Reticulon-1C Involvement in Muscle Regeneration
Abstract
:1. Introduction
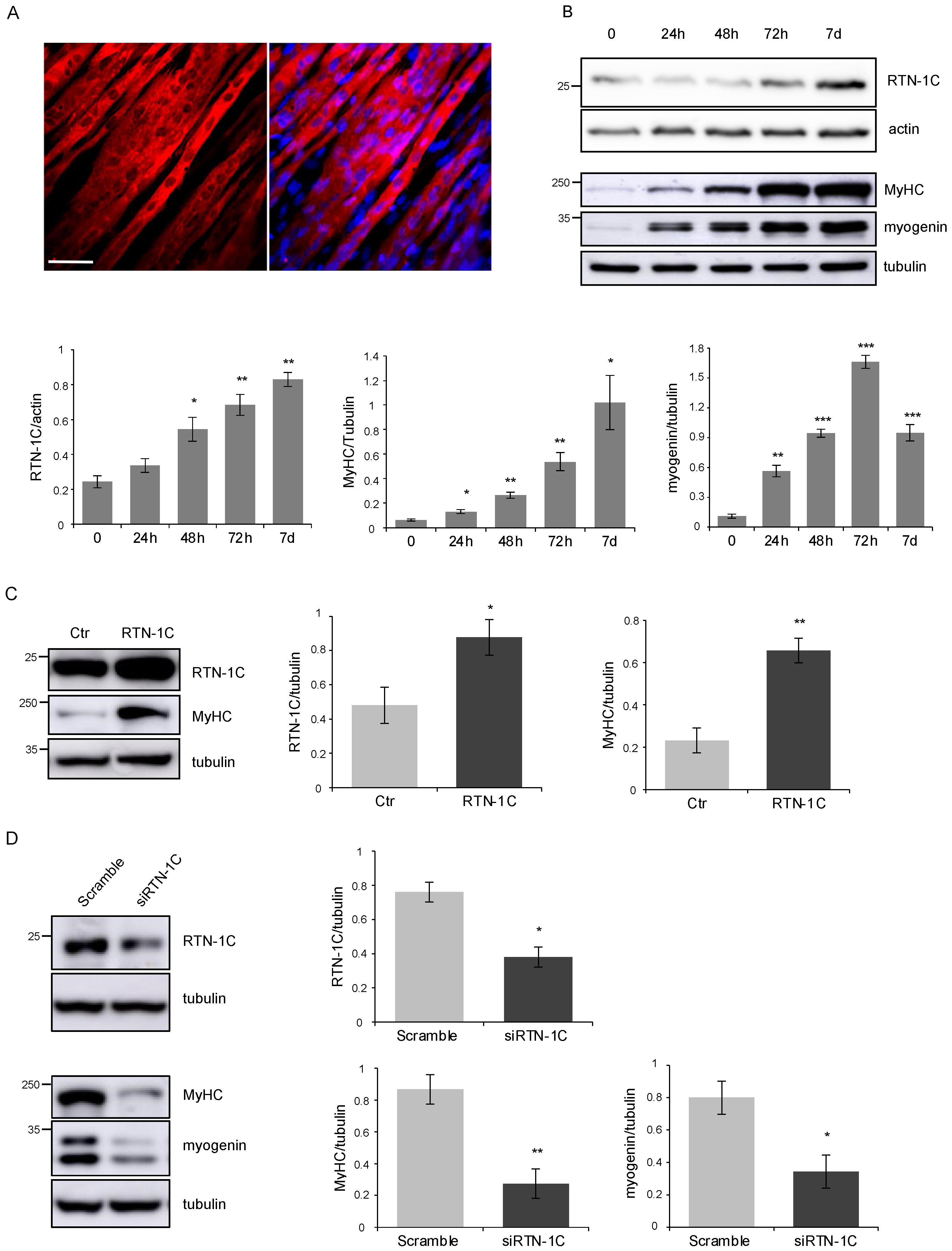
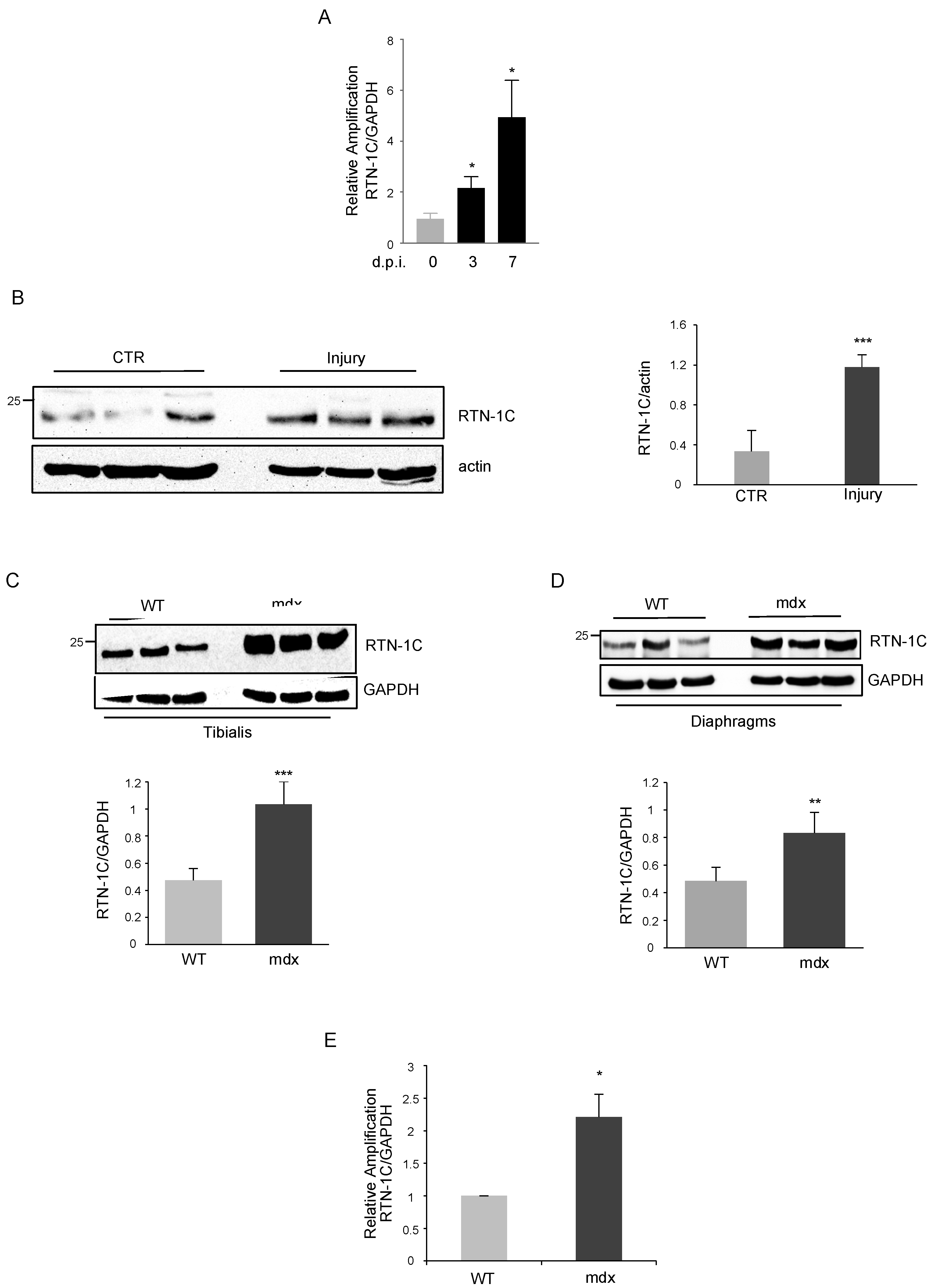
2. Results
3. Discussion
4. Materials and Methods
4.1. Cells
4.2. Mice
4.3. Cell Preparation and Isolation by FACS
4.4. Western Blot Analysis
4.5. Antibodies
4.6. Immunofluorescence
- Mouse anti-MyHC (produced in our laboratory by hybridoma cells) diluted 1:2.
- Anti-mouse Alexa555 (Molecular Probes) diluted 1:100.
4.7. Quantitative-RT PCR
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sui, S.X.; Williams, L.J.; Holloway-Kew, K.L.; Hyde, N.K.; Pasco, J.A. Skeletal Muscle Health and Cognitive Function: A Narrative Review. Int. J. Mol. Sci. 2020, 22, 255. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Cai, Z.; Li, D.; Zhang, Y.; He, M.; Yang, Y.; Liu, D.; Xie, W.; Li, Y.; Xiao, W. Myogenic Differentiation of Stem Cells for Skeletal Muscle Regeneration. Stem Cells Int. 2021, 2021, 8884283. [Google Scholar] [CrossRef] [PubMed]
- Reilly, B.D.; Franklin, C. Prevention of muscle wasting and osteoporosis: The value of examining novel animal models. J. Exp. Biol. 2016, 219, 2582–2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Malhotra, S.; Kumar, A. Nuclear factor-kappa B signaling in skeletal muscle atrophy. J. Mol. Med. 2008, 86, 1113–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamakawa, H.; Kusumoto, D.; Hashimoto, H.; Yuasa, S. Stem Cell Aging in Skeletal Muscle Regeneration and Disease. Int. J. Mol. Sci. 2020, 21, 1830. [Google Scholar] [CrossRef] [Green Version]
- Bohnert, K.R.; McMillan, J.D.; Kumar, A. Emerging roles of ER stress and unfolded protein response pathways in skeletal muscle health and disease. J. Cell. Physiol. 2018, 233, 67–78. [Google Scholar] [CrossRef]
- Deldicque, L.; Hespel, P.; Francaux, M. Endoplasmic reticulum stress in skeletal muscle: Origin and metabolic consequences. Exerc. Sport Sci. Rev. 2012, 40, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Afroze, D.; Kumar, A. ER stress in skeletal muscle remodeling and myopathies. FEBS J. 2019, 286, 379–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rayavarapu, S.; Coley, W.; Nagaraju, K. Endoplasmic Reticulum Stress in Skeletal Muscle Homeostasis and Disease. Curr. Rheumatol. Rep. 2012, 14, 238–243. [Google Scholar] [CrossRef] [Green Version]
- Hetz, C.; Saxena, S. ER stress and the unfolded protein response in neurodegeneration. Nat. Rev. Neurol. 2017, 13, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Pant, M.; Sopariwala, D.H.; Bal, N.C.; Lowe, J.; Delfín, D.A.; Rafael-Fortney, J.; Periasamy, M. Metabolic Dysfunction and Altered Mitochondrial Dynamics in the Utrophin-Dystrophin Deficient Mouse Model of Duchenne Muscular Dystrophy. PLoS ONE 2015, 10, e0123875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Strittmatter, S.M. The reticulons: A family of proteins with diverse functions. Genome Biol. 2007, 8, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Sano, F.; Fazi, B.; Citro, G.; Lovat, P.; Cesareni, G.; Piacentini, M. Glucosylceramide synthase and its functional interaction with RTN-1C regulate chemotherapeutic-induced apoptosis in neuroepithelioma cells. Cancer Res. 2003, 63, 3860–3865. [Google Scholar] [PubMed]
- Di Sano, F.; Fazi, B.; Tufi, R.; Nardacci, R.; Piacentini, M. Reticulon-1C acts as a molecular switch between endoplasmic reticulum stress and genotoxic cell death pathway in human neuroblastoma cells. J. Neurochem. 2007, 102, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Reali, V.; Mehdawy, B.; Nardacci, R.; Filomeni, G.; Risuglia, A.; Rossin, F.; Antonioli, M.; Marsella, C.; Fimia, G.M.; Piacentini, M.; et al. Reticulon protein-1C is a key component of MAMs. Biochim. Biophys. Acta Bioenerget. 2015, 1853, 733–745. [Google Scholar] [CrossRef]
- D’Eletto, M.; Risuglia, A.; Oliverio, S.; Mehdawy, B.; Nardacci, R.; Bordi, M.; Di Sano, F. Modulation of autophagy by RTN-1C: Role in autophagosome biogenesis. Cell Death Dis. 2019, 10, 868. [Google Scholar] [CrossRef] [PubMed]
- Fiacco, E.; Castagnetti, F.; Bianconi, V.; Madaro, L.; De Bardi, M.; Nazio, F.; D’Amico, A.; Bertini, E.; Cecconi, F.; Puri, P.L.; et al. Autophagy regulates satellite cell ability to regenerate normal and dystrophic muscles. Cell Death Differ. 2016, 23, 1839–1849. [Google Scholar] [CrossRef] [Green Version]
- Sartori, R.; Romanello, V.; Sandri, M. Mechanisms of muscle atrophy and hypertrophy: Implications in health and disease. Nat. Commun. 2021, 12, 330. [Google Scholar] [CrossRef] [PubMed]
- Paolini, A.; Omairi, S.; Mitchell, R.; Vaughan, D.; Matsakas, A.; Vaiyapuri, S.; Ricketts, T.; Rubinsztein, D.C.; Patel, K. Attenuation of autophagy impacts on muscle fibre development, starvation induced stress and fibre regeneration following acute injury. Sci. Rep. 2018, 8, 9062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandoná, M.; Consalvi, S.; Tucciarone, L.; De Bardi, M.; Scimeca, M.; Angelini, D.F.; Buffa, V.; D’Amico, A.; Bertini, E.S.; Cazzaniga, S.; et al. HDAC inhibitors tune miRNAs in extracellular vesicles of dystrophic muscle-resident mesenchymal cells. EMBO Rep. 2020, 21, e50863. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.A.; Olsen, I.; Zammit, P.S.; Heslop, L.; Petrie, A.; Partridge, T.A.; Morgan, J.E. Stem Cell Function, Self-Renewal, and Behavioral Heterogeneity of Cells from the Adult Muscle Satellite Cell Niche. Cell 2005, 122, 289–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, T.H.T.; Rando, T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013, 14, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Bentzinger, C.F.; Wang, Y.X.; Dumont, N.A.; Rudnicki, M.A. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 2013, 14, 1062–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrotta, C.; Cattaneo, M.G.; Molteni, R.; De Palma, C. Autophagy in the Regulation of Tissue Differentiation and Homeostasis. Front. Cell Dev. Biol. 2020, 8, 602901. [Google Scholar] [CrossRef] [PubMed]
- Lepper, C.; Partridge, T.A.; Fan, C.-M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 2011, 138, 3639–3646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hens, J.; Nuydens, R.; Geerts, H.; Senden, N.H.M.; Van De Ven, W.J.M.; Roebroek, A.J.M.; Van De Velde, H.J.K.; Ramaekers, F.C.S.; Broers, J.L.V. Neuronal differentiation is accompanied by NSP-C expression. Cell Tissue Res. 1998, 292, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Roebroek, A.J.; Contreras, B.; Pauli, I.G.; Van de Ven, W.J. cDNA Cloning, Genomic Organization, and Expression of the HumanRTN2Gene, a Member of a Gene Family Encoding Reticulons. Genomics 1998, 51, 98–106. [Google Scholar] [CrossRef]
- Fazi, B.; Melino, S.M.; De Rubeis, S.; Bagni, C.; Paci, M.; Piacentini, M.; Di Sano, F. Acetylation of RTN-1C regulates the induction of ER stress by the inhibition of HDAC activity in neuroectodermal tumors. Oncogene 2009, 28, 3814–3824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marinkovic, M.; Fuoco, C.; Sacco, F.; Perpetuini, A.C.; Giuliani, G.; Micarelli, E.; Pavlidou, T.; Petrilli, L.L.; Reggio, A.; Riccio, F.; et al. Fibro-adipogenic pro-genitors of dystrophic mice are insensitive to NOTCH regulation of adipogenesis. Life Sci. Alliance 2019, 2, e201900437. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossin, F.; Avitabile, E.; Catarinella, G.; Fornetti, E.; Testa, S.; Oliverio, S.; Gargioli, C.; Cannata, S.; Latella, L.; Di Sano, F. Reticulon-1C Involvement in Muscle Regeneration. Metabolites 2021, 11, 855. https://doi.org/10.3390/metabo11120855
Rossin F, Avitabile E, Catarinella G, Fornetti E, Testa S, Oliverio S, Gargioli C, Cannata S, Latella L, Di Sano F. Reticulon-1C Involvement in Muscle Regeneration. Metabolites. 2021; 11(12):855. https://doi.org/10.3390/metabo11120855
Chicago/Turabian StyleRossin, Federica, Elena Avitabile, Giorgia Catarinella, Ersilia Fornetti, Stefano Testa, Serafina Oliverio, Cesare Gargioli, Stefano Cannata, Lucia Latella, and Federica Di Sano. 2021. "Reticulon-1C Involvement in Muscle Regeneration" Metabolites 11, no. 12: 855. https://doi.org/10.3390/metabo11120855
APA StyleRossin, F., Avitabile, E., Catarinella, G., Fornetti, E., Testa, S., Oliverio, S., Gargioli, C., Cannata, S., Latella, L., & Di Sano, F. (2021). Reticulon-1C Involvement in Muscle Regeneration. Metabolites, 11(12), 855. https://doi.org/10.3390/metabo11120855





