clin. Chem. 1984, 30, 1631–1636. [Google Scholar] [CrossRef] [PubMed]
- Bales, J.R.; Nicholson, J.K.; Sadler, P.J. Two-dimensional proton nuclear magnetic resonance ’maps’ of acetaminophen metabolites in human urine. Clin. Chem. 1985, 31, 757–762. [Google Scholar] [CrossRef]
- Wilson, I.D.; Fromson, J.; Ismail, I.M.; Nicholson, J.K. Proton magnetic resonance spectroscopy of human urine: Excretion of 1-(3′-carboxypropyl)-3,7-dimethylxanthine by man after dosing with oxpentifylline. J. Pharm. Biomed. Anal. 1987, 5, 157–163. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Wilson, I.D. High resolution nuclear magnetic resonance spectroscopy of biological fluids as an aid to drug development. Prog. Drug Res. 1987, 31, 427–479. [Google Scholar]
- Plumb, R.S.; Stumpf, C.L.; Granger, J.H.; Castro-Perez, J.; Haselden, J.N.; Dear, G.J. Use of liquid chromatography/time-of-flight mass spectrometry and multivariate statistical analysis shows promise for the detection of drug metabolites in biological fluids. Rapid. Commun. Mass Spectrom. 2003, 17, 2632–2638. [Google Scholar] [CrossRef]
- Chen, C.; Gonzalez, F.J.; Idle, J.R. LC–MS-based metabolomics in drug metabolism. Drug Metab. Rev. 2007, 39, 581–597. [Google Scholar] [CrossRef]
- Chen, C.; Krausz, K.W.; Idle, J.R.; Gonzalez, F.J. Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. J. Biol. Chem. 2008, 283, 4543–4559. [Google Scholar] [CrossRef]
- Gray, N.; Adesina-Georgiadis, K.; Chekmeneva, E.; Plumb, R.S.; Wilson, I.D.; Nicholson, J.K. Development of a Rapid Microbore Metabolic Profiling Ultraperformance Liquid Chromatography−Mass Spectrometry Approach for High-Throughput Phenotyping Studies. Anal. Chem. 2016, 88, 5742–5751. [Google Scholar] [CrossRef]
- Adesina-Georgiadis, K.N.; Gray, N.; Plumb, R.S.; Thompson, D.F.; Holmes, E.; Nicholson, J.K.; Wilson, I.D. The metabolic fate and effects of 2-Bromophenol in male Sprague-Dawley rats. Xenobiotica 2019, 49, 1352–1359. [Google Scholar] [CrossRef]
- Zhoua, Z.-M.; Wang, Y.-K.; Yana, D.-M.; Fanga, J.-H.; Xiao, X.-R.; Zhang, T.; Cheng, Y.; Xue, K.-P.; Li, F. Metabolic profiling of tyrosine kinase inhibitor nintedanib using metabolomics. J. Pharm. Biomed. Anal. 2020, 180, 113045. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lu, Y.-F.; Guan, X.; Zhao, M.; Wang, J.; Li, F. Characterizing novel metabolic pathways of melatonin receptor agonist agomelatine using metabolomic approaches. Biochem. Pharmacol. 2016, 109, 70–82. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Mairinger, T.J.; Causon, S.; Hann, S. The potential of ion mobility–mass spectrometry for non-targeted metabolomics. Curr. Opin. Chem. Biol. 2018, 42, 9–15. [Google Scholar] [CrossRef]
- Zhang, X.; Quinn, K.; Cruickshank-Quinn, C.; Reisdorph, R.; Reisdorph, N. The application of ion mobility mass spectrometry to metabolomics. Curr. Opin. Chem. Biol. 2018, 42, 60–66. [Google Scholar] [CrossRef]
- Nichols, C.M.; Dodds, J.N.; Rose, B.S.; Picache, J.A.; Morris, C.B.; Codreanu, S.G.; May, J.C.S.; Sherrod, D.; McLean, J.A. Untargeted molecular discovery in primary metabolism: Collision cross section as a molecular descriptor in ion mobility-mass spectrometry. Anal. Chem. 2018, 90, 14484–14492. [Google Scholar] [CrossRef]
- Harry, E.L.; Weston, D.J.; Bristow, A.W.T.; Wilson, I.D.; Creaser, C.S. An approach to enhancing coverage of the urinary metabonome using liquid chromatography-ion mobility-mass spectrometry. J. Chromatogr. B 2008, 871, 357–361. [Google Scholar] [CrossRef]
- Letertre, M.; Munjoma, M.C.; Slade, S.E.; Plumb, R.; Swann, J.; Coen, M.; Nicholson, J.K.; Wilson, I.D. Metabolic phenotyping using UPLC–MS and rapid microbore UPLC–IM–MS: Determination of the effect of different dietary regimes on the urinary metabolome of the rat. Chromatographia 2020, 83, 853–861. [Google Scholar] [CrossRef]
- Nye, L.C.; Williams, J.P.; Munjoma, N.C.; Letertre, M.P.M.; Coen, M.; Bouwmeester, R.; Martens, L.; Swann, J.R.; Nicholson, J.K.; Plumb, R.S.; et al. A comparison of collision cross section values obtained via travelling wave ion mobility-mass spectrometry and ultra high performance liquid chromatography-ion mobility-mass spectrometry: Application to the characterisation of metabolites in rat urine. J. Chromatogr. A 2019, 1602, 386–396. [Google Scholar] [CrossRef]
- Valavanidis, A.; Vlachogianni, T.; Fiotakis, C. 8-Hydroxy-2’-deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Health Part C 2009, 27, 120–139. [Google Scholar] [CrossRef]
- Poliaková, M.; Aebersold, D.M.; Zimmer, Y.; Medová, M. The relevance of tyrosine kinase inhibitors for global metabolic pathways in cancer. Mol. Cancer 2018, 17, 27. [Google Scholar] [CrossRef]
- Jensen, B.C.; Parry, T.L.; Huang, W.; Ilaiwy, A.; Bain, J.R.; Muehlbauer, M.; O’Neal, S.K.; Patterson, C.; Johnson, C.L.; Willis, M.S. Non-Targeted Metabolomics Analysis of the Effects of Tyrosine Kinase Inhibitors Sunitinib and Erlotinib on Heart, Muscle, Liver and Serum Metabolism In Vivo. Metabolites 2017, 7, 31. [Google Scholar] [CrossRef]
- Sangster, T.; Major, H.; Plumb, R.; Wilson, A.J.; Wilson, I.D. A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst 2006, 131, 1075–1078. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Wingate, J.E.; Wilson, I.D. Within-Day Reproducibility of an HPLC−MS-Based Method for Metabonomic Analysis: Application to Human Urine. J. Proteome Res. 2007, 6, 3291–3303. [Google Scholar] [CrossRef]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Suarez, E.; Hughes, C.; Gethings, L.; Giles, K.; Wildgoose, J.; Stapels, M.; Fadgen, K.E.; Geromanos, S.J.; Vissers, J.P.C.; Elortza, F.; et al. An Ion Mobility Assisted Data Independent LC-MS Strategy for the Analysis of Complex Biological Samples. Curr. Anal. Chem. 2013, 9, 199–211. [Google Scholar]
- Rainville, P.D.; Wilson, I.D.; Nicholson, J.K.; Isaac, G.; Mullin, L.; Langridge, J.I.; Plumb, R.S. Ion mobility spectrometry combined with ultra performance liquid chromatography/mass spectrometry for metabolic phenotyping of urine: Effects of column length, gradient duration and ion mobility spectrometry on metabolite detection. Anal. Chim. Acta 2017, 982, 1–8. [Google Scholar] [CrossRef] [PubMed]



| Compound | Adduct | Experimental RT (min) | Authentic Standard RT (min) | Experimental CCS (Å) | Predicted CCS (Å) | ΔCCS Experimental vs. Predicted (%) | Authentic Standard CCS (Å) | ΔCCS Measured vs. Predicted (%) |
|---|---|---|---|---|---|---|---|---|
| Tryptophan | [M + H] | 3.59 | 3.22 | 144.1 | 141.9 | 1.5 | 143.8 | 0.3 |
| Taurocholic acid | [M − H] | 5.77 | 5.94 | 205.9 | 205.9 | 0 | 207 | 0.5 |
| Arginyl-lysine | [M + H] | 5.04 | n/a | 166.0 | 173.0 | 3.0 | n/a | - |
| Arginyl-lysine | [M − H] | 5.03 | n/a | 167.2 | 172.8 | 3.2 | n/a | - |
| Deoxyguanosine | [M + H] | 0.69 | 0.7 | 155.7 | 154.1 | 1.0 | 153.7 | 1.3 |
| Asparaginyl-histidine | [M + H] | 2.68 | n/a | 157.9 | 156.7 | 0.8 | n/a | - |
| 8-hydroxy-deoxyguanosine | [M + H] | 2.24 | 2.51 | 159.8 | 158.4 | 0.9 | n/a | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molloy, B.; Mullin, L.; King, A.; Gethings, L.A.; Plumb, R.S.; Wilson, I.D. The Pharmacometabodynamics of Gefitinib after Intravenous Administration to Mice: A Preliminary UPLC–IM–MS Study. Metabolites 2021, 11, 379. https://doi.org/10.3390/metabo11060379
Molloy B, Mullin L, King A, Gethings LA, Plumb RS, Wilson ID. The Pharmacometabodynamics of Gefitinib after Intravenous Administration to Mice: A Preliminary UPLC–IM–MS Study. Metabolites. 2021; 11(6):379. https://doi.org/10.3390/metabo11060379
Chicago/Turabian StyleMolloy, Billy, Lauren Mullin, Adam King, Lee A. Gethings, Robert S. Plumb, and Ian D. Wilson. 2021. "The Pharmacometabodynamics of Gefitinib after Intravenous Administration to Mice: A Preliminary UPLC–IM–MS Study" Metabolites 11, no. 6: 379. https://doi.org/10.3390/metabo11060379
APA StyleMolloy, B., Mullin, L., King, A., Gethings, L. A., Plumb, R. S., & Wilson, I. D. (2021). The Pharmacometabodynamics of Gefitinib after Intravenous Administration to Mice: A Preliminary UPLC–IM–MS Study. Metabolites, 11(6), 379. https://doi.org/10.3390/metabo11060379






