Integrated Metabolomics and Proteomics Dynamics of Serum Samples Reveals Dietary Zeolite Clinoptilolite Supplementation Restores Energy Balance in High Yielding Dairy Cows
Abstract
:1. Introduction
2. Results
2.1. Confirmation of NEB by NEFA and BHB Levels
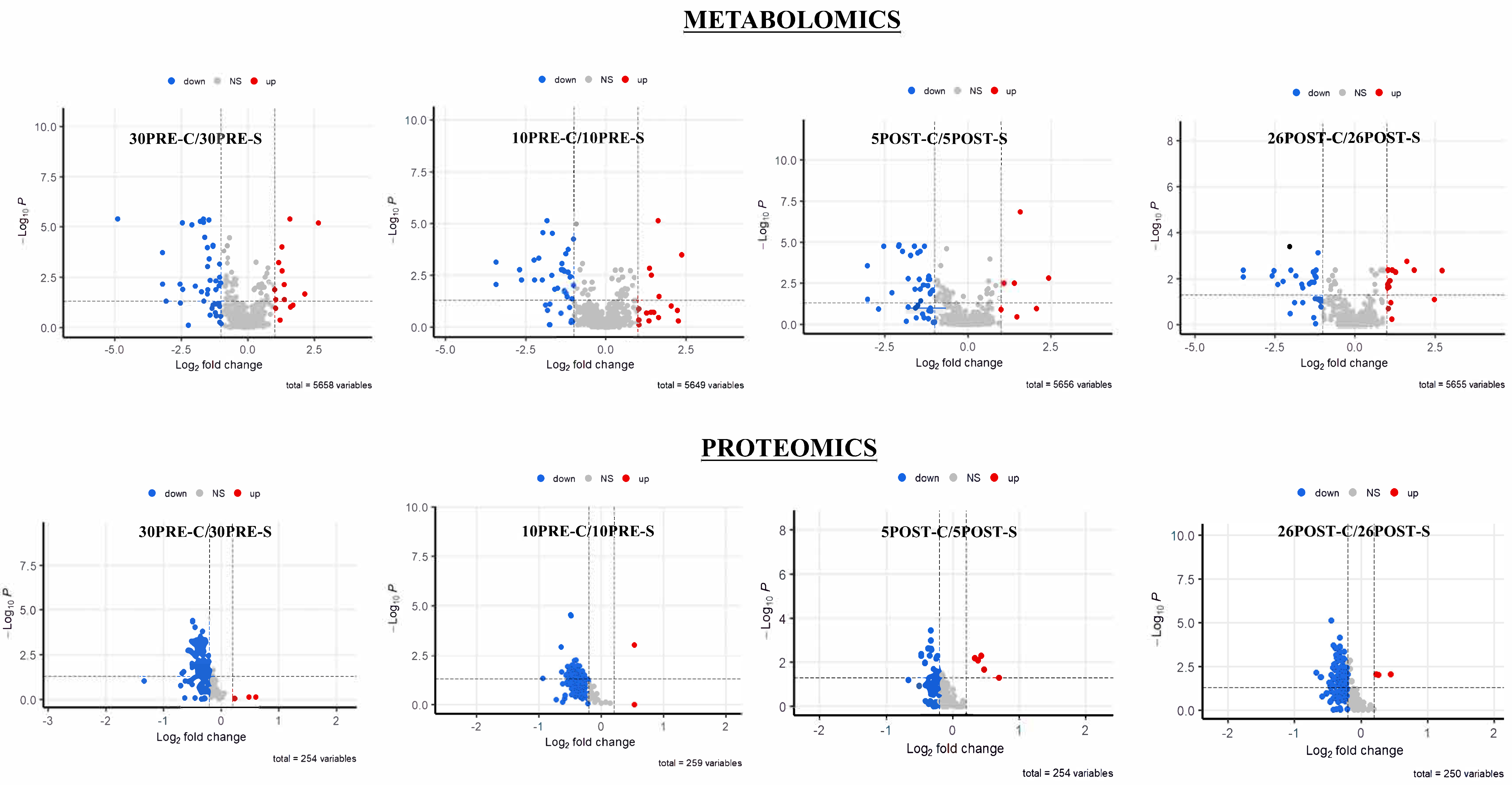
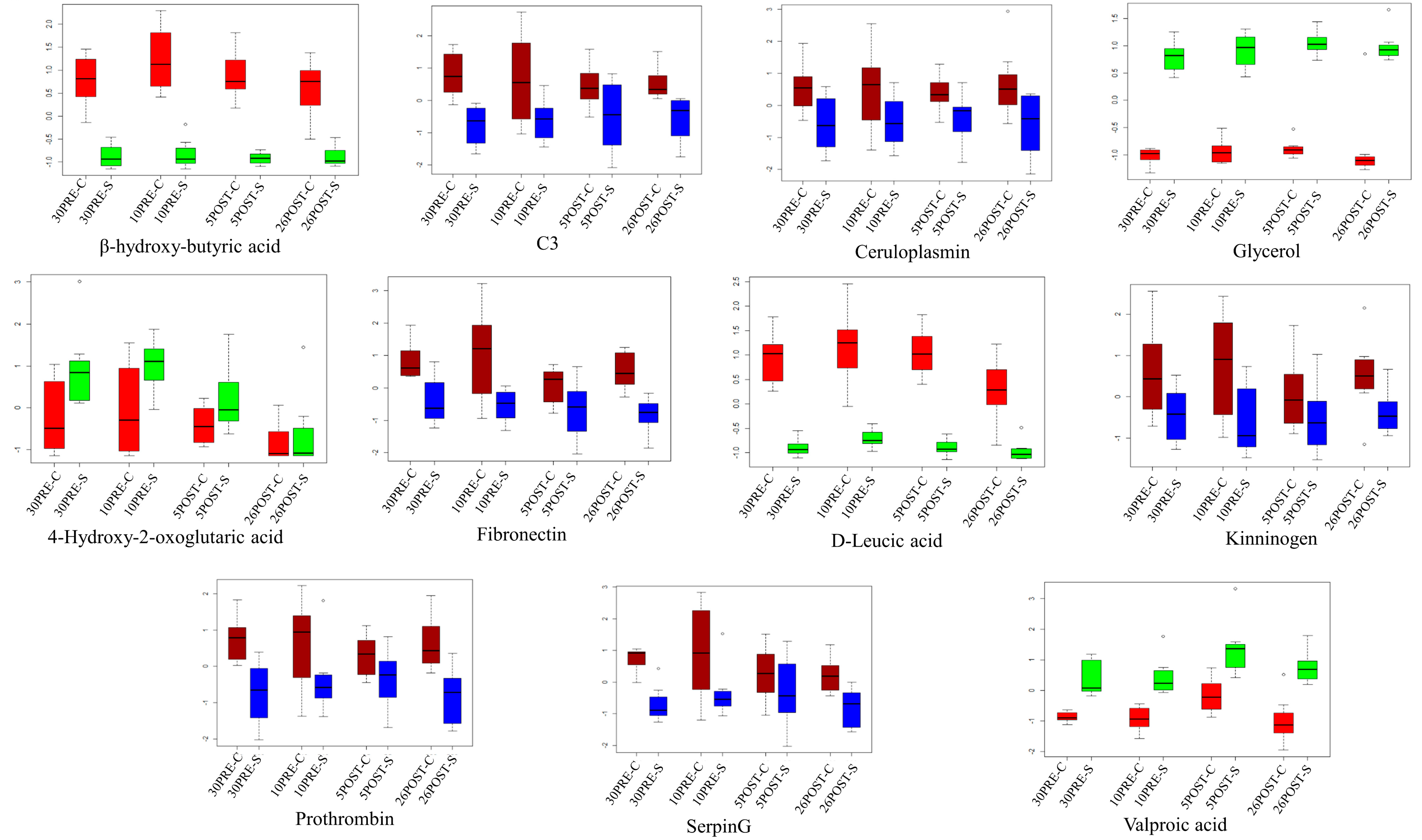
2.2. Metabolomics and Proteomics Statistical Analysis
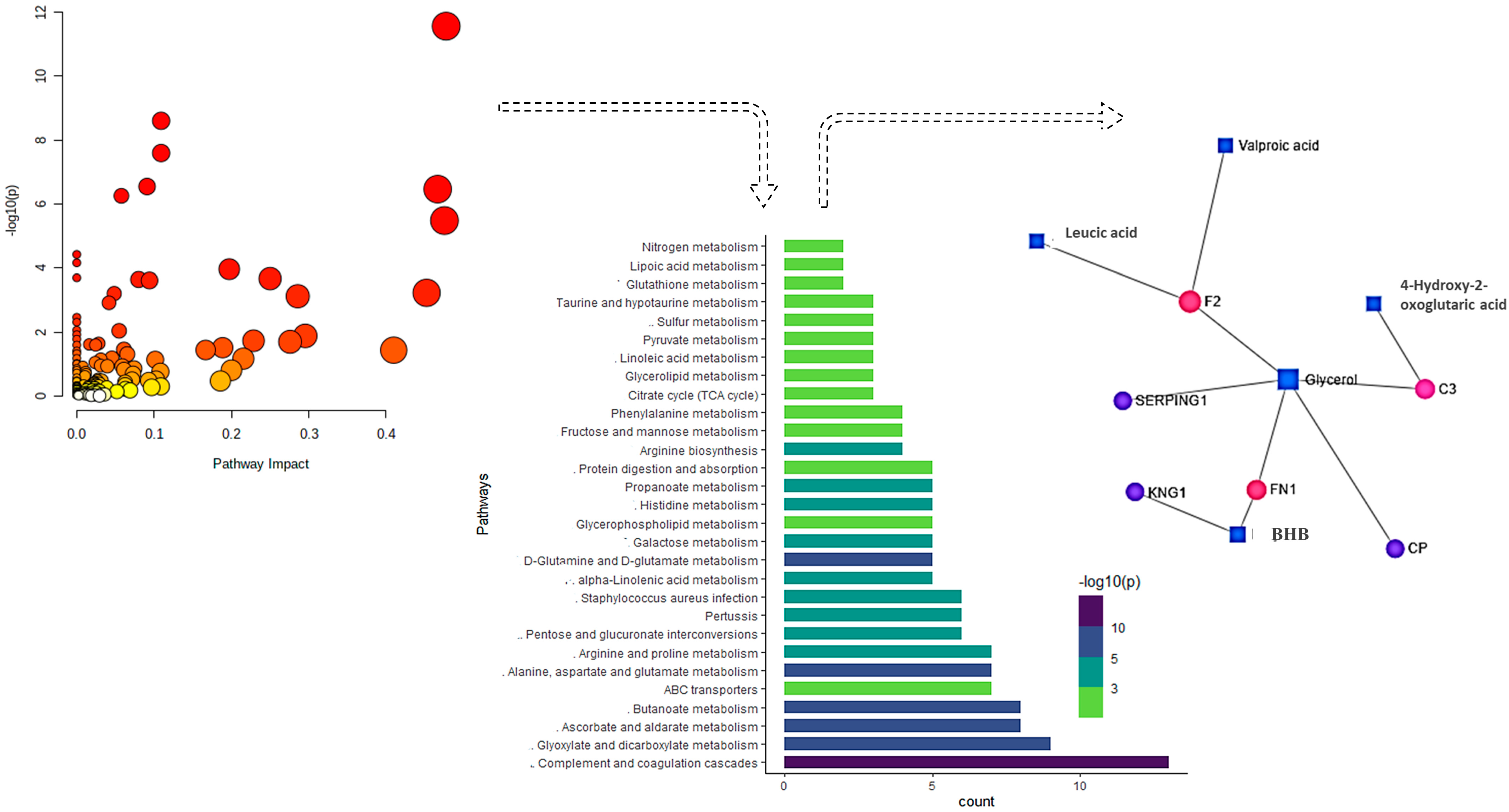
2.3. GO and KEGG Pathway Analysis
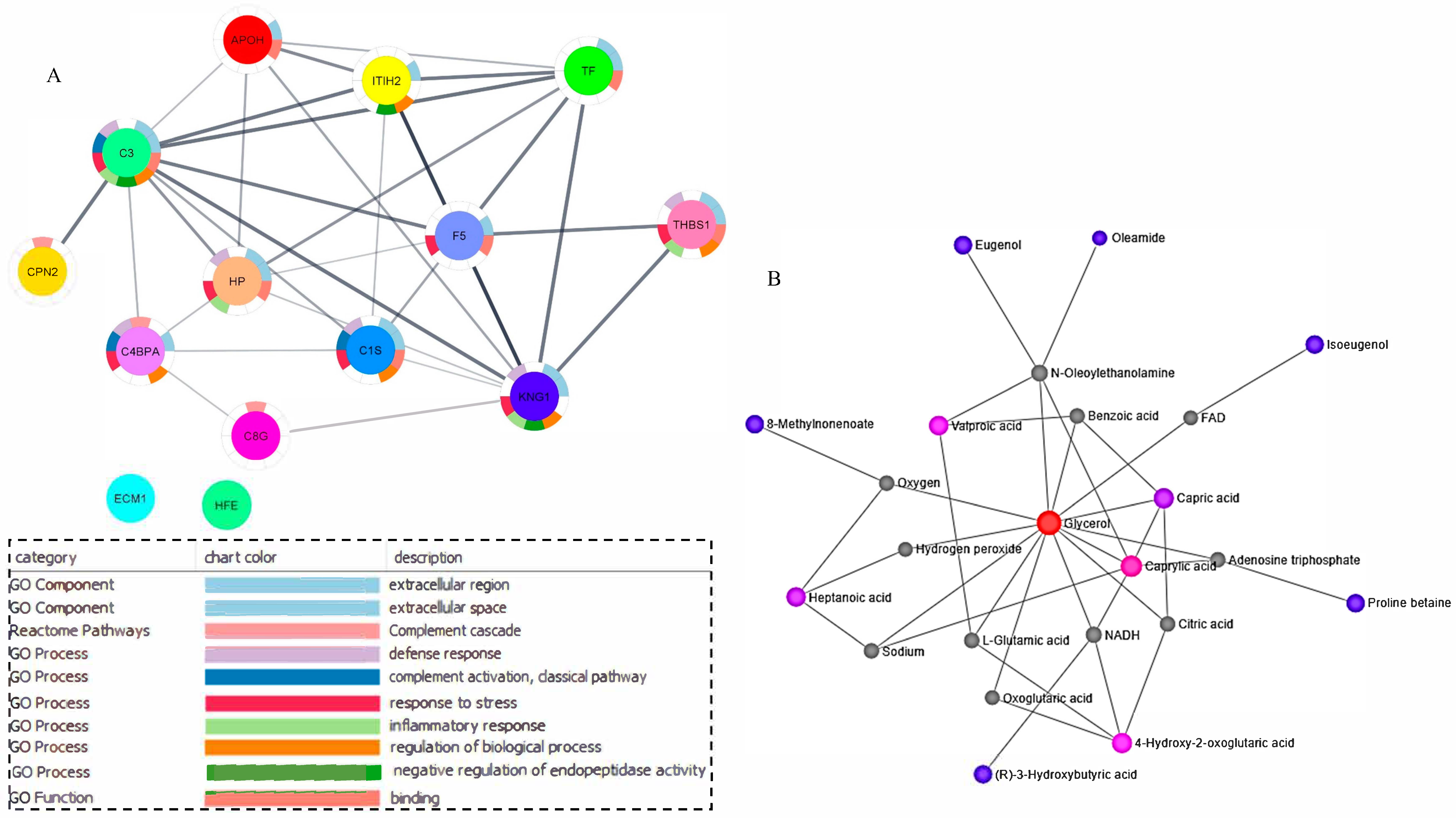
2.4. Hierarchical Clustering and Correlation Analysis
2.5. Networking and Joint Pathway Analysis
3. Discussion
4. Materials and Methods
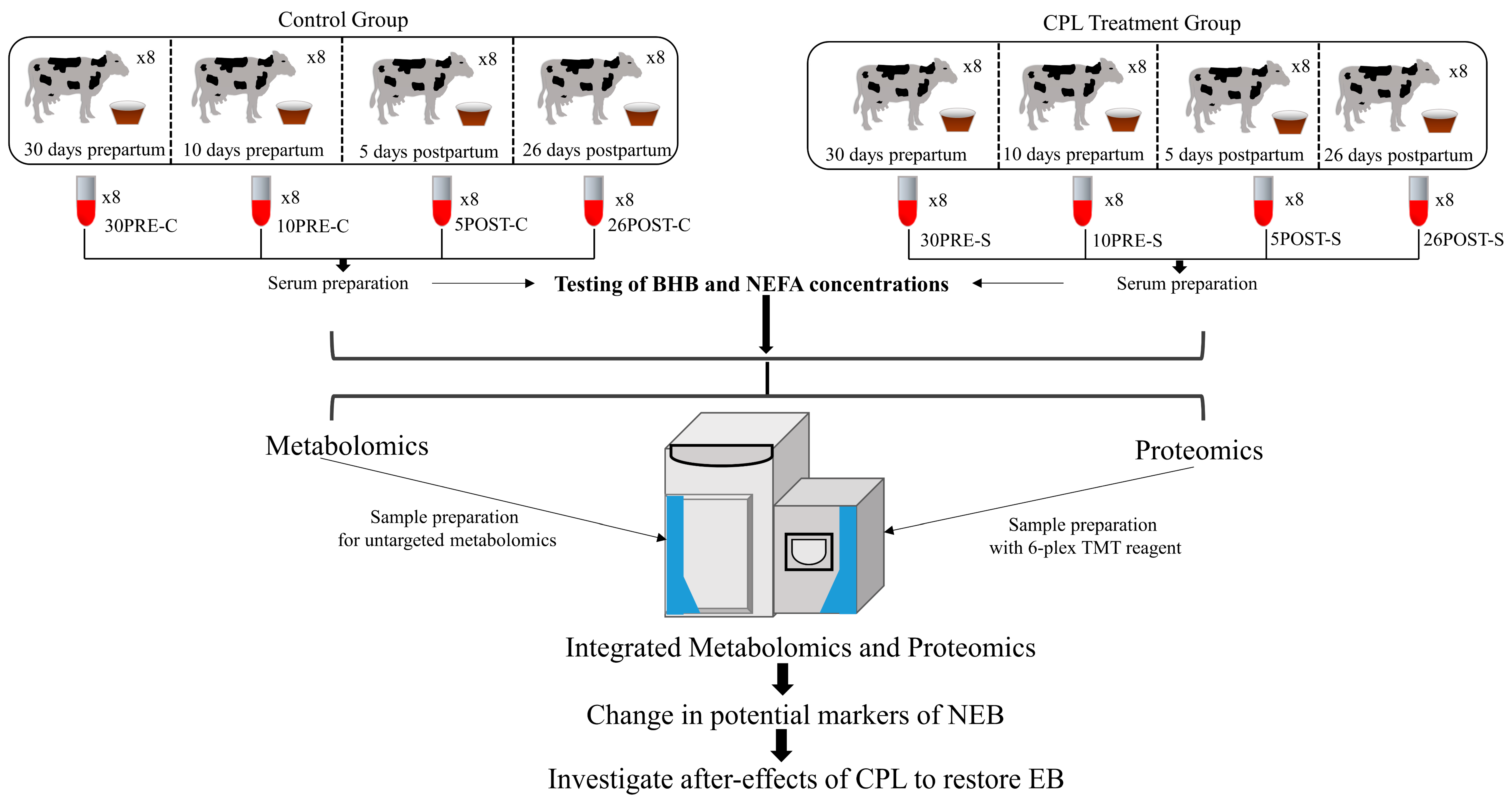
4.1. Animals and Housing
4.2. Dietary Supplementation and Animal Group Assignment
4.3. Serum Collection and Sampling Strategy
4.4. Sample Preparation for Metabolomics and Proteomics
4.5. LC-MS Measurements
4.6. Database Searching
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, J.; Antunes Fernandes, E.; Páez Cano, A.E.; Vinitwatanakhun, J.; Boeren, S.; Van Hooijdonk, T.; Van Knegsel, A.; Vervoort, J.; Hettinga, K.A. Changes in milk proteome and metabolome associated with dry period length, energy balance, and lactation stage in postparturient dairy cows. J. Proteome Res. 2013, 12, 3288–3296. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Vervoort, J.; Saccenti, E.; van Hoeij, R.; Kemp, B.; van Knegsel, A. Milk Metabolomics Data Reveal the Energy Balance of Individual Dairy Cows in Early Lactation. Sci. Rep. 2018, 8, 15828. [Google Scholar] [CrossRef] [Green Version]
- Sundrum, A. Metabolic disorders in the transition period indicate that the dairy cows’ ability to adapt is overstressed. Animals 2015, 5, 978–1020. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Hametner, C.; Tuchscherer, A.; Losand, B.; Kanitz, E.; Otten, W.; Singh, S.P.; Bruckmaier, R.M.; Becker, F.; Kanitz, W.; et al. Variation in fat mobilization during early lactation differently affects feed intake, body condition, and lipid and glucose metabolism in high-yielding dairy cows. J. Dairy Sci. 2013, 96, 165–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares, R.A.N.; Vargas, G.; Muniz, M.M.M.; Soares, M.A.M.; Cánovas, A.; Schenkel, F.; Squires, E.J. Differential gene expression in dairy cows under negative energy balance and ketosis: A systematic review and meta-analysis. J. Dairy Sci. 2021, 104, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Pascottini, O.B.; Van Schyndel, S.J.; Spricigo, J.F.W.; Carvalho, M.R.; Mion, B.; Ribeiro, E.S.; LeBlanc, S.J. Effect of anti-inflammatory treatment on systemic inflammation, immune function, and endometrial health in postpartum dairy cows. Sci. Rep. 2020, 10, 5236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abuelo, A.; Hernández, J.; Benedito, J.L.; Castillo, C. Redox biology in transition periods of dairy cattle: Role in the health of periparturient and neonatal animals. Antioxidants 2019, 8, 20. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; van Hoeij, R.J.; Bruckmaier, R.M.; Kok, A.; Lam, T.J.G.M.; Kemp, B.; van Knegsel, A.T.M. Consequences of Transition Treatments on Fertility and Associated Metabolic Status for Dairy Cows in Early Lactation. Animals 2020, 10, 1100. [Google Scholar] [CrossRef]
- Wankhade, P.R.; Manimaran, A.; Kumaresan, A.; Jeyakumar, S.; Ramesha, K.P.; Sejian, V.; Rajendran, D.; Varghese, M.R. Metabolic and immunological changes in transition dairy cows: A review. Vet. World 2017, 10, 1367–1377. [Google Scholar] [CrossRef] [Green Version]
- Sheehy, M.R.; Fahey, A.G.; Aungier, S.P.M.; Carter, F.; Crowe, M.A.; Mulligan, F.J. A comparison of serum metabolic and production profiles of dairy cows that maintained or lost body condition 15 days before calving. J. Dairy Sci. 2017, 100, 536–547. [Google Scholar] [CrossRef] [Green Version]
- Wathes, D.C.; Cheng, Z.; Chowdhury, W.; Fenwick, M.A.; Fitzpatrick, R.; Morris, D.G.; Patton, J.; Murphy, J.J. Negative energy balance alters global gene expression and immune responses in the uterus of postpartum dairy cows. Physiol. Genom. 2009, 39, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.J.; Kessler, E.C.; Albrecht, C.; Bruckmaier, R.M. Response of the Cholesterol Metabolism to a Negative Energy Balance in Dairy Cows Depends on the Lactational Stage. PLoS ONE 2015, 10, e0121956. [Google Scholar] [CrossRef] [Green Version]
- Sangeetha, C.; Baskar, P. Zeolite and its potential uses in agriculture: A critical review. Agric. Rev. 2016, 37, 101–108. [Google Scholar] [CrossRef]
- Nadziakiewicza, M.; Kehoe, S.; Micek, P. Physico-Chemical Properties of Clay Minerals and Their Use as a Health Promoting Feed Additive. Animals 2019, 9, 714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavelić, S.K.; Medica, J.S.; Gumbarević, D.; Filošević, A.; Pržulj, N.; Pavelić, K. Critical review on zeolite clinoptilolite safety and medical applications in vivo. Front. Pharmacol. 2018, 9, 1350. [Google Scholar] [CrossRef] [PubMed]
- Mumpton, F.A. La roca magica: Uses of natural zeolites in agriculture and industry. Proc. Natl. Acad. Sci. USA 1999, 96, 3463–3470. [Google Scholar] [CrossRef] [Green Version]
- Mastinu, A.; Kumar, A.; Maccarinelli, G.; Bonini, S.A.; Premoli, M.; Aria, F.; Gianoncelli, A.; Memo, M. Zeolite Clinoptilolite: Therapeutic Virtues of an Ancient Mineral. Molecules 2019, 24, 1517. [Google Scholar] [CrossRef] [Green Version]
- Dschaak, C.M.; Eun, J.S.; Young, A.J.; Stott, R.D.; Peterson, S. Effects of Supplementation of Natural Zeolite on Intake, Digestion, Ruminal Fermentation, and Lactational Performance of Dairy Cows. Prof. Anim. Sci. 2010, 26, 647–654. [Google Scholar] [CrossRef]
- Simona, M.; Camelia, T. Zeolites Applications in Veterinary Medicine. In Zeolites-New Challenges; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef] [Green Version]
- Cerbu, C.; Ilaș, V.A.; Czopowicz, M.; Potârniche, A.V.; Bodart-Nieva, E.P.; Mureșan, E.A.; Kaba, J.; Spinu, M.; Pall, E. The Use of Activated Micronized Zeolite Clinoptilolite as a Possible Alternative to Antibiotics and Chestnut Extract for the Control of Undifferentiated Calf Diarrhea: An In Vitro and In Vivo Study. Animals 2020, 10, 2284. [Google Scholar] [CrossRef]
- Grądzki, Z.; Jarosz, Ł.; Stępień-Pyśniak, D.; Marek, A. The effect of feed supplementation with Transcarpathian zeolite (clinoptilolite) on the concentrations of acute phase proteins and cytokines in the serum and hepatic tissue of chickens. Poult. Sci. 2020, 99, 2424–2437. [Google Scholar] [CrossRef]
- Caixeta, L.S.; Omontese, B.O. Monitoring and Improving the Metabolic Health of Dairy Cows during the Transition Period. Animals 2021, 11, 352. [Google Scholar] [CrossRef]
- Karatzia, M.A.; Katsoulos, P.D.; Karatzias, H. Diet supplementation with clinoptilolite improves energy status, reproductive efficiency and increases milk yield in dairy heifers. Anim. Prod. Sci. 2013, 53, 234. [Google Scholar] [CrossRef]
- Khachlouf, K.; Hamed, H.; Gdoura, R.; Gargouri, A. Effects of dietary Zeolite supplementation on milk yield and composition and blood minerals status in lactating dairy cows. J. Appl. Anim. Res. 2019, 47, 54–62. [Google Scholar] [CrossRef]
- Đuričić, D.; Sukalić, T.; Marković, F.; Kočila, P.; Žaja, I.Ž.; Menčik, S.; Dobranić, T.; Benić, M.; Samardžija, M. Effects of dietary vibroactivated clinoptilolite supplementation on the intramammary microbiological findings in dairy cows. Animals 2020, 10, 202. [Google Scholar] [CrossRef] [Green Version]
- Folnožić, I.; Samardžija, M.; Đuričić, D.; Vince, S.; Perkov, S.; Jelušić, S.; Valpotić, H.; Ljubić, B.B.; Lojkić, M.; Gračner, D.; et al. Effects of in-feed clinoptilolite treatment on serum metabolic and antioxidative biomarkers and acute phase response in dairy cows during pregnancy and early lactation. Res. Vet. Sci. 2019, 127, 57–64. [Google Scholar] [CrossRef]
- Valpotić, H.; Gračner, D.; Turk, R.; Đuričić, D.; Vince, S.; Folnožić, I.; Lojkić, M.; Žaja, I.Ž.; Bedrica, L.; Maćešić, N.; et al. Zeolite clinoptilolite nanoporous feed additive for animals of veterinary importance: Potentials and limitations. Period. Biol. 2017, 119, 159–172. [Google Scholar] [CrossRef]
- Mellouk, N.; Rame, C.; Naquin, D.; Jaszczyszyn, Y.; Touzé, J.L.; Briant, E.; Guillaume, D.; Ntallaris, T.; Humblot, P.; Dupont, J. Impact of the severity of negative energy balance on gene expression in the subcutaneous adipose tissue of periparturient primiparous Holstein dairy cows: Identification of potential novel metabolic signals for the reproductive system. PLoS ONE 2019, 14. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Gautam, V.; Naseem, S. Acute-phase proteins: As diagnostic tool. J. Pharm. Bioallied Sci. 2011, 3, 118–127. [Google Scholar] [CrossRef]
- Kaya, S.; Merhan, O.; Kacar, C.; Colak, A.; Bozukluhan, K. Determination of ceruloplasmin, some other acute phase proteins, and biochemical parameters in cows with endometritis. Vet. World 2016, 9, 1056–1062. [Google Scholar] [CrossRef] [PubMed]
- Fenwick, M.A.; Llewellyn, S.; Fitzpatrick, R.; Kenny, D.A.; Murphy, J.J.; Patton, J.; Wathes, D.C. Negative energy balance in dairy cows in associated with specific changes in IGF-binding protein expression in the oviduct. Reproduction 2008, 135, 63–75. [Google Scholar] [CrossRef] [Green Version]
- Bae, O.N.; Wang, J.M.; Baek, S.H.; Wang, Q.; Yuan, H.; Chen, A.F. Oxidative stress-mediated thrombospondin-2 upregulation impairs bone marrow-derived angiogenic cell function in diabetes mellitus. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1920–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tezel, G.; Yang, X.; Luo, C.; Kain, A.D.; Powell, D.W.; Kuehn, M.H.; Kaplan, H.J. Oxidative stress and the regulation of complement activation in human glaucoma. Investig. Ophthalmol. Vis. Sci. 2010, 51, 5071–5082. [Google Scholar] [CrossRef] [PubMed]
- Lord, M.S.; Melrose, J.; Day, A.J.; Whitelock, J.M. The Inter-α-Trypsin Inhibitor Family: Versatile Molecules in Biology and Pathology. J. Histochem. Cytochem. 2020, 68, 907–927. [Google Scholar] [CrossRef]
- Yang, J.; Ahn, H.N.; Chang, M.; Narasimhan, P.; Chan, P.H.; Song, Y.S. Complement component 3 inhibition by an antioxidant is neuroprotective after cerebral ischemia and reperfusion in mice. J. Neurochem. 2013, 124, 523–535. [Google Scholar] [CrossRef]
- Chandra, G.; Aggarwal, A.; Singh, A.K.; Kumar, M.; Upadhyay, R.C. Effect of Vitamin E and Zinc Supplementation on Energy Metabolites, Lipid Peroxidation, and Milk Production in Peripartum Sahiwal Cows. Asian-Australas. J. Anim. Sci. 2013, 26, 1569. [Google Scholar] [CrossRef] [Green Version]
- Ren, R.; Hashimoto, T.; Mizuno, M.; Takigawa, H.; Yoshida, M.; Azuma, T.; Kanazawa, K. A lipid peroxidation product 9-oxononanoic acid induces phospholipase A2 activity and thromboxane A2 production in human blood. J. Clin. Biochem. Nutr. 2013, 52, 228–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herdt, T.H. Ruminant adaptation to negative energy balance. Influences on the etiology of ketosis and fatty liver. Vet. Clin. N. Am. Food Anim. Pract. 2000, 16. [Google Scholar] [CrossRef]
- Weston, A.L.; Weinstein, A.M.; Barton, C.; Yaffe, K. Potentially inappropriate medication use in older adults with mild cognitive impairment. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2010, 65A, 318–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Snowden, S.; Suvitaival, T.; Ali, A.; Merkler, D.J.; Ahmad, T.; Westwood, S.; Baird, A.; Proitsi, P.; Nevado-Holgado, A.; et al. Primary fatty amides in plasma associated with brain amyloid burden, hippocampal volume, and memory in the European Medical Information Framework for Alzheimer’s Disease biomarker discovery cohort. Alzheimer’s Dement. 2019, 15, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Kholif, A.E. Glycerol use in dairy diets: A systemic review. Anim. Nutr. 2019, 5, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.D.; Stockman, M.C.; Yu, L.; Meshulam, T.; McCarthy, A.C.; Ionson, A.; Burritt, N.; Deeney, J.; Cabral, H.; Corkey, B.; et al. Effects of medium chain triglycerides supplementation on insulin sensitivity and beta cell function: A feasibility study. PLoS ONE 2019, 14, e0226200. [Google Scholar] [CrossRef] [Green Version]
- Zuo, S.; Fries, B.E.; Szafara, K.; Regal, R. Valproic acid as a potentiator of metabolic syndrome in institutionalized residents on concomitant antipsychotics: Fat chance, or slim to none? Pharm. Ther. 2015, 40, 126–132. [Google Scholar]
- Oh, Y.T.; Lee, J.Y.; Lee, J.; Lee, J.H.; Kim, J.E.; Ha, J.; Kang, I. Oleamide suppresses lipopolysaccharide-induced expression of iNOS and COX-2 through inhibition of NF-κB activation in BV2 murine microglial cells. Neurosci. Lett. 2010, 474, 148–153. [Google Scholar] [CrossRef]
- Johny, A.K.; Darre, M.J.; Donoghue, A.M.; Donoghue, D.J.; Venkitanarayanan, K. Antibacterial effect of trans-cinnamaldehyde, eugenol, carvacrol, and thymol on salmonella enteritidis and Campylobacter jejuni in chicken cecal contents in vitro. J. Appl. Poult. Res. 2010, 19, 237–244. [Google Scholar] [CrossRef]
- Marth, E.H. Salmonellae and Salmonellosis Associated with Milk and Milk Products. A Review. J. Dairy Sci. 1969, 52, 283–315. [Google Scholar] [CrossRef]
- Hyldgaard, M.; Mygind, T.; Piotrowska, R.; Foss, M.; Meyer, R.L. Isoeugenol has a non-disruptive detergent-like mechanism of action. Front. Microbiol. 2015, 6, 754. [Google Scholar] [CrossRef] [PubMed]
- Solis de los Santos, F.; Donoghue, A.M.; Venkitanarayanan, K.; Metcalf, J.H.; Reyes-Herrera, I.; Dirain, M.L.; Aguiar, V.F.; Blore, P.J.; Donoghue, D.J. The natural feed additive caprylic acid decreases Campylobacter jejuni colonization in market-aged broiler chickens. Poult. Sci. 2009, 88, 61–64. [Google Scholar] [CrossRef]
- Zutz, C.; Bacher, M.; Parich, A.; Kluger, B.; Gacek-Matthews, A.; Schuhmacher, R.; Wagner, M.; Rychli, K.; Strauss, J. Valproic acid induces antimicrobial compound production in doratomyces microspores. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Atsumi, T.; Fujisawa, S.; Tonosaki, K. A comparative study of the antioxidant/prooxidant activities of eugenol and isoeugenol with various concentrations and oxidation conditions. Toxicol. In Vitro 2005, 19, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Baddela, V.S.; Becker, F.; Dannenberger, D.; Viergutz, T.; Vanselow, J. Elevated free fatty acids affect bovine granulosa cell function: A molecular cue for compromised reproduction during negative energy balance. Endocr. Connect. 2019, 8, 493–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Sun, L.; Zheng, X.; Liu, J.; Zheng, R.; Yang, R.; Wang, Y. Alterations in complement and coagulation pathways of human placentae subjected to in vitro fertilization and embryo transfer in the first trimester. Medicine 2019, 98, e17031. [Google Scholar] [CrossRef]
- Rui, L. Energy metabolism in the liver. Compr. Physiol. 2014, 4, 177–197. [Google Scholar] [CrossRef] [Green Version]
- Ruderman, N.B. Muscle Amino Acid Metabolism and Gluconeogenesis. Annu. Rev. Med. 1975, 26, 245–258. [Google Scholar] [CrossRef]
- Khan, S.; Kumar, S.; Jena, G. Valproic acid reduces insulin-resistance, fat deposition and FOXO1-mediated gluconeogenesis in type-2 diabetic rat. Biochimie 2016, 125, 42–52. [Google Scholar] [CrossRef]
- Rogiers, V.; Vandenberghe, Y.; Vercruysse, A. Inhibition of gluconeogenesis by sodium valproate and its metabolites in isolated rat hepatocytes. Xenobiotica 1985, 15, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Horvatić, A.; Guillemin, N.; Kaab, H.; McKeegan, D.; O’Reilly, E.; Bain, M.; Kuleš, J.; Eckersall, P.D. Quantitative proteomics using tandem mass tags in relation to the acute phase protein response in chicken challenged with Escherichia coli lipopolysaccharide endotoxin. J. Proteom. 2019, 192, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Gloaguen, Y.; Morton, F.; Daly, R.; Gurden, R.; Rogers, S.; Wandy, J.; Wilson, D.; Barrett, M.; Burgess, K. PiMP my metabolome: An integrated, web-based tool for LC-MS metabolomics data. Bioinformatics 2017, 33, 4007–4009. [Google Scholar] [CrossRef] [Green Version]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Acc Id | Gene | Protein | Log 30pre-C/S | Log 10pre-C/S | Log 5post-C/S | Log 26post-C/S | Q |
|---|---|---|---|---|---|---|---|
| Q28178 | THBS1 | Thrombospondin-1 | −0.28 | −0.51 | −0.21 | −0.37 | 0.001181 |
| Q2UVX4 | C3 | Complement C3 | −0.36 | −0.30 | −0.22 | −0.17 | 0.001302 |
| P00735 | F2 | Prothrombin | −0.33 | −0.34 | −0.13 | −0.27 | 0.00257 |
| Q28065 | C4BPA | C4b-binding protein alpha chain | −0.41 | −0.43 | −0.25 | −0.35 | 0.003106 |
| A5D7R6 | ITIH2 | ITIH2 protein | −0.29 | −0.37 | −0.23 | −0.38 | 0.003999 |
| A5PJT7 | ECM1 | ECM1 protein | −0.52 | −0.39 | −0.19 | −0.42 | 0.004807 |
| G3X6K8 | HP | Haptoglobin | −1.34 | −0.73 | −0.06 | −0.02 | 0.006941 |
| Q29443 | TF | Serotransferrin | −0.39 | −0.12 | −0.19 | −0.25 | 0.007474 |
| G5E604 | HFE | Ig-like domain-containing protein | 0.78 | 0.14 | 0.38 | −0.18 | 0.007474 |
| Q0VCX1 | C1S | Complement C1s subcomponent | −0.40 | −0.24 | −0.25 | −0.42 | 0.009913 |
| A8YXZ2 | C8G | C8G protein | −0.21 | −0.29 | −0.21 | −0.22 | 0.019858 |
| P17690 | APOH | Beta-2-glycoprotein 1 | −0.22 | −0.34 | −0.25 | −0.25 | 0.019858 |
| Q28107 | F5 | Coagulation factor V | −0.31 | −0.35 | −0.16 | −0.29 | 0.024159 |
| A6QP30 | CPN2 | CPN2 protein | −0.15 | −0.24 | −0.18 | −0.36 | 0.02606 |
| P01044 | KNG1 | Kininogen-1 | −0.25 | −0.56 | −0.18 | −0.29 | 0.03451 |
| Metabolites | KEGG | Log 30pre-C/S | Log 10pre-C/S | Log 5post-C/S | Log 26post-C/S | Q |
|---|---|---|---|---|---|---|
| (-)-Bornyl acetate | NA | −1.46 | −1.36 | −1.44 | −1.42 | 0.000308 |
| (S)-Homostachydrine | C08283 | 0.84 | 0.98 | −0.42 | 0.98 | 0.018369 |
| 1,4-beta-D-Glucan | C00760 | 1.28 | 0.39 | 0.70 | 0.99 | 1.26 × 10-8 |
| 4-Hydroxy-2-oxoglutaric acid | C01127 | 1.60 | 1.40 | 0.65 | 4.09 | 0.000308 |
| 9-oxo-nonanoic acid | C16322 | −1.32 | −1.18 | −1.15 | −0.64 | 0.000957 |
| Butabarbital | C07827 | −1.06 | −0.92 | −0.65 | −1.23 | 4.92 × 10-13 |
| Capric acid | C01571 | −1.68 | −1.34 | −1.30 | −0.16 | 0.011789 |
| Caprylic acid | C06423 | 0.65 | 0.77 | 0.74 | 1.59 | 5.24 × 10-11 |
| D(-)-beta-hydroxy butyric acid | C01089 | −3.20 | −3.43 | −3.03 | −2.07 | 0.04743 |
| D-Leucic acid | C03264 | −1.52 | −1.39 | −1.51 | −1.10 | 0.011789 |
| Eugenol | C10453 | −1.62 | −1.84 | −0.56 | −1.06 | 2.20 × 10-6 |
| Glycerol | C00116 | 1.57 | 1.63 | 1.58 | 1.91 | 0.000339 |
| Heptanoic acid | C17714 | 0.53 | 0.83 | 0.67 | 1.73 | 9.61 × 10-8 |
| Isoeugenol | C10469 | −1.62 | −1.84 | −0.56 | −1.06 | 2.20 × 10-6 |
| L-(+)-Anaferine | C06183 | −0.78 | −1.20 | −0.54 | −0.39 | 0.000175 |
| L-Aspartate | C00049 | −0.13 | −0.52 | −0.24 | −0.10 | 0.035791 |
| Methylmalonate | C02170 | −0.68 | −0.44 | −0.81 | −0.40 | 2.09 × 10-12 |
| Oleamide | C19670 | −1.21 | −0.67 | −1.13 | −0.50 | 0.00019 |
| Valproic acid | C07185 | 0.65 | 0.77 | 0.74 | 1.59 | 5.24 × 10-11 |
| Botrydial | C09622 | 0.00 | 0.00 | −1.43 | −1.70 | 7.21 × 10-15 |
| cis-1,2-Cyclohexanediol | C12313 | 1.15 | 1.36 | 1.41 | 2.32 | 1.31 × 10-14 |
| 1-(3,4-Dihydroxyphenyl)-5-hydroxy-3-decanone | C17748 | −0.75 | −1.12 | −0.76 | −0.09 | 1.02× 10-5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maity, S.; Rubić, I.; Kuleš, J.; Horvatić, A.; Đuričić, D.; Samardžija, M.; Ljubić, B.B.; Turk, R.; Gračner, D.; Maćešić, N.; et al. Integrated Metabolomics and Proteomics Dynamics of Serum Samples Reveals Dietary Zeolite Clinoptilolite Supplementation Restores Energy Balance in High Yielding Dairy Cows. Metabolites 2021, 11, 842. https://doi.org/10.3390/metabo11120842
Maity S, Rubić I, Kuleš J, Horvatić A, Đuričić D, Samardžija M, Ljubić BB, Turk R, Gračner D, Maćešić N, et al. Integrated Metabolomics and Proteomics Dynamics of Serum Samples Reveals Dietary Zeolite Clinoptilolite Supplementation Restores Energy Balance in High Yielding Dairy Cows. Metabolites. 2021; 11(12):842. https://doi.org/10.3390/metabo11120842
Chicago/Turabian StyleMaity, Sudipa, Ivana Rubić, Josipa Kuleš, Anita Horvatić, Dražen Đuričić, Marko Samardžija, Blanka Beer Ljubić, Romana Turk, Damjan Gračner, Nino Maćešić, and et al. 2021. "Integrated Metabolomics and Proteomics Dynamics of Serum Samples Reveals Dietary Zeolite Clinoptilolite Supplementation Restores Energy Balance in High Yielding Dairy Cows" Metabolites 11, no. 12: 842. https://doi.org/10.3390/metabo11120842
APA StyleMaity, S., Rubić, I., Kuleš, J., Horvatić, A., Đuričić, D., Samardžija, M., Ljubić, B. B., Turk, R., Gračner, D., Maćešić, N., Valpotić, H., & Mrljak, V. (2021). Integrated Metabolomics and Proteomics Dynamics of Serum Samples Reveals Dietary Zeolite Clinoptilolite Supplementation Restores Energy Balance in High Yielding Dairy Cows. Metabolites, 11(12), 842. https://doi.org/10.3390/metabo11120842











