Lung Adenocarcinoma Transcriptomic Analysis Predicts Adenylate Kinase Signatures Contributing to Tumor Progression and Negative Patient Prognosis
Abstract
1. Introduction
2. Results
2.1. Baseline Patient Sample Characteristics
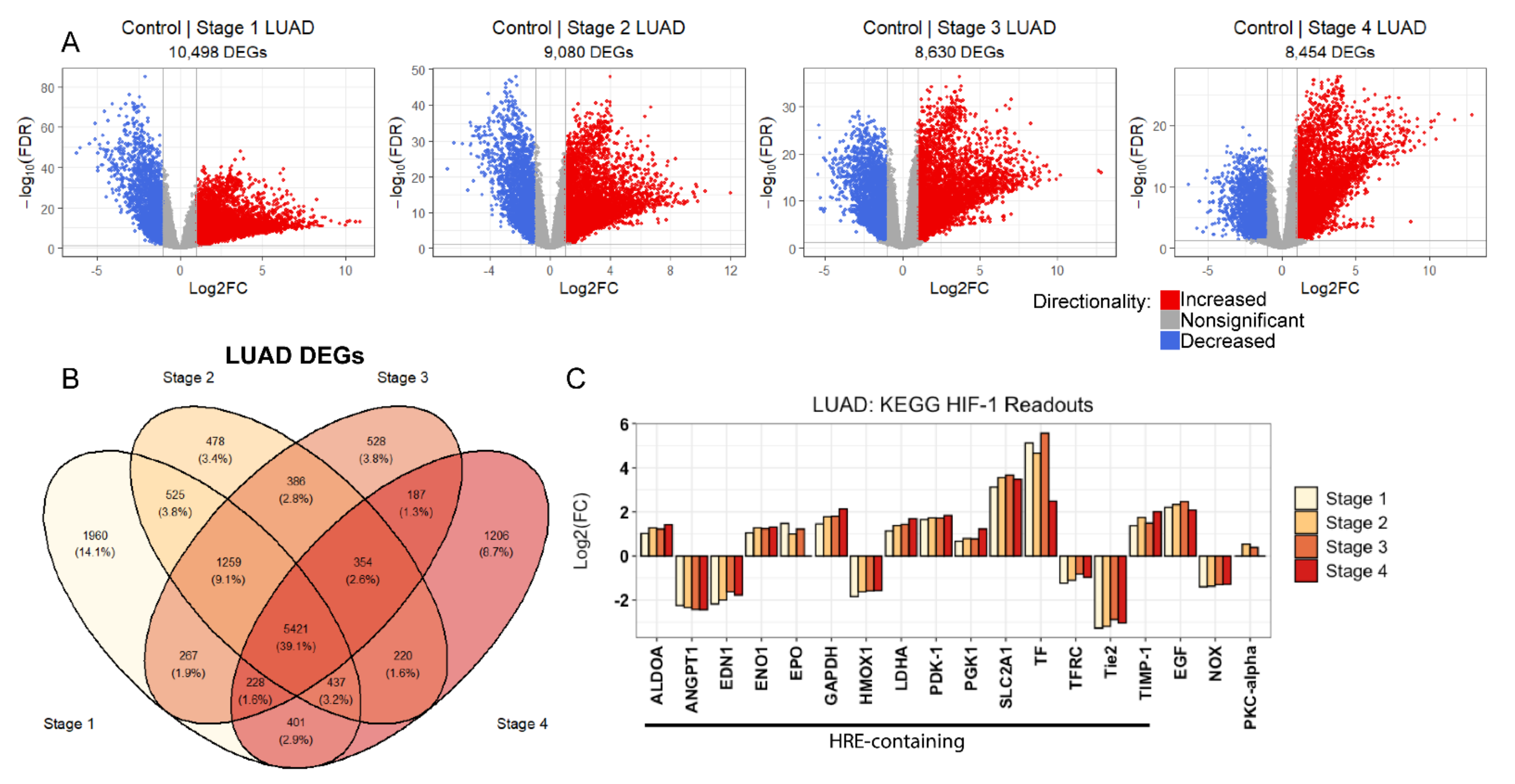
2.2. Evidence of Sustained Hypoxic Response through HIF-1 Signaling in LUAD
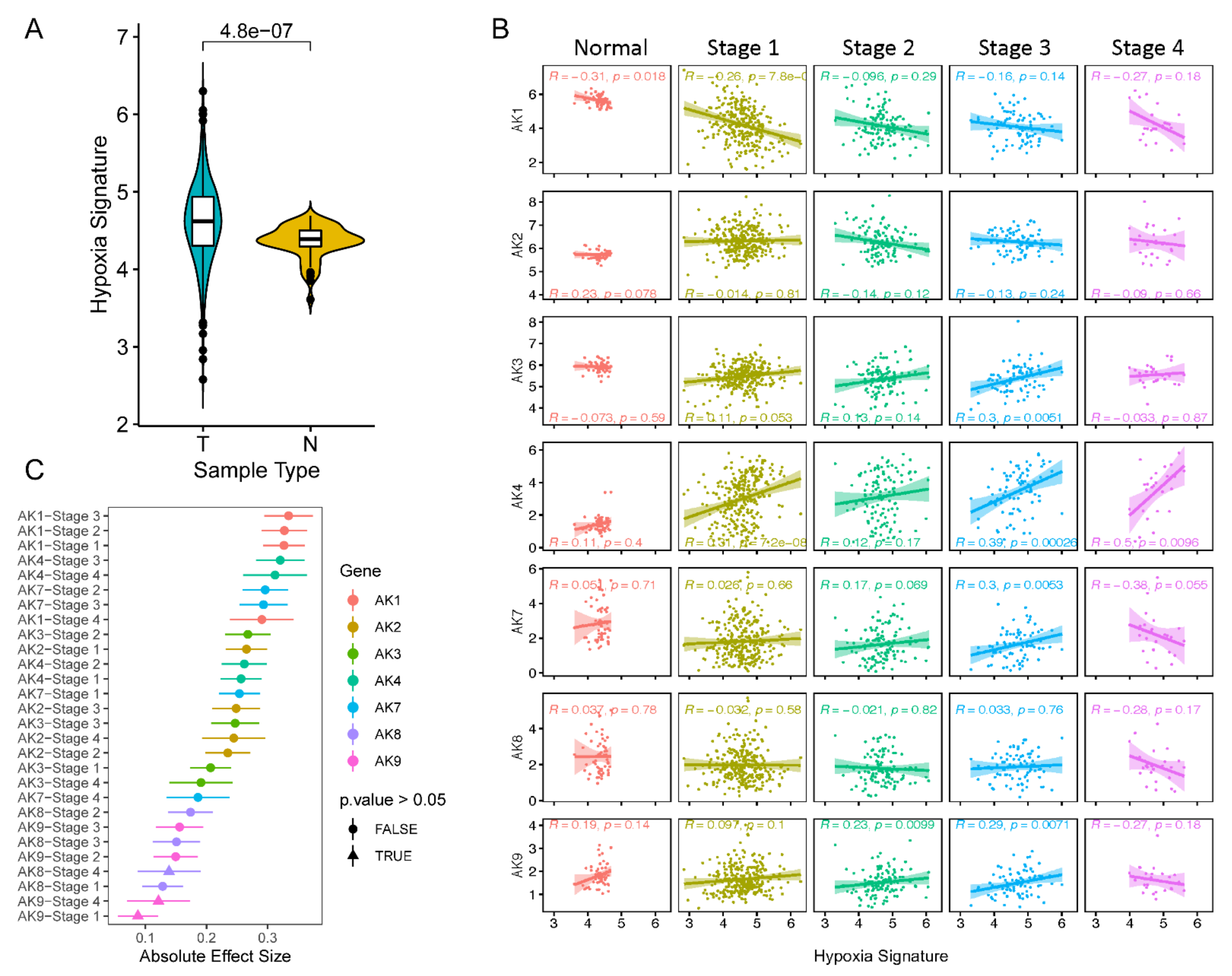
2.3. AK Levels Positively Correlate with Hypoxia Scores throughout LUAD Tumor Progression
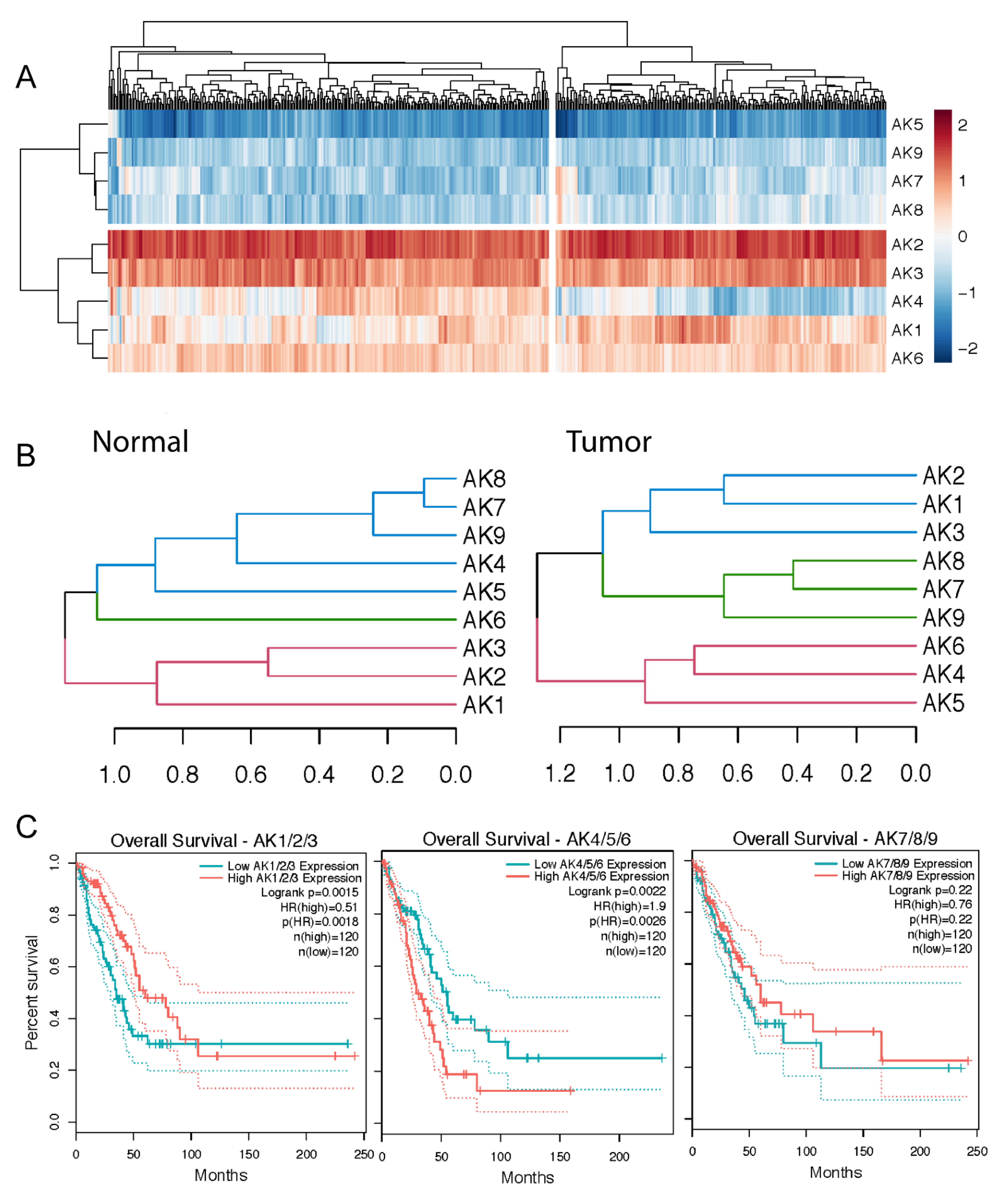
2.4. Two AK Clusters Predict Poor LUAD Patient Prognosis
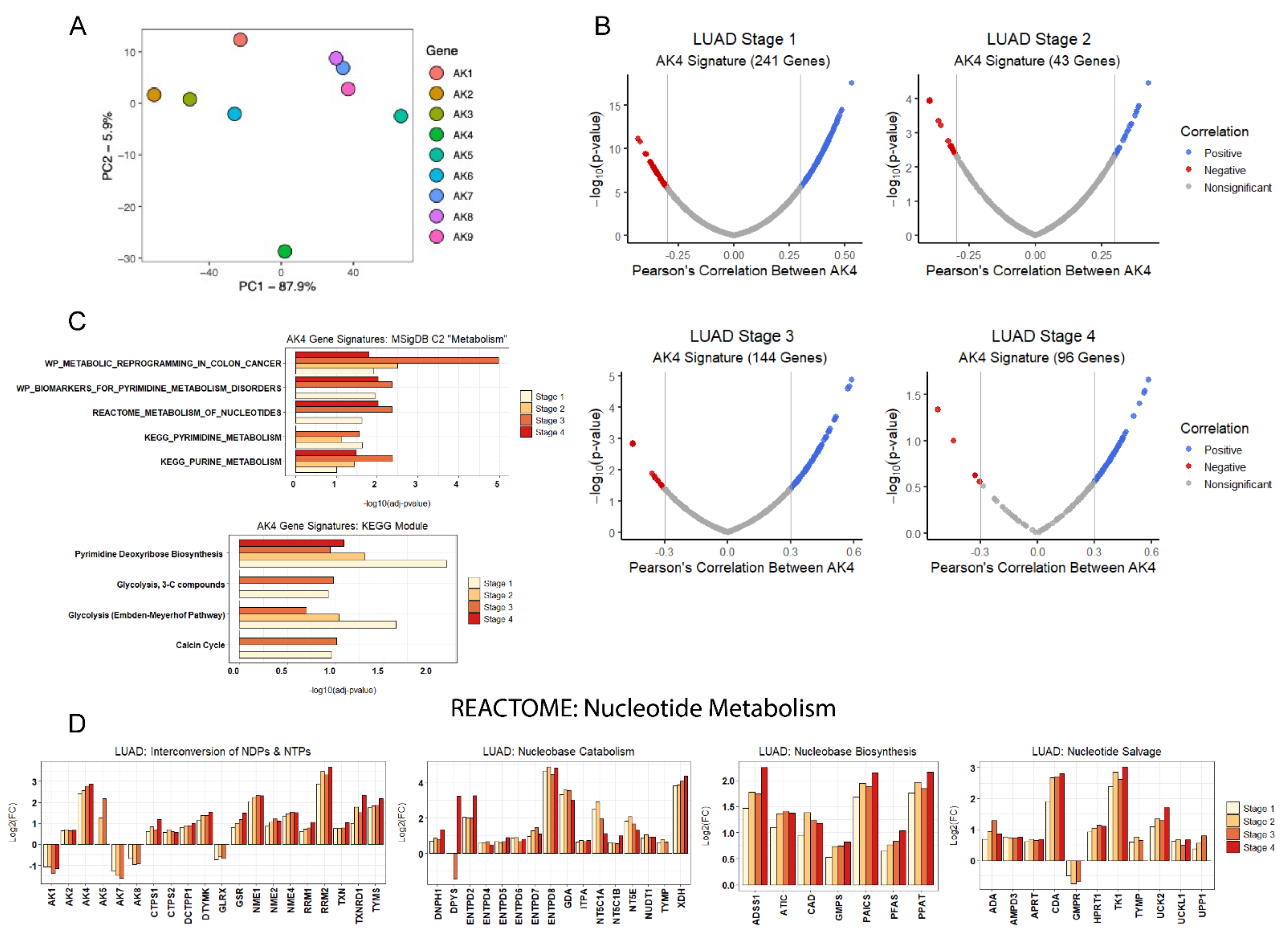
2.5. AK4 Expression Network Comprised of Perturbed Nucleotide Metabolism in LUAD
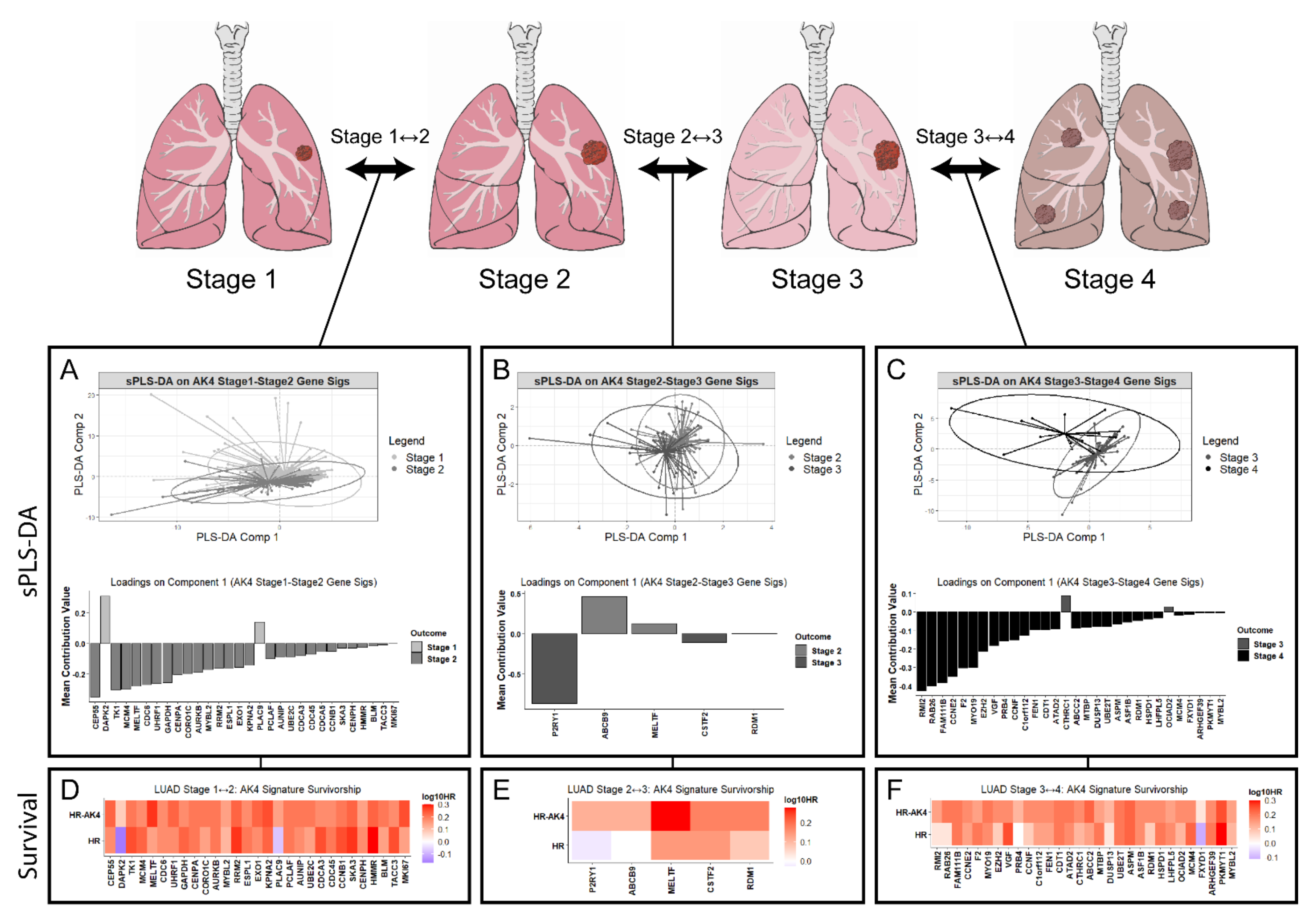
2.6. AK4 Co-Expression Networks Identify Potential Drivers of LUAD Tumor Progression
3. Discussion
4. Materials and Methods
4.1. TCGA Patient Tumor Data Curation and Consolidation
4.2. Gene Expression Query, Normalization, and Quantification
4.3. Statistical Analysis
4.4. Hypoxia Signature
4.5. Differential Gene Expression Analysis
4.6. Co-expression Network Creation and Over-Representation Analysis
4.7. Sparse Partial Least Squares-Discriminant Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schito, L.; Rey, S. Hypoxia: Turning vessels into vassals of cancer immunotolerance. Cancer Lett. 2020, 487, 74–84. [Google Scholar] [CrossRef]
- Schito, L.; Rey, S. Cell-Autonomous Metabolic Reprogramming in Hypoxia. Trends Cell Biol. 2018, 28, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Masson, N.; Ratcliffe, P.J. Hypoxia signaling pathways in cancer metabolism: The importance of co-selecting interconnected physiological pathways. Cancer Metab. 2014, 2, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Schito, L.; Semenza, G.L. Hypoxia-Inducible Factors: Master Regulators of Cancer Progression. Trends Cancer 2016, 2, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Masoud, G.N.; Li, W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 2015, 5, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Ziello, J.E.; Jovin, I.S.; Huang, Y. Hypoxia-Inducible Factor (HIF)-1 Regulatory Pathway and its Potential for Therapeutic Intervention in Malignancy and Ischemia. Yale J. Biol. Med. 2007, 80, 51–60. [Google Scholar]
- Hong, S.-S.; Lee, H.; Kim, K.-W. HIF-1α: A Valid Therapeutic Target for Tumor Therapy. Cancer Res. Treat. 2004, 36, 343–353. [Google Scholar] [CrossRef]
- Bhandari, V.; Li, C.H.; Bristow, R.G.; Boutros, P.C.; Consortium, P. Divergent mutational processes distinguish hypoxic and normoxic tumours. Nat. Commun. 2020, 11, 737. [Google Scholar] [CrossRef]
- Bhandari, V.; Hoey, C.; Liu, L.Y.; Lalonde, E.; Ray, J.; Livingstone, J.; Lesurf, R.; Shiah, Y.-J.; Vujcic, T.; Huang, X.; et al. Molecular landmarks of tumor hypoxia across cancer types. Nat. Genet. 2019, 51, 308–318. [Google Scholar] [CrossRef]
- Klepinin, A.; Zhang, S.; Klepinina, L.; Rebane-Klemm, E.; Terzic, A.; Kaambre, T.; Dzeja, P. Adenylate Kinase and Metabolic Signaling in Cancer Cells. Front. Oncol. 2020, 10, 660. [Google Scholar] [CrossRef]
- Ionescu, M.I. Adenylate Kinase: A Ubiquitous Enzyme Correlated with Medical Conditions. Protein J. 2019, 38, 120–133. [Google Scholar] [CrossRef]
- Jan, Y.H.; Lai, T.C.; Yang, C.J.; Huang, M.S.; Hsiao, M. A co-expressed gene status of adenylate kinase 1/4 reveals prognostic gene signature associated with prognosis and sensitivity to EGFR targeted therapy in lung adenocarcinoma. Sci. Rep. 2019, 9, 12329. [Google Scholar] [CrossRef]
- Jan, Y.-H.; Lai, T.-C.; Yang, C.-J.; Lin, Y.-F.; Huang, M.-S.; Hsiao, M. Adenylate kinase 4 modulates oxidative stress and stabilizes HIF-1α to drive lung adenocarcinoma metastasis. J. Hematol. Oncol. 2019, 12, 12. [Google Scholar] [CrossRef]
- Chin, W.-Y.; He, C.-Y.; Chow, T.W.; Yu, Q.-Y.; Lai, L.-C.; Miaw, S.-C. Adenylate Kinase 4 Promotes Inflammatory Gene Expression via Hif1α and AMPK in Macrophages. Front. Immunol. 2021, 12, 630318. [Google Scholar] [CrossRef]
- Semenza, G.L.; Agani, F.; Booth, G.; Forsythe, J.; Iyer, N.; Jiang, B.H.; Leung, S.; Roe, R.; Wiener, C.; Yu, A. Structural and functional analysis of hypoxia-inducible factor 1. Kidney Int. 1997, 51, 553–555. [Google Scholar] [CrossRef]
- Jiang, B.-H.; Rue, E.; Wang, G.L.; Roe, R.; Semenza, G.L. Dimerization, DNA Binding, and Transactivation Properties of Hypoxia-inducible Factor 1. J. Biol. Chem. 1996, 271, 17771–17778. [Google Scholar] [CrossRef] [PubMed]
- Berra, E.; Benizri, E.; Ginouvès, A.; Volmat, V.; Roux, D.; Pouysségur, J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003, 22, 4082–4090. [Google Scholar] [CrossRef] [PubMed]
- Bruick, R.K.; McKnight, S.L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 2001, 294, 1337–1340. [Google Scholar] [CrossRef] [PubMed]
- Ivan, M.; Kondo, K.; Yang, H.; Kim, W.; Valiando, J.; Ohh, M.; Salic, A.; Asara, J.M.; Lane, W.S.; Kaelin, W.G.J. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 2001, 292, 464–468. [Google Scholar] [CrossRef]
- Kaelin, W.G.J. Cancer and Altered Metabolism: Potential Importance of HIF and 2-Oxoglutarate-dependent Dioxygenases. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 335–345. [Google Scholar] [CrossRef]
- Paltoglou, S.; Roberts, B.J. HIF-1α and EPAS ubiquitination mediated by the VHL tumour suppressor involves flexibility in the ubiquitination mechanism, similar to other RING E3 ligases. Oncogene 2007, 26, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Groulx, I.; Lee, S. Oxygen-Dependent Ubiquitination and Degradation of Hypoxia-Inducible Factor Requires Nuclear-Cytoplasmic Trafficking of the von Hippel-Lindau Tumor Suppressor Protein. Mol. Cell. Biol. 2002, 22, 5319–5336. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Hypoxia-Inducible Factors in Physiology and Medicine. Cell 2012, 148, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Qingdong, K.; Max, C. Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 2006, 70, 1469–1480. [Google Scholar] [CrossRef]
- Kietzmann, T.; Mennerich, D.; Dimova, E.Y. Hypoxia-Inducible Factors (HIFs) and Phosphorylation: Impact on Stability, Localization, and Transactivity. Front. Cell Dev. Biol. 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Tal, R.; Shaish, A.; Bangio, L.; Peled, M.; Breitbart, E.; Harats, D. Activation of C-transactivation domain is essential for optimal HIF-1 alpha-mediated transcriptional and angiogenic effects. Microvasc. Res. 2008, 76, 1–6. [Google Scholar] [CrossRef]
- Semenza, G.L. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 2001, 13, 167–171. [Google Scholar] [CrossRef]
- Wenger, R.H.; Stiehl, D.P.; Camenisch, G. Integration of Oxygen Signaling at the Consensus HRE. Sci. STKE 2005, 2005. [Google Scholar] [CrossRef]
- Wong, B.W.; Kuchnio, A.; Bruning, U.; Carmeliet, P. Emerging novel functions of the oxygen-sensing prolyl hydroxylase domain enzymes. Trends Biochem. Sci. 2013, 38, 3–11. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; Chandel, N.S. Mitochondrial reactive oxygen species regulate hypoxic signaling. Curr. Opin. Cell Biol. 2009, 21, 894–899. [Google Scholar] [CrossRef]
- Richard, D.E.; Berra, E.; Pouysségur, J. Nonhypoxic Pathway Mediates the Induction of Hypoxia-inducible Factor 1α in Vascular Smooth Muscle Cells. J. Biol. Chem. 2000, 275, 26765–26771. [Google Scholar] [CrossRef]
- Finley, L.W.S.; Carracedo, A.; Lee, J.; Souza, A.; Egia, A.; Zhang, J.; Teruya-Feldstein, J.; Moreira, P.I.; Cardoso, S.M.; Clish, C.B.; et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 2011, 19, 416–428. [Google Scholar] [CrossRef]
- Gerald, D.; Berra, E.; Frapart, Y.M.; Chan, D.A.; Giaccia, A.J.; Mansuy, D.; Pouysségur, J.; Yaniv, M.; Mechta-Grigoriou, F. JunD Reduces Tumor Angiogenesis by Protecting Cells from Oxidative Stress. Cell 2004, 118, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Ghaloul-Gonzalez, L.; Mohsen, A.-W.; Karunanidhi, A.; Seminotti, B.; Chong, H.; Madan-Khetarpal, S.; Sebastian, J.; Vockley, C.W.; Reyes-Múgica, M.; Vander Lugt, M.T.; et al. Reticular Dysgenesis and Mitochondriopathy Induced by Adenylate Kinase 2 Deficiency with Atypical Presentation. Sci. Rep. 2019, 9, 15739. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Ravi, R.; Pham, V.; Bedi, A.; Chatterjee, A.; Sidransky, D. Adenylate kinase 3 sensitizes cells to cigarette smoke condensate vapor induced cisplatin resistance. PLoS ONE 2011, 6, e20806. [Google Scholar] [CrossRef]
- Ji, Y.; Yang, C.; Tang, Z.; Yang, Y.; Tian, Y.; Yao, H.; Zhu, X.; Zhang, Z.; Ji, J.; Zheng, X. Adenylate kinase hCINAP determines self-renewal of colorectal cancer stem cells by facilitating LDHA phosphorylation. Nat. Commun. 2017, 8, 15308. [Google Scholar] [CrossRef]
- Liu, X.; Gonzalez, G.; Dai, X.; Miao, W.; Yuan, J.; Huang, M.; Bade, D.; Li, L.; Sun, Y.; Wang, Y. Adenylate Kinase 4 Modulates the Resistance of Breast Cancer Cells to Tamoxifen through an m6A-Based Epitranscriptomic Mechanism. Mol. Ther. 2020, 28, 2593–2604. [Google Scholar] [CrossRef]
- Hu, C.-J.; Iyer, S.; Sataur, A.; Covello, K.L.; Chodosh, L.A.; Simon, M.C. Differential Regulation of the Transcriptional Activities of Hypoxia-Inducible Factor 1 Alpha (HIF-1α) and HIF-2α in Stem Cells. Mol. Cell. Biol. 2006, 26, 3514–3526. [Google Scholar] [CrossRef]
- Fujisawa, K.; Terai, S.; Takami, T.; Yamamoto, N.; Yamasaki, T.; Matsumoto, T.; Yamaguchi, K.; Owada, Y.; Nishina, H.; Noma, T.; et al. Modulation of anti-cancer drug sensitivity through the regulation of mitochondrial activity by adenylate kinase 4. J. Exp. Clin. Cancer Res. 2016, 35, 48. [Google Scholar] [CrossRef]
- Jan, Y.H.; Tsai, H.Y.; Yang, C.J.; Huang, M.S.; Yang, Y.F.; Lai, T.C.; Lee, C.H.; Jeng, Y.M.; Huang, C.Y.; Su, J.L.; et al. Adenylate kinase-4 is a marker of poor clinical outcomes that promotes metastasis of lung cancer by downregulating the transcription factor ATF3. Cancer Res. 2012, 72, 5119–5129. [Google Scholar] [CrossRef] [PubMed]
- Xin, F.; Yao, D.-W.; Fan, L.; Liu, J.-H.; Liu, X.-D. Adenylate kinase 4 promotes bladder cancer cell proliferation and invasion. Clin. Exp. Med. 2019, 19, 525–534. [Google Scholar] [CrossRef]
- Everson, W.V.; Flaim, K.E.; Susco, D.M.; Kimball, S.R.; Jefferson, L.S. Effect of amino acid deprivation on initiation of protein synthesis in rat hepatocytes. Am. J. Physiol.-Cell Physiol. 1989, 256, C18–C27. [Google Scholar] [CrossRef] [PubMed]
- Higashimura, Y.; Nakajima, Y.; Yamaji, R.; Harada, N.; Shibasaki, F.; Nakano, Y.; Inui, H. Up-regulation of glyceraldehyde-3-phosphate dehydrogenase gene expression by HIF-1 activity depending on Sp1 in hypoxic breast cancer cells. Arch. Biochem. Biophys. 2011, 509, 1–8. [Google Scholar] [CrossRef]
- Lu, S.; Gu, X.; Hoestje, S.; Epner, D.E. Identification of an additional hypoxia responsive element in the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Biochim. Biophys. Acta Gene Struct. Expr. 2002, 1574, 152–156. [Google Scholar] [CrossRef]
- Jiang, Z.; Wang, X.; Li, J.; Yang, H.; Lin, X. Aldolase A as a prognostic factor and mediator of progression via inducing epithelial–mesenchymal transition in gastric cancer. J. Cell. Mol. Med. 2018, 22, 4377–4386. [Google Scholar] [CrossRef]
- Minchenko, O.; Opentanova, I.; Minchenko, D.; Ogura, T.; Esumi, H. Hypoxia induces transcription of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase-4 gene via hypoxia-inducible factor-1α activation. FEBS Lett. 2004, 576, 14–20. [Google Scholar] [CrossRef]
- Mo, Z.; Yu, L.; Cao, Z.; Hu, H.; Luo, S.; Zhang, S. Identification of a Hypoxia-Associated Signature for Lung Adenocarcinoma. Front. Genet. 2020, 11, 647. [Google Scholar] [CrossRef] [PubMed]
- Serganova, I.; Cohen, I.J.; Vemuri, K.; Shindo, M.; Maeda, M.; Mane, M.; Moroz, E.; Khanin, R.; Satagopan, J.; Koutcher, J.A.; et al. LDH-A regulates the tumor microenvironment via HIF-signaling and modulates the immune response. PLoS ONE 2018, 13, e0203965. [Google Scholar] [CrossRef]
- Semenza, G.L. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem. J. 2007, 405, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Han, Z.; He, S.; Wu, X.; Chen, T.; Shao, C.; Chen, D.; Su, N.; Chen, Y.; Wang, T.; et al. HIF1/2α mediates hypoxia-induced LDHA expression in human pancreatic cancer cells. Oncotarget 2017, 8, 24840–24852. [Google Scholar] [CrossRef]
- Semenza, G.L.; Jiang, B.-H.; Leung, S.W.; Passantino, R.; Concordet, J.-P.; Maire, P.; Giallongo, A. Hypoxia Response Elements in the Aldolase A, Enolase 1, and Lactate Dehydrogenase A Gene Promoters Contain Essential Binding Sites for Hypoxia-inducible Factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef]
- Fu, D.; He, C.; Wei, J.; Zhang, Z.; Luo, Y.; Tan, H.; Ren, C. PGK1 is a Potential Survival Biomarker and Invasion Promoter by Regulating the HIF-1α–Mediated Epithelial-Mesenchymal Transition Process in Breast Cancer. Cell. Physiol. Biochem. 2018, 51, 2434–2444. [Google Scholar] [CrossRef]
- Javan, B.; Shahbazi, M. Hypoxia-inducible tumour-specific promoters as a dual-targeting transcriptional regulation system for cancer gene therapy. Ecancermedicalscience 2017, 11. [Google Scholar] [CrossRef]
- Chateauvieux, S.; Grigorakaki, C.; Morceau, F.; Dicato, M.; Diederich, M. Erythropoietin, erythropoiesis and beyond. Biochem. Pharmacol. 2011, 82, 1291–1303. [Google Scholar] [CrossRef]
- Reed, M.J.; Koike, T.; Sadoun, E.; Sage, E.H.; Puolakkainen, P. Inhibition of TIMP1 enhances angiogenesis in vivo and cell migration in vitro. Microvasc. Res. 2003, 65, 9–17. [Google Scholar] [CrossRef]
- van Cruijsen, H.; Giaccone, G.; Hoekman, K. Epidermal growth factor receptor and angiogenesis: Opportunities for combined anticancer strategies. Int. J. Cancer 2005, 117, 883–888. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Tchernyshyov, I.; Semenza, G.L.; Dang, C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006, 3, 177–185. [Google Scholar] [CrossRef]
- Park, J.H.; Choi, H.; Kim, Y.B.; Kim, Y.S.; Sheen, S.S.; Choi, J.-H.; Lee, H.L.; Lee, K.S.; Chung, W.Y.; Lee, S.; et al. Serum angiopoietin-1 as a prognostic marker in resected early stage lung cancer. Lung Cancer 2009, 66, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-I.; Hsu, Y.-C.; Lien, C.-F.; Lin, J.-H.; Chiu, H.-W.; Yang, K.-T. Non-Lethal Levels of Oxidative Stress in Response to Short-Term Intermittent Hypoxia Enhance Ca2+ Handling in Neonatal Rat Cardiomyocytes. Cell. Physiol. Biochem. 2014, 33, 513–527. [Google Scholar] [CrossRef]
- Ding, H.L.; Zhu, H.F.; Dong, J.W.; Zhu, W.Z.; Zhou, Z.N. Intermittent hypoxia protects the rat heart against ischemia/reperfusion injury by activating protein kinase C. Life Sci. 2004, 75, 2587–2603. [Google Scholar] [CrossRef]
- Greig, A.V.H.; Burnstock, G.; Linge, C.; Healy, V.; Lim, P.; Clayton, E.; Rustin, M.H.A.; McGrouther, D.A. Expression of Purinergic Receptors in Non-melanoma Skin Cancers and Their Functional Roles in A431 Cells. J. Investig. Dermatol. 2003, 121, 315–327. [Google Scholar] [CrossRef]
- Tong, L.; Liu, J.; Yan, W.; Cao, W.; Shen, S.; Li, K.; Li, L.; Niu, G. RDM1 plays an oncogenic role in human lung adenocarcinoma cells. Sci. Reports 2018, 8. [Google Scholar] [CrossRef]
- Saxena, K.; Jolly, M.K. Acute vs. Chronic vs. Cyclic Hypoxia: Their Differential Dynamics, Molecular Mechanisms, and Effects on Tumor Progression. Biomolecules 2019, 9, 339. [Google Scholar] [CrossRef]
- Reiterer, M.; Colaço, R.; Emrouznejad, P.; Jensen, A.; Rundqvist, H.; Johnson, R.S.; Branco, C. Acute and chronic hypoxia differentially predispose lungs for metastases. Sci. Rep. 2019, 9, 10246. [Google Scholar] [CrossRef] [PubMed]
- Nanduri, J.; Vaddi, D.R.; Khan, S.A.; Wang, N.; Makarenko, V.; Semenza, G.L.; Prabhakar, N.R. HIF-1α Activation by Intermittent Hypoxia Requires NADPH Oxidase Stimulation by Xanthine Oxidase. PLoS ONE 2015, 10, e0119762. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yuan, G.; Khan, S.A.; Luo, W.; Nanduri, J.; Semenza, G.L.; Prabhakar, N.R. Hypoxia-Inducible Factor 1 Mediates Increased Expression of NADPH Oxidase-2 in Response to Intermittent Hypoxia. J. Cell. Physiol. 2011, 226, 2925–2933. [Google Scholar] [CrossRef]
- Singleton, D.C.; Macann, A.; Wilson, W.R. Therapeutic targeting of the hypoxic tumour microenvironment. Nat. Rev. Clin. Oncol. 2021, 18, 751–772. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Vanhove, K.; Graulus, G.J.; Mesotten, L.; Thomeer, M.; Derveaux, E.; Noben, J.P.; Guedens, W.; Adriaensens, P. The Metabolic Landscape of Lung Cancer: New Insights in a Disturbed Glucose Metabolism. Front. Oncol. 2019, 9, 1215. [Google Scholar] [CrossRef]
- Louis, E.; Adriaensens, P.; Guedens, W.; Bigirumurame, T.; Baeten, K.; Vanhove, K.; Vandeurzen, K.; Darquennes, K.; Vansteenkiste, J.; Dooms, C.; et al. Detection of lung cancer through metabolic changes measured in blood plasma. J. Thorac. Oncol. 2016, 11, 516–523. [Google Scholar] [CrossRef]
- Jagarlamudi, K.K.; Shaw, M. Thymidine kinase 1 as a tumor biomarker: Technical advances offer new potential to an old biomarker. Biomark. Med. 2018, 12, 1035–1048. [Google Scholar] [CrossRef]
- Malvi, P.; Janostiak, R.; Nagarajan, A.; Cai, G.; Wajapeyee, N. Loss of thymidine kinase 1 inhibits lung cancer growth and metastatic attributes by reducing GDF15 expression. PLoS Genet. 2019, 15, e1008439. [Google Scholar] [CrossRef]
- Lim, Y.T.; Prabhu, N.; Dai, L.; Go, K.D.; Chen, D.; Sreekumar, L.; Egeblad, L.; Eriksson, S.; Chen, L.; Veerappan, S.; et al. An efficient proteome-wide strategy for discovery and characterization of cellular nucleotide-protein interactions. PLoS ONE 2018, 13, e0208273. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, baw100. [Google Scholar] [CrossRef]
- Qian, J.; Rankin, E.B. Hypoxia-Induced Phenotypes that Mediate Tumor Heterogeneity. Adv. Exp. Med. Biol. 2019, 1136, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Lukovic, J.; Han, K.; Pintilie, M.; Chaudary, N.; Hill, R.P.; Fyles, A.; Milosevic, M. Intratumoral heterogeneity and hypoxia gene expression signatures: Is a single biopsy adequate? Clin. Transl. Radiat. Oncol. 2019, 19, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Ramos, M.; Schiffer, L.; Re, A.; Azhar, R.; Basunia, A.; Rodriguez, C.; Chan, T.; Chapman, P.; Davis, S.R.; Gomez-Cabrero, D.; et al. Software for the Integration of Multiomics Experiments in Bioconductor. Cancer Res. 2017, 77, e39–e42. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Lun, A.T.L.; Chen, Y.; Smyth, G.K. It’s DE-Licious: A Recipe for Differential Expression Analyses of RNA-Seq Experiments using Quasi-Likelihood Methods in EdgeR. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2016; Volume 1418, pp. 391–416. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Contreras-López, O.; Moyano, T.C.; Soto, D.C.; Gutiérrez, R.A. Step-by-Step Construction of Gene Co-Expression Networks from High-Throughput Arabidopsis RNA Sequencing Data. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2018; Volume 1761, pp. 275–301. [Google Scholar]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Chung, D.; Keles, S. Sparse partial least squares classification for high dimensional data. Stat. Appl. Genet. Mol. Biol. 2010, 9. [Google Scholar] [CrossRef] [PubMed]
- Lê Cao, K.A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
| Variable | TCGA-LUAD Cohort (n = 526 Patients) |
|---|---|
| Age | |
| <60 | 128 |
| ≥60 | 295 |
| Not reported | 103 |
| Sex | |
| Female | 282 |
| Male | 244 |
| Vitals | |
| Deceased | 190 |
| Alive | 336 |
| Tumor Stage | |
| 1 | 316 |
| 2 | 135 |
| 3 | 97 |
| 4 | 28 |
| Not reported | 9 |
| Sample Type | |
| Solid tissue normal (N) | 59 |
| Primary tumor (T) | 526 |
| Race | |
| American Indian or Alaska Native | 1 |
| Asian | 7 |
| Black or African American | 54 |
| White | 398 |
| Not reported | 66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chacon-Barahona, J.A.; Salladay-Perez, I.A.; Lanning, N.J. Lung Adenocarcinoma Transcriptomic Analysis Predicts Adenylate Kinase Signatures Contributing to Tumor Progression and Negative Patient Prognosis. Metabolites 2021, 11, 859. https://doi.org/10.3390/metabo11120859
Chacon-Barahona JA, Salladay-Perez IA, Lanning NJ. Lung Adenocarcinoma Transcriptomic Analysis Predicts Adenylate Kinase Signatures Contributing to Tumor Progression and Negative Patient Prognosis. Metabolites. 2021; 11(12):859. https://doi.org/10.3390/metabo11120859
Chicago/Turabian StyleChacon-Barahona, Jonathan A., Ivan A. Salladay-Perez, and Nathan James Lanning. 2021. "Lung Adenocarcinoma Transcriptomic Analysis Predicts Adenylate Kinase Signatures Contributing to Tumor Progression and Negative Patient Prognosis" Metabolites 11, no. 12: 859. https://doi.org/10.3390/metabo11120859
APA StyleChacon-Barahona, J. A., Salladay-Perez, I. A., & Lanning, N. J. (2021). Lung Adenocarcinoma Transcriptomic Analysis Predicts Adenylate Kinase Signatures Contributing to Tumor Progression and Negative Patient Prognosis. Metabolites, 11(12), 859. https://doi.org/10.3390/metabo11120859






