Lipid Profiling in Cancer Diagnosis with Hand-Held Ambient Mass Spectrometry Probes: Addressing the Late-Stage Performance Concerns
Abstract
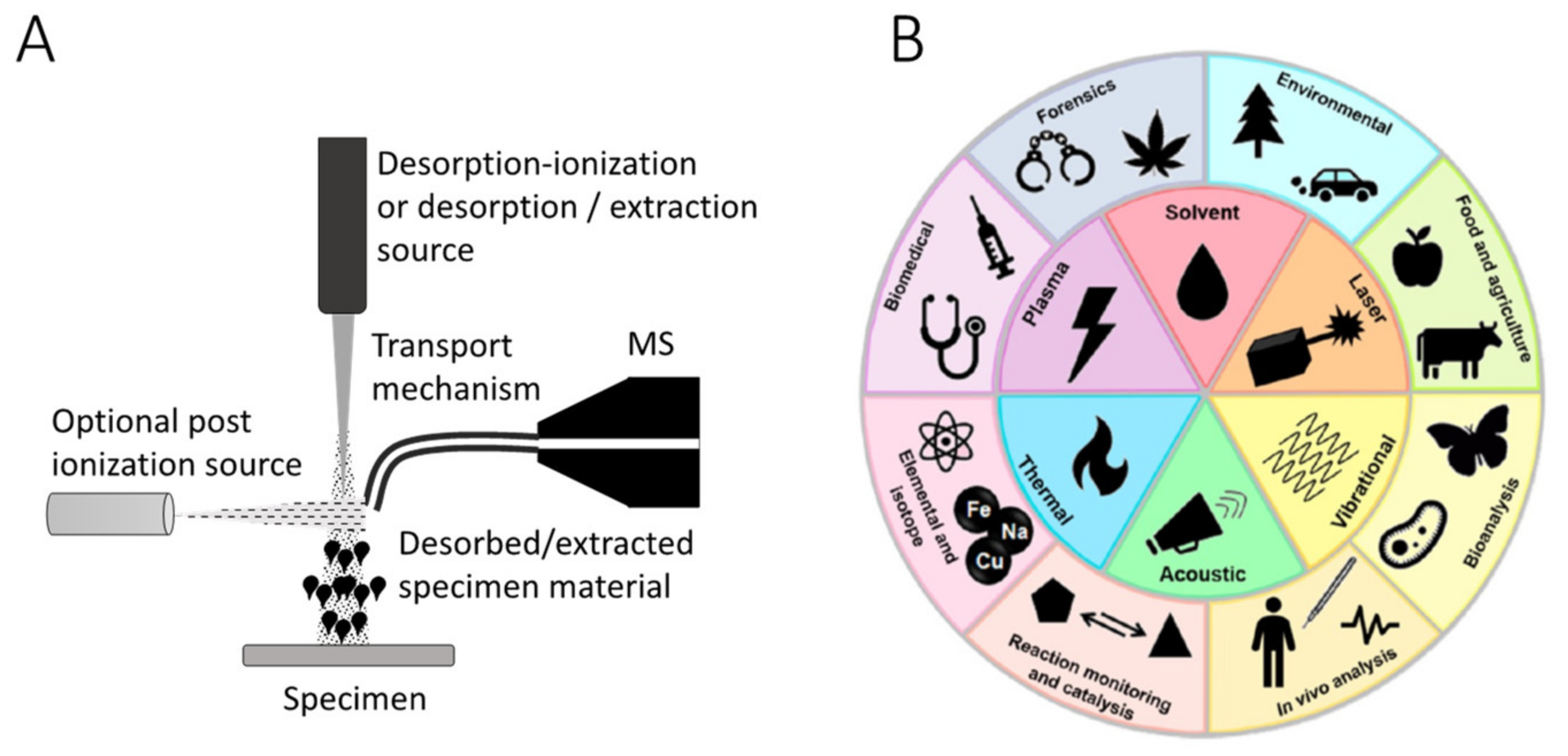
1. Introduction and Problem Statement
2. State of the Art
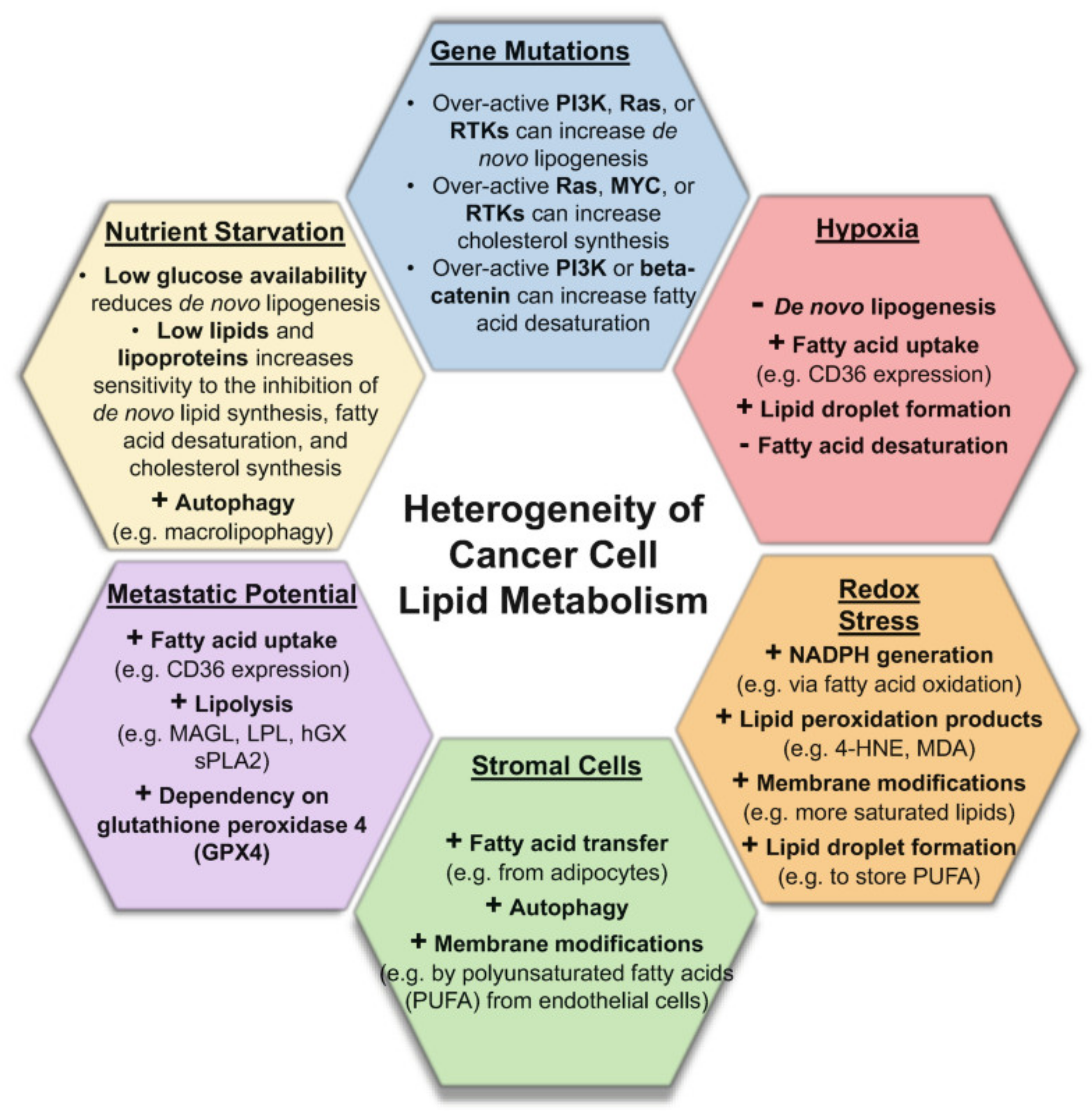
2.1. Molecular Heterogeneities in Biological Tissues Impact Metabolome and Lipidome Profiles
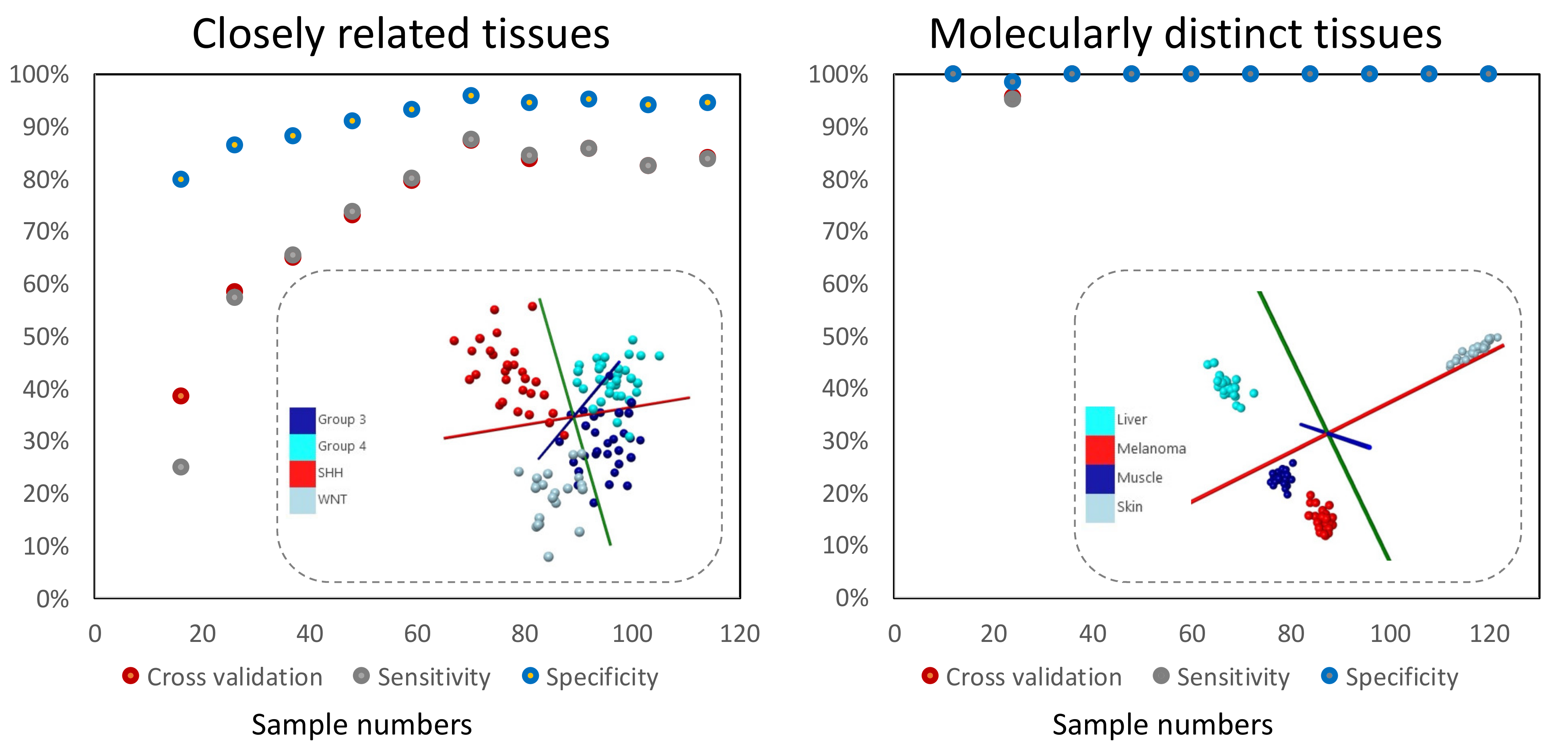
2.2. Initial Statistical Modeling Should Be Based on Sufficient Sample Numbers
2.3. Lessons Learned from Metabolomic and Proteomic Biomarker Discovery and Food Sciences
2.4. Moving beyond Statistical Associations
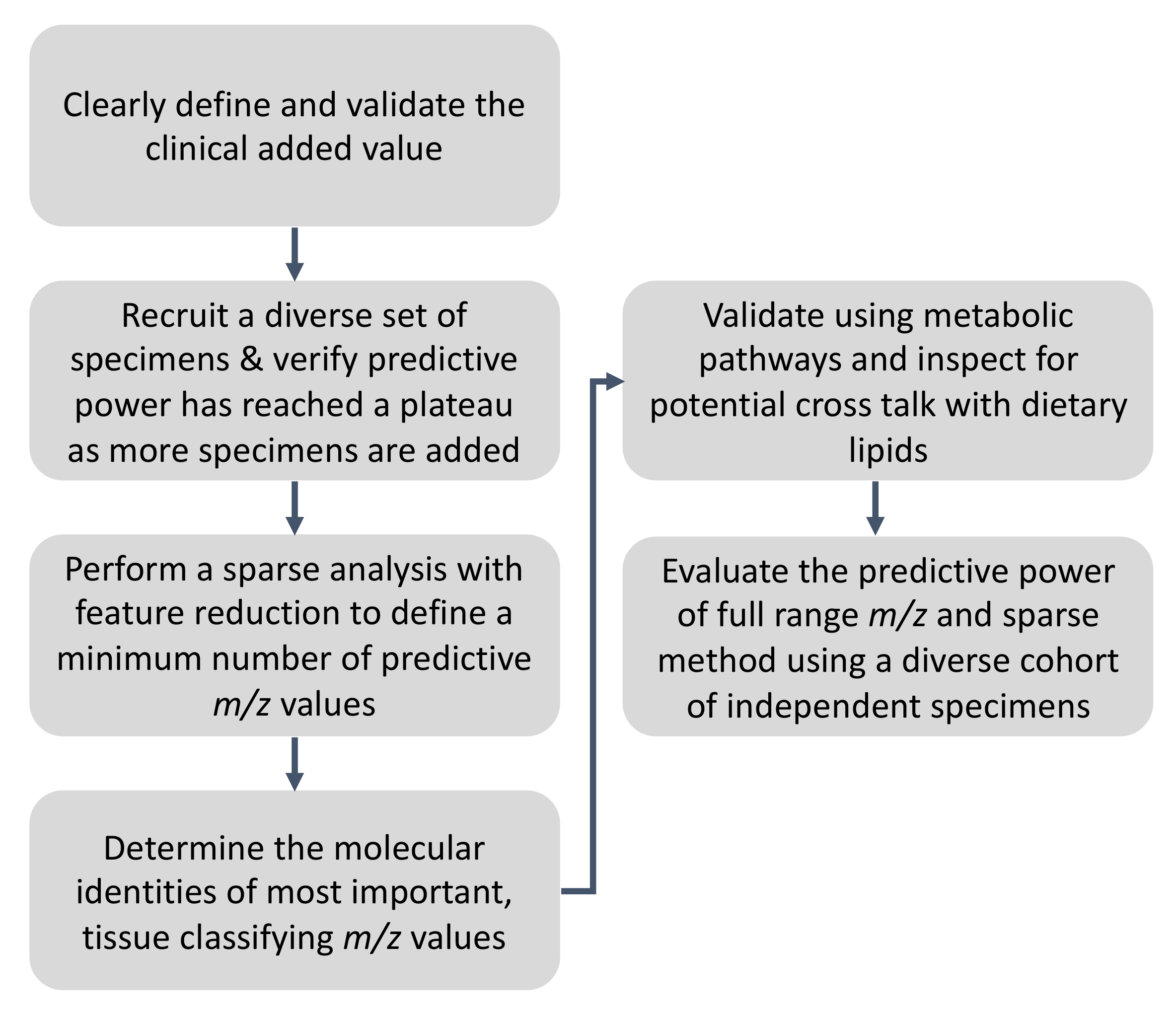
3. Recommendations and the Proposed Workflow for Ambient MS Method Validation for Rapid Pathology Determination
4. Conclusions and Caveats
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sans, M.; Garza, K.Y.; Eberlin, L.S. Mass spectrometry technologies to advance care for cancer patients in clinical and intraoperative use. Mass Spectrom. Rev. 2020, 40, 692–720. [Google Scholar] [CrossRef] [PubMed]
- Feider, C.L.; Krieger, A.C.; DeHoog, R.J.; Eberlin, L.S. Ambient Ionization Mass Spectrometry: Recent Developments and Applications. Anal. Chem. 2019, 91, 4266–4290. [Google Scholar] [CrossRef]
- Takats, Z.; Strittmatter, N.; McKenzie, J. Ambient Mass Spectrometry in Cancer Research. Methods Mol. Biol. 2017, 134, 231–256. [Google Scholar] [CrossRef]
- Ifa, D.R.; Eberlin, L.S. Ambient Ionization Mass Spectrometry for Cancer Diagnosis and Surgical Margin Evaluation. Clin. Chem. 2016, 62, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.; Calligaris, D.; Regan, M.S.; Krummel, D.P.; Agar, J.N.; Kallay, L.; Macdonald, T.; Schniederjan, M.; Santagata, S.; Pomeroy, S.L.; et al. Rapid discrimination of pediatric brain tumors by mass spectrometry imaging. J. Neuro-Oncol. 2018, 140, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Eberlin, L.S. DESI-MS Imaging of Lipids and Metabolites from Biological Samples. In Mass Spectrometry in Metabolomics; Humana Press: New York, NY, USA, 2014; Volume 1198, pp. 299–311. [Google Scholar] [CrossRef]
- Pirro, V.; Jarmusch, A.K.; Ferreira, C.R.; Cooks, R.G. Ambient Lipidomic Analysis of Brain Tissue Using Desorption Electrospray Ionization (DESI) Mass Spectrometry. Neuromethods 2017, 125, 187–210. [Google Scholar] [CrossRef]
- Woolman, M.; Kuzan-Fischer, C.M.; Ferry, I.; Kiyota, T.; Luu, B.; Wu, M.; Munoz, D.G.; Das, S.; Aman, A.; Taylor, M.D.; et al. Picosecond Infrared Laser Desorption Mass Spectrometry Identifies Medulloblastoma Subgroups on Intrasurgical Timescales. Cancer Res. 2019, 79, 2426–2434. [Google Scholar] [CrossRef]
- Santagata, S.; Eberlin, L.S.; Norton, I.; Calligaris, D.; Feldman, D.R.; Ide, J.L.; Liu, X.; Wiley, J.S.; Vestal, M.L.; Ramkissoon, S.H.; et al. Intraoperative mass spectrometry mapping of an onco-metabolite to guide brain tumor surgery. Proc. Natl. Acad. Sci. USA 2014, 111, 11121–11126. [Google Scholar] [CrossRef]
- Calligaris, D.; Norton, I.; Feldman, D.R.; Ide, J.L.; Dunn, I.F.; Eberlin, L.S.; Cooks, R.G.; Jolesz, F.A.; Golby, A.J.; Santagata, S.; et al. Mass spectrometry imaging as a tool for surgical decision-making. J. Mass Spectrom. 2013, 48, 1178–1187. [Google Scholar] [CrossRef]
- Eberlin, L.S.; Norton, I.; Orringer, D.; Dunn, I.F.; Liu, X.; Ide, J.L.; Jarmusch, A.K.; Ligon, K.L.; Jolesz, F.A.; Golby, A.J.; et al. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc. Natl. Acad. Sci. USA 2013, 110, 1611–1616. [Google Scholar] [CrossRef]
- Saudemont, P.; Quanico, J.; Robin, Y.-M.; Baud, A.; Balog, J.; Fatou, B.; Tierny, D.; Pascal, Q.; Minier, K.; Pottier, M.; et al. Real-Time Molecular Diagnosis of Tumors Using Water-Assisted Laser Desorption/Ionization Mass Spectrometry Technology. Cancer Cell 2018, 34, 840–851.e4. [Google Scholar] [CrossRef]
- Balog, J.; Sasi-Szabó, L.; Kinross, J.; Lewis, M.R.; Muirhead, L.J.; Veselkov, K.; Mirnezami, R.; Dezso, B.; Damjanovich, L.; Darzi, A.; et al. Intraoperative Tissue Identification Using Rapid Evaporative Ionization Mass Spectrometry. Sci. Transl. Med. 2013, 5, 194ra93. [Google Scholar] [CrossRef]
- Schäfer, K.-C.; Balog, J.; Szaniszló, T.; Szalay, D.; Mezey, G.; Dénes, J.; Bognár, L.; Oertel, M.; Takáts, Z. Real Time Analysis of Brain Tissue by Direct Combination of Ultrasonic Surgical Aspiration and Sonic Spray Mass Spectrometry. Anal. Chem. 2011, 83, 7729–7735. [Google Scholar] [CrossRef]
- Schäfer, K.-C.; Szaniszló, T.; Günther, S.; Balog, J.; Dénes, J.; Keserű, M.; Dezso, B.; Tóth, M.; Spengler, B.; Takáts, Z. In Situ, Real-Time Identification of Biological Tissues by Ultraviolet and Infrared Laser Desorption Ionization Mass Spectrometry. Anal. Chem. 2011, 83, 1632–1640. [Google Scholar] [CrossRef]
- Zhang, J.; Rector, J.; Lin, J.; Young, J.H.; Sans, M.; Katta, N.; Giese, N.; Yu, W.; Nagi, C.; Suliburk, J.; et al. Nondestructive tissue analysis for Ex Vivo and In Vivo cancer diagnosis using a handheld mass spectrometry system. Sci. Transl. Med. 2017, 9, eaan3968. [Google Scholar] [CrossRef]
- Kiritani, S.; Yoshimura, K.; Arita, J.; Kokudo, T.; Hakoda, H.; Tanimoto, M.; Ishizawa, T.; Akamatsu, N.; Kaneko, J.; Takeda, S.; et al. A new rapid diagnostic system with ambient mass spectrometry and machine learning for colorectal liver metastasis. BMC Cancer 2021, 21, 262. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-C.; Chung, H.-H.; Dutkiewicz, E.P.; Chen, C.-L.; Hsieh, H.-Y.; Chen, B.-R.; Wang, M.-Y.; Hsu, C.-C. Predicting Breast Cancer by Paper Spray Ion Mobility Spectrometry Mass Spectrometry and Machine Learning. Anal. Chem. 2020, 92, 1653–1657. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zare, R.N. Personal Information from Latent Fingerprints Using Desorption Electrospray Ionization Mass Spectrometry and Machine Learning. Anal. Chem. 2017, 89, 1369–1372. [Google Scholar] [CrossRef]
- Tata, A.; Woolman, M.; Ventura, M.; Bernards, N.; Ganguly, M.; Gribble, A.; Shrestha, B.; Bluemke, E.; Ginsberg, H.J.; Vitkin, A.; et al. Rapid Detection of Necrosis in Breast Cancer with Desorption Electrospray Ionization Mass Spectrometry. Sci. Rep. 2016, 6, 35374. [Google Scholar] [CrossRef] [PubMed]
- Koundouros, N.; Karali, E.; Tripp, A.; Valle, A.; Inglese, P.; Perry, N.J.; Magee, D.J.; Virmouni, S.A.; Elder, G.A.; Tyson, A.; et al. Metabolic Fingerprinting Links Oncogenic PIK3CA with Enhanced Arachidonic Acid-Derived Eicosanoids. Cell 2020, 181, 1596–1611.e27. [Google Scholar] [CrossRef]
- Pirro, V.; Alfaro, C.M.; Jarmusch, A.K.; Hattab, E.M.; Cohen-Gadol, A.A.; Cooks, R.G. Intraoperative assessment of tumor margins during glioma resection by desorption electrospray ionization-mass spectrometry. Proc. Natl. Acad. Sci. USA 2017, 114, 6700–6705. [Google Scholar] [CrossRef] [PubMed]
- Yannell, K.E.; Smith, K.; Alfaro, C.M.; Jarmusch, A.K.; Pirro, V.; Cooks, R.G. N-Acetylaspartate and 2-Hydroxyglutarate Assessed in Human Brain Tissue by Mass Spectrometry as Neuronal Markers of Oncogenesis. Clin. Chem. 2017, 63, 1766–1767. [Google Scholar] [CrossRef] [PubMed]
- Jarmusch, A.K.; Pirro, V.; Baird, Z.; Hattab, E.; Cohen-Gadol, A.; Cooks, R.G. Lipid and metabolite profiles of human brain tumors by desorption electrospray ionization-MS. Proc. Natl. Acad. Sci. USA 2016, 113, 1486–1491. [Google Scholar] [CrossRef] [PubMed]
- Reitman, Z.; Jin, G.; Karoly, E.D.; Spasojevic, I.; Yang, J.; Kinzler, K.W.; He, Y.; Bigner, D.D.; Vogelstein, B.; Yan, H. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc. Natl. Acad. Sci. USA 2011, 108, 3270–3275. [Google Scholar] [CrossRef] [PubMed]
- Eberlin, L.S.; Tibshirani, R.J.; Zhang, J.; Longacre, T.A.; Berry, G.J.; Bingham, D.B.; Norton, J.A.; Zare, R.N.; Poultsides, G.A. Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc. Natl. Acad. Sci. USA 2014, 111, 2436–2441. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, Y.; Jijiwa, M.; Nasu, M.; Ai, J.; Dai, S.; Jiang, B.; Zhang, J.; Huang, G.; Deng, Y. Identification of plasma lipid species as promising diagnostic markers for prostate cancer. BMC Med. Inform. Decis. Mak. 2020, 20, 223. [Google Scholar] [CrossRef]
- Yan, F.; Zhao, H.; Zeng, Y. Lipidomics: A promising cancer biomarker. Clin. Transl. Med. 2018, 7, 21. [Google Scholar] [CrossRef]
- Tata, A.; Woolman, M.; Bluemke, E.; Zarrine-Afsar, A. Chapter 5—Ambient laser-based mass spectrometry analysis methods: A survey of core technologies and reported applications. In Ambient Ionization Mass Spectrometry in Life Sciences; Zaitsu, K., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 119–169. [Google Scholar]
- Fatou, B.; Saudemont, P.; Leblanc, E.; Vinatier, D.; Mesdag, V.; Wisztorski, M.; Focsa, C.; Salzet, M.; Ziskind, M.; Fournier, I. In Vivo Real-Time Mass Spectrometry for Guided Surgery Application. Sci. Rep. 2016, 6, 25919. [Google Scholar] [CrossRef]
- Woolman, M.; Ferry, I.; Kuzan-Fischer, C.M.; Wu, M.; Zou, J.; Kiyota, T.; Isik, S.; Dara, D.; Aman, A.; Das, S.; et al. Rapid determination of medulloblastoma subgroup affiliation with mass spectrometry using a handheld picosecond infrared laser desorption probe. Chem. Sci. 2017, 8, 6508–6519. [Google Scholar] [CrossRef]
- Diamandis, E. The failure of protein cancer biomarkers to reach the clinic: Why, and what can be done to address the problem? BMC Med. 2012, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Drucker, E.; Krapfenbauer, K. Pitfalls and limitations in translation from biomarker discovery to clinical utility in predictive and personalised medicine. EPMA J. 2013, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Hanash, S.M. Why have protein biomarkers not reached the clinic? Genome Med. 2011, 3, 66. [Google Scholar] [CrossRef] [PubMed]
- Raudys, S.; Jain, A.K. Small sample size effects in statistical pattern recognition: Recommendations for practitioners. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 13, 252–264. [Google Scholar] [CrossRef]
- Seddiki, K.; Saudemont, P.; Precioso, F.; Ogrinc, N.; Wisztorski, M.; Salzet, M.; Fournier, I.; Droit, A. Cumulative learning enables convolutional neural network representations for small mass spectrometry data classification. Nat. Commun. 2020, 11, 5595. [Google Scholar] [CrossRef] [PubMed]
- Othman, R. Dietary Lipids and Cancer. Libyan J. Med. 2007, 2, 180–184. [Google Scholar] [CrossRef]
- Peck, B.; Schulze, A. Lipid Metabolism at the Nexus of Diet and Tumor Microenvironment. Trends Cancer 2019, 5, 693–703. [Google Scholar] [CrossRef]
- Xia, S.; Lin, R.; Jin, L.; Zhao, L.; Kang, H.-B.; Pan, Y.; Liu, S.; Qian, G.; Qian, Z.; Konstantakou, E.; et al. Prevention of Dietary-Fat-Fueled Ketogenesis Attenuates BRAF V600E Tumor Growth. Cell Metab. 2017, 25, 358–373. [Google Scholar] [CrossRef]
- Dahdah, N.; Gonzalez-Franquesa, A.; Samino, S.; Gama-Perez, P.; Herrero, L.; Perales, J.; Yanes, O.; Malagón, M.; Garcia-Roves, P. Effects of Lifestyle Intervention in Tissue-Specific Lipidomic Profile of Formerly Obese Mice. Int. J. Mol. Sci. 2021, 22, 3694. [Google Scholar] [CrossRef]
- Slenter, D.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Mélius, J.; Cirillo, E.; Coort, S.L.; Digles, D.; et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef]
- Park, J.K.; Coffey, N.J.; Limoges, A.; Le, A. The Heterogeneity of Lipid Metabolism in Cancer. Adv. Exp. Med. Biol. 2021, 1311, 39–56. [Google Scholar] [CrossRef]
- Blaise, B.J.; Correia, G.D.S.; Tin, A.; Young, J.H.; Vergnaud, A.-C.; Lewis, M.; Pearce, J.; Elliott, P.; Nicholson, J.; Holmes, E.; et al. Power Analysis and Sample Size Determination in Metabolic Phenotyping. Anal. Chem. 2016, 88, 5179–5188. [Google Scholar] [CrossRef]
- Blaise, B.J. Data-Driven Sample Size Determination for Metabolic Phenotyping Studies. Anal. Chem. 2013, 85, 8943–8950. [Google Scholar] [CrossRef]
- Xia, J.; Broadhurst, D.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef]
- Guo, Y.; Graber, A.; McBurney, R.N.; Balasubramanian, R. Sample size and statistical power considerations in high-dimensionality data settings: A comparative study of classification algorithms. BMC Bioinform. 2010, 11, 447. [Google Scholar] [CrossRef]
- Sun, T.; Wang, X.; Cong, P.; Xu, J.; Xue, C. Mass spectrometry-based lipidomics in food science and nutritional health: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2530–2558. [Google Scholar] [CrossRef]
- McGrath, T.F.; Haughey, S.A.; Patterson, J.; Fauhl-Hassek, C.; Donarski, J.; Alewijn, M.; van Ruth, S.; Elliott, C.T. What are the scientific challenges in moving from targeted to non-targeted methods for food fraud testing and how can they be addressed? Spectroscopy case study. Trends Food Sci. Technol. 2018, 76, 38–55. [Google Scholar] [CrossRef]
- Esslinger, S.; Riedl, J.; Fauhl-Hassek, C. Potential and limitations of non-targeted fingerprinting for authentication of food in official control. Food Res. Int. 2014, 60, 189–204. [Google Scholar] [CrossRef]
- Cavanna, D.; Righetti, L.; Elliott, C.; Suman, M. The scientific challenges in moving from targeted to non-targeted mass spectrometric methods for food fraud analysis: A proposed validation workflow to bring about a harmonized approach. Trends Food Sci. Technol. 2018, 80, 223–241. [Google Scholar] [CrossRef]
- U.S. Pharmacopeia. Appendix XVIII: Guidance on Developing and Validating Non-Targeted Methods for Adulteration Detection; U.S. Pharmacopeial Convention: Rockville, MA, USA, 2018. [Google Scholar]
- Marton, M.J.; Weiner, R. Practical Guidance for Implementing Predictive Biomarkers into Early Phase Clinical Studies. BioMed Res. Int. 2013, 2013, 891391. [Google Scholar] [CrossRef]
- Ioannidis, J.P.A. Comparison of Effect Sizes Associated with Biomarkers Reported in Highly Cited Individual Articles and in Subsequent Meta-analyses. JAMA 2011, 305, 2200–2210. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, J.P.A.; Bossuyt, P.M.M. Waste, Leaks, and Failures in the Biomarker Pipeline. Clin. Chem. 2017, 63, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Goossens, N.; Nakagawa, S.; Sun, X.; Hoshida, Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015, 4, 256–269. [Google Scholar] [CrossRef]
- Woolman, M.; Katz, L.; Tata, A.; Basu, S.S.; Zarrine-Afsar, A. Breaking Through the Barrier: Regulatory Considerations Relevant to Ambient Mass Spectrometry at the Bedside. Clin. Lab. Med. 2021, 41, 221–246. [Google Scholar] [CrossRef]
- Kuo, T.-H.; Dutkiewicz, E.P.; Pei, J.; Hsu, C.-C. Ambient Ionization Mass Spectrometry Today and Tomorrow: Embracing Challenges and Opportunities. Anal. Chem. 2020, 92, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Draft Guidance for Industry, Food and Drug Administration Staff, and Clinical Laboratories. Available online: https://www.fda.gov/media/89841/download (accessed on 25 September 2021).
- Kennedy, A.D.; Wittmann, B.M.; Evans, A.M.; Miller, L.A.; Toal, D.R.; Lonergan, S.; Elsea, S.H.; Pappan, K.L. Metabolomics in the clinic: A review of the shared and unique features of untargeted metabolomics for clinical research and clinical testing. J. Mass Spectrom. 2018, 53, 1143–1154. [Google Scholar] [CrossRef]
- 510(k) Substantial Equivalence Determination Decision Summary, Number: K130831. Available online: https://www.accessdata.fda.gov/cdrh_docs/reviews/K130831.pdf (accessed on 25 September 2021).
- 510(k) Substantial Equivalence Determination Decision Summary, Number: KK162950. Available online: https://www.accessdata.fda.gov/cdrh_docs/reviews/K162950.pdf (accessed on 25 September 2021).
- USP Pharmacopeial Convention. Appendix XVIII 2016: Guidance on Developing and Validating Nontargeted Methods for Adulteration Detection; U.S. Pharmacopeial Convention: Rockville, MA, USA, 2016. [Google Scholar]
- Bioanalytical Method Validation Guidance for Industry. Available online: https://www.fda.gov/files/drugs/published/Bioanalytical-Method-Validation-Guidance-for-Industry.pdf (accessed on 25 September 2021).
- Franceschi, P.; Vrhovsek, U.; Mattivi, F.; Wehrens, R. Metabolic Biomarker Identification with Few Samples; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef]
- Kreidler, S.M.; Muller, K.E.; Grunwald, G.K.; Ringham, B.M.; Coker-Dukowitz, Z.T.; Sakhadeo, U.R.; Baron, A.E.; Glueck, D.H. GLIMMPSE: Online Power Computation for Linear Models with and without a Baseline Covariate. J. Stat. Softw. 2013, 54, 1–26. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Woolman, M.; Tata, A.; Zarrine-Afsar, A. Potential impact of tissue molecular heterogeneity on ambient mass spectrometry profiles: A note of caution in choosing the right disease model. Anal. Bioanal. Chem. 2021, 413, 2655–2664. [Google Scholar] [CrossRef]
- Shevchenko, A.; Simons, K. Lipidomics: Coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 2010, 11, 593–598. [Google Scholar] [CrossRef]
- Yang, K.; Han, X. Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem. Sci. 2016, 41, 954–969. [Google Scholar] [CrossRef]
- Milgraum, L.Z.; Witters, L.A.; Pasternack, G.R.; Kuhajda, F.P. Enzymes of the fatty acid synthesis pathway are highly expressed in In Situ breast carcinoma. Clin. Cancer Res. 1997, 3, 2115–2120. [Google Scholar]
- Swinnen, J.V.; Vanderhoydonc, F.; Elgamal, A.A.; Eelen, M.; Vercaeren, I.; Joniau, S.; Van Poppel, H.; Baert, L.; Goossens, K.; Heyns, W.; et al. Selective activation of the fatty acid synthesis pathway in human prostate cancer. Int. J. Cancer 2000, 88, 176–179. [Google Scholar] [CrossRef]
- Jung, Y.Y.; Kim, H.M.; Koo, J.S. Expression of Lipid Metabolism-Related Proteins in Metastatic Breast Cancer. PLoS ONE 2015, 10, e0137204. [Google Scholar] [CrossRef] [PubMed]
- Avantaggiati, M.L. Cancer metabolism as a therapeutic target: Finding the right target(s) in the context of tumor heterogeneity, evolution, and metabolic plasticity. Oncology 2013, 27, 427–432. [Google Scholar]
- Camarda, R.; Zhou, A.; Kohnz, R.A.; Balakrishnan, S.; Mahieu, C.; Anderton, B.; Eyob, H.; Kajimura, S.; Tward, A.; Krings, G.; et al. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat. Med. 2016, 22, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V.; Hamaker, M.; Sun, P.; Le, A.; Gao, P. Therapeutic targeting of cancer cell metabolism. J. Mol. Med. 2011, 89, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Ellsworth, R.E.; Blackburn, H.L.; Shriver, C.D.; Soon-Shiong, P.; Ellsworth, D.L. Molecular heterogeneity in breast cancer: State of the science and implications for patient care. Semin. Cell Dev. Biol. 2017, 64, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Cajal, S.R.Y.; Sesé, M.; Capdevila, C.; Aasen, T.; De Mattos-Arruda, L.; Diaz-Cano, S.J.; Hernández-Losa, J.; Castellvi, J. Clinical implications of intratumor heterogeneity: Challenges and opportunities. J. Mol. Med. 2020, 98, 161–177. [Google Scholar] [CrossRef]
- Antonio, M.J.; Zhang, C.; Le, A. Different Tumor Microenvironments Lead to Different Metabolic Phenotypes. Adv. Exp. Med. Biol. 2021, 1311, 137–147. [Google Scholar] [CrossRef]
- Nabi, K.; Le, A. The Intratumoral Heterogeneity of Cancer Metabolism. Adv. Exp. Med. Biol. 2021, 1311, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.; Le, A. The Heterogeneity of Liver Cancer Metabolism. Adv. Exp. Med. Biol. 2021, 1311, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Le, A. The Heterogeneity of Breast Cancer Metabolism. Adv. Exp. Med. Biol. 2021, 1311, 89–101. [Google Scholar] [CrossRef]
- Sharick, J.T.; Walsh, C.M.; Sprackling, C.M.; Pasch, C.A.; Pham, D.L.; Esbona, K.; Choudhary, A.; Varela, R.G.; Burkard, M.E.; McGregor, S.M.; et al. Metabolic Heterogeneity in Patient Tumor-Derived Organoids by Primary Site and Drug Treatment. Front. Oncol. 2020, 10, 553. [Google Scholar] [CrossRef] [PubMed]
- Balaban, S.; Lee, L.S.; Varney, B.; Aishah, A.; Gao, Q.; Shearer, R.F.; Saunders, D.N.; Grewal, T.; Hoy, A.J. Heterogeneity of fatty acid metabolism in breast cancer cells underlies differential sensitivity to palmitate-induced apoptosis. Mol. Oncol. 2018, 12, 1623–1638. [Google Scholar] [CrossRef] [PubMed]
- Gentric, G.; Mieulet, V.; Mechta-Grigoriou, F. Heterogeneity in Cancer Metabolism: New Concepts in an Old Field. Antioxid. Redox Signal. 2017, 26, 462–485. [Google Scholar] [CrossRef]
- Hensley, C.T.; Faubert, B.; Yuan, Q.; Lev-Cohain, N.; Jin, E.; Kim, J.; Jiang, L.; Ko, B.; Skelton, R.; Loudat, L.; et al. Metabolic Heterogeneity in Human Lung Tumors. Cell 2016, 164, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Commander, R.; Wei, C.; Sharma, A.; Mouw, J.K.; Burton, L.J.; Summerbell, E.; Mahboubi, D.; Peterson, R.J.; Konen, J.; Zhou, W.; et al. Subpopulation targeting of pyruvate dehydrogenase and GLUT1 decouples metabolic heterogeneity during collective cancer cell invasion. Nat. Commun. 2020, 11, 2091–2105. [Google Scholar] [CrossRef]
- Havas-Cavalletti, K.; Milchevskaya, V.; Radic, K.; Alladin, A.; Kafkia, E.; Garcia, M.; Stolte, J.; Klaus, B.; Rotmensz, N.; Gibson, T.J.; et al. Metabolic shifts in residual breast cancer drive tumor recurrence. J. Clin. Investig. 2017, 127, 2091–2105. [Google Scholar] [CrossRef]
- Kim, J.; DeBerardinis, R.J. Mechanisms and Implications of Metabolic Heterogeneity in Cancer. Cell Metab. 2019, 30, 434–446. [Google Scholar] [CrossRef]
- Benfeitas, R.; Uhlen, M.; Nielsen, J.; Mardinoglu, A. New Challenges to Study Heterogeneity in Cancer Redox Metabolism. Front. Cell Dev. Biol. 2017, 5, 65. [Google Scholar] [CrossRef] [PubMed]
- Angerer, T.B.; Magnusson, Y.; Landberg, G.; Fletcher, J.S. Lipid Heterogeneity Resulting from Fatty Acid Processing in the Human Breast Cancer Microenvironment Identified by GCIB-ToF-SIMS Imaging. Anal. Chem. 2016, 88, 11946–11954. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Gao, D.; Zhang, Y.; Wu, D.; Shen, L.; Wang, X. Heterogeneity of lipidomic profiles among lung cancer subtypes of patients. J. Cell. Mol. Med. 2018, 22, 5155–5159. [Google Scholar] [CrossRef]
- Li, Z.; Cheng, S.; Lin, Q.; Cao, W.; Yang, J.; Zhang, M.; Shen, A.; Zhang, W.; Xia, Y.; Ma, X.; et al. Single-cell lipidomics with high structural specificity by mass spectrometry. Nat. Commun. 2021, 12, 1237–1250. [Google Scholar] [CrossRef]
- Blomme, A.; Van Simaeys, G.; Doumont, G.; Costanza, B.; Bellier, J.; Otaka, Y.; Sherer, F.; Lovinfosse, P.; Boutry, S.; Palacios, A.P.; et al. Murine stroma adopts a human-like metabolic phenotype in the PDX model of colorectal cancer and liver metastases. Oncogene 2018, 37, 1237–1250. [Google Scholar] [CrossRef]
- Agar, N.Y.; Golby, A.J.; Ligon, K.L.; Norton, I.; Mohan, V.; Wiseman, J.M.; Tannenbaum, A.; Jolesz, F.A. Development of Stereotactic Mass Spectrometry for Brain Tumor Surgery. Neurosurgery 2011, 68, 280–290. [Google Scholar] [CrossRef]
- Brown, H.M.; Alfaro, C.M.; Pirro, V.; Dey, M.; Hattab, E.M.; Cohen-Gadol, A.A.; Cooks, R.G. Intraoperative Mass Spectrometry Platform for IDH Mutation Status Prediction, Glioma Diagnosis, and Estimation of Tumor Cell Infiltration. J. Appl. Lab. Med. 2021, 6, 902–916. [Google Scholar] [CrossRef]
- Wolff, A.; Hammond, M.E.H.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Update. Arch. Pathol. Lab. Med. 2014, 138, 241–256. [Google Scholar] [CrossRef]
- Rauser, S.; Marquardt, C.; Balluff, B.; Deininger, S.-O.; Albers, C.; Belau, E.; Hartmer, R.; Suckau, D.; Specht, K.; Ebert, M.P.; et al. Classification of HER2 Receptor Status in Breast Cancer Tissues by MALDI Imaging Mass Spectrometry. J. Proteome Res. 2010, 9, 1854–1863. [Google Scholar] [CrossRef] [PubMed]
- Porcari, A.; Zhang, J.; Garza, K.Y.; Peres, R.M.R.; Lin, J.; Young, J.H.; Tibshirani, R.; Nagi, C.; Paiva, G.R.; Carter, S.A.; et al. Multicenter Study Using Desorption-Electrospray-Ionization-Mass-Spectrometry Imaging for Breast-Cancer Diagnosis. Anal. Chem. 2018, 90, 11324–11332. [Google Scholar] [CrossRef] [PubMed]
- Buckley, N.; Forde, C.; McArt, D.G.; Boyle, D.P.; Mullan, P.B.; James, J.A.; Maxwell, P.; McQuaid, S.; Salto-Tellez, M. Quantification of HER2 heterogeneity in breast cancer–implications for identification of sub-dominant clones for personalised treatment. Sci. Rep. 2016, 6, 23383. [Google Scholar] [CrossRef]
- Sans, M.; Gharpure, K.; Tibshirani, R.; Zhang, J.; Liang, L.; Liu, J.; Young, J.H.; Dood, R.L.; Sood, A.K.; Eberlin, L.S. Metabolic Markers and Statistical Prediction of Serous Ovarian Cancer Aggressiveness by Ambient Ionization Mass Spectrometry Imaging. Cancer Res. 2017, 77, 2903–2913. [Google Scholar] [CrossRef]
- Thompson, D.O.; Karmali, R.A.; Cohen, L.A.; Choi, K.; Rose, D.P. Dietary Fat and Mammary Cancer. II. Modulation of Serum and Tumor Lipid Composition and Tumor Prostaglandins by Different Dietary Fats: Association with Tumor Incidence Patterns. J. Natl. Cancer Inst. 1986, 77, 43–51. [Google Scholar] [CrossRef]
- O’Keefe, S.J.D.; Li, J.; Lahti, L.; Ou, J.; Carbonero, F.; Mohammed, K.; Posma, J.M.; Kinross, J.; Wahl, E.; Ruder, E.; et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015, 6, 6342. [Google Scholar] [CrossRef]
- Ellis, J.K.; Athersuch, T.J.; Thomas, L.D.K.; Teichert, F.; Pérez-Trujillo, M.; Svendsen, C.; Spurgeon, D.J.; Singh, R.; Järup, L.; Bundy, J.G.; et al. Metabolic profiling detects early effects of environmental and lifestyle exposure to cadmium in a human population. BMC Med. 2012, 10, 61. [Google Scholar] [CrossRef] [PubMed]
- Finicle, B.T.; Jayashankar, V.; Edinger, A.L. Nutrient scavenging in cancer. Nat. Rev. Cancer 2018, 18, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Bruce, K.D.; Zsombok, A.; Eckel, R.H. Lipid Processing in the Brain: A Key Regulator of Systemic Metabolism. Front. Endocrinol. 2017, 8, 60. [Google Scholar] [CrossRef]
- Woolman, M.; Katz, L.; Gopinath, G.; Kiyota, T.; Kuzan-Fischer, C.M.; Ferry, I.; Zaidi, M.; Peters, K.; Aman, A.; McKee, T.; et al. Mass Spectrometry Imaging Reveals a Gradient of Cancer-like Metabolic States in the Vicinity of Cancer Not Seen in Morphometric Margins from Microscopy. Anal. Chem. 2021, 93, 4408–4416. [Google Scholar] [CrossRef]
- Sasaki, A.; Nagatake, T.; Egami, R.; Gu, G.; Takigawa, I.; Ikeda, W.; Nakatani, T.; Kunisawa, J.; Fujita, Y. Obesity Suppresses Cell-Competition-Mediated Apical Elimination of RasV12-Transformed Cells from Epithelial Tissues. Cell Rep. 2018, 23, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Evans, K.; Burdge, G.C.; Wootton, S.A.; Clark, M.L.; Frayn, K.N. Regulation of dietary fatty acid entrapment in subcutaneous adipose tissue and skeletal muscle. Diabetes 2002, 51, 2684–2690. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, E.; Makowski, L.; DiGiovanni, J.; Kolonin, M.G. Cancer as a Matter of Fat: The Crosstalk between Adipose Tissue and Tumors. Trends Cancer 2018, 4, 374–384. [Google Scholar] [CrossRef]
- Zhao, C.; Wu, M.; Zeng, N.; Xiong, M.; Hu, W.; Lv, W.; Yi, Y.; Zhang, Q.; Wu, Y. Cancer-associated adipocytes: Emerging supporters in breast cancer. J. Exp. Clin. Cancer Res. 2020, 39, 156. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Li, Y.; Xing, L.; Tan, Y.; Sun, J.; Zeng, B.; Xiang, T.; Tan, J.; Ren, G.; Wang, Y. Utilization of adipocyte-derived lipids and enhanced intracellular trafficking of fatty acids contribute to breast cancer progression. Cell Commun. Signal. 2018, 16, 32. [Google Scholar] [CrossRef] [PubMed]
- Volden, P.A.; Skor, M.N.; Johnson, M.B.; Singh, P.; Patel, F.N.; McClintock, M.K.; Brady, M.J.; Conzen, S.D. Mammary Adipose Tissue-Derived Lysophospholipids Promote Estrogen Receptor–Negative Mammary Epithelial Cell Proliferation. Cancer Prev. Res. 2016, 9, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Attané, C.; Milhas, D.; Hoy, A.; Muller, C. Metabolic Remodeling Induced by Adipocytes: A New Achilles’ Heel in Invasive Breast Cancer? Curr. Med. Chem. 2020, 27, 3984–4001. [Google Scholar] [CrossRef]
- Munteanu, R.; Onaciu, A.; Moldovan, C.; Zimta, A.-A.; Gulei, D.; Paradiso, A.V.; Lazar, V.; Berindan-Neagoe, I. Adipocyte-Based Cell Therapy in Oncology: The Role of Cancer-Associated Adipocytes and Their Reinterpretation as Delivery Platforms. Pharmaceutics 2020, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Zhao, Y.; Tang, Y.; Wei, X.; Shi, X.; Sun, W.; Sun, X.; Yin, X.; Sun, X.; Kim, S.; et al. Chronic Alcohol Exposure Stimulates Adipose Tissue Lipolysis in Mice: Role of Reverse Triglyceride Transport in the Pathogenesis of Alcoholic Steatosis. Am. J. Pathol. 2012, 180, 998–1007. [Google Scholar] [CrossRef]
- Pravdova, E.; Fickova, M. Alcohol intake modulates hormonal activity of adipose tissue. Endocr. Regul. 2006, 40, 91–104. [Google Scholar]
- Wei, X.; Shi, X.; Zhong, W.; Zhao, Y.; Tang, Y.; Sun, W.; Yin, X.; Bogdanov, B.; Kim, S.; McClain, C.; et al. Chronic Alcohol Exposure Disturbs Lipid Homeostasis at the Adipose Tissue-Liver Axis in Mice: Analysis of Triacylglycerols Using High-Resolution Mass Spectrometry in Combination with In Vivo Metabolite Deuterium Labeling. PLoS ONE 2013, 8, e55382. [Google Scholar] [CrossRef]
- Sales, S.; Graessler, J.; Ciucci, S.; Al-Atrib, R.; Vihervaara, T.; Schuhmann, K.; Kauhanen, D.; Sysi-Aho, M.; Bornstein, S.R.; Bickle, M.; et al. Gender, Contraceptives and Individual Metabolic Predisposition Shape a Healthy Plasma Lipidome. Sci. Rep. 2016, 6, 27710. [Google Scholar] [CrossRef]
- Slade, E.; Irvin, M.R.; Xie, K.; Arnett, D.K.; Claas, S.A.; Kind, T.; Fardo, D.W.; Graf, G.A. Age and sex are associated with the plasma lipidome: Findings from the GOLDN study. Lipids Health Dis. 2021, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Audano, M.; Maldini, M.; De Fabiani, E.; Mitro, N.; Caruso, D. Gender-related metabolomics and lipidomics: From experimental animal models to clinical evidence. J. Proteom. 2018, 178, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Chandrasekaran, B. 39 Dimensionality and sample size considerations in pattern recognition practice. In Handbook of Statistics; Elsevier: Amsterdam, The Netherlands, 1982; Volume 2, pp. 835–855. [Google Scholar] [CrossRef]
- Kalayeh, H.M.; Landgrebe, D.A. Predicting the Required Number of Training Samples. IEEE Trans. Pattern Anal. Mach. Intell. 1983, PAMI-5, 664–667. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Tamayo, P.; Rogers, S.; Rifkin, R.; Engle, A.; Campbell, C.; Golub, T.R.; Mesirov, J.P. Estimating Dataset Size Requirements for Classifying DNA Microarray Data. J. Comput. Biol. 2003, 10, 119–142. [Google Scholar] [CrossRef] [PubMed]
- Dobbin, K.K.; Zhao, Y.; Simon, R.M. How Large a Training Set is Needed to Develop a Classifier for Microarray Data? Clin. Cancer Res. 2008, 14, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, R.L.; Zeng-Treitler, Q.; Kandula, S.; Ngo, L.H. Predicting sample size required for classification performance. BMC Med. Inform. Decis. Mak. 2012, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Beleites, C.; Neugebauer, U.; Bocklitz, T.; Krafft, C.; Popp, J. Sample size planning for classification models. Anal. Chim. Acta 2013, 760, 25–33. [Google Scholar] [CrossRef]
- Lee, P.Y.; Yeoh, Y.; Omar, N.; Pung, Y.-F.; Lim, L.C.; Low, T.Y. Molecular tissue profiling by MALDI imaging: Recent progress and applications in cancer research. Crit. Rev. Clin. Lab. Sci. 2021, 1–17. [Google Scholar] [CrossRef]
- Mascini, N.E.; Teunissen, J.; Noorlag, R.; Willems, S.M.; Heeren, R.M. Tumor classification with MALDI-MSI data of tissue microarrays: A case study. Methods 2018, 151, 21–27. [Google Scholar] [CrossRef]
- Klein, O.; Kanter, F.; Kulbe, H.; Jank, P.; Denkert, C.; Nebrich, G.; Schmitt, W.; Wu, Z.; Kunze, C.A.; Sehouli, J.; et al. MALDI-Imaging for Classification of Epithelial Ovarian Cancer Histotypes from a Tissue Microarray Using Machine Learning Methods. Proteom. Clin. Appl. 2019, 13, e1700181. [Google Scholar] [CrossRef]
- Kassuhn, W.; Klein, O.; Darb-Esfahani, S.; Lammert, H.; Handzik, S.; Taube, E.; Schmitt, W.; Keunecke, C.; Horst, D.; Dreher, F.; et al. Classification of Molecular Subtypes of High-Grade Serous Ovarian Cancer by MALDI-Imaging. Cancers 2021, 13, 1512. [Google Scholar] [CrossRef] [PubMed]
- Cairns, D.A.; Barrett, J.; Billingham, L.; Stanley, A.J.; Xinarianos, G.; Field, J.; Johnson, P.J.; Selby, P.J.; Banks, R. Sample size determination in clinical proteomic profiling experiments using mass spectrometry for class comparison. Proteomics 2009, 9, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Demidenko, E. Sample size determination for logistic regression revisited. Stat. Med. 2007, 26, 3385–3397. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K.; Michaud, J.; Scott, H. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 2005, 21, 2067–2075. [Google Scholar] [CrossRef] [PubMed]
- Nyangoma, S.O.; Collins, S.I.; Altman, D.G.; Johnson, P.; Billingham, L. Sample Size Calculations for Designing Clinical Proteomic Profiling Studies Using Mass Spectrometry. Stat. Appl. Genet. Mol. Biol. 2012, 11. [Google Scholar] [CrossRef]
- Alfaro, C.M.; Jarmusch, A.K.; Pirro, V.; Kerian, K.S.; Masterson, T.A.; Cheng, L.; Cooks, R.G. Ambient ionization mass spectrometric analysis of human surgical specimens to distinguish renal cell carcinoma from healthy renal tissue. Anal. Bioanal. Chem. 2016, 408, 5407–5414. [Google Scholar] [CrossRef]
- Margulis, K.; Chiou, A.S.; Aasi, S.Z.; Tibshirani, R.J.; Tang, J.Y.; Zare, R.N. Distinguishing malignant from benign microscopic skin lesions using desorption electrospray ionization mass spectrometry imaging. Proc. Natl. Acad. Sci. USA 2018, 115, 6347–6352. [Google Scholar] [CrossRef] [PubMed]
- Giordano, S.; Takeda, S.; Donadon, M.; Saiki, H.; Brunelli, L.; Pastorelli, R.; Cimino, M.; Soldani, C.; Franceschini, B.; Di Tommaso, L.; et al. Rapid automated diagnosis of primary hepatic tumour by mass spectrometry and artificial intelligence. Liver Int. 2020, 40, 3117–3124. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Cameron, S.J.; Whelan, E.; Bowden, S.; Tzafetas, M.; Mitra, A.; Semertzidou, A.; Athanasiou, A.; Bennett, P.R.; MacIntyre, D.A.; et al. Laser-assisted rapid evaporative ionisation mass spectrometry (LA-REIMS) as a metabolomics platform in cervical cancer screening. EBioMedicine 2020, 60, 103017. [Google Scholar] [CrossRef]
- Van Kersbergen, J.; Ghazvinian Zanjani, F.; Zinger, S.; Van der Sommen, F.; Balluff, B.; Vos, N.; Ellis, S.; Heeran, R.; Lucas, M.; Marquering, H.; et al. Cancer detection in mass spectrometry imaging data by dilated convolutional neural networks. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; p. 16. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2012, 25, 84–90. [Google Scholar] [CrossRef]
- Forman, G.; Cohen, I. Learning from little: Comparison of classifiers given little training. In European Conference on Principles of Data Mining and Knowledge Discovery; Springer: Berlin/Heidelberg, Germany, 2004; Volume 3202. [Google Scholar]
- Shaikhina, T.; Khovanova, N.A. Handling limited datasets with neural networks in medical applications: A small-data approach. Artif. Intell. Med. 2017, 75, 51–63. [Google Scholar] [CrossRef]
- Bodai, Z.; Cameron, S.; Bolt, F.; Simon, D.; Schaffer, R.; Karancsi, T.; Balog, J.; Rickards, T.; Burke, A.; Hardiman, K.; et al. Effect of Electrode Geometry on the Classification Performance of Rapid Evaporative Ionization Mass Spectrometric (REIMS) Bacterial Identification. J. Am. Soc. Mass Spectrom. 2018, 29, 26–33. [Google Scholar] [CrossRef]
- Johnson, C.; Gonzalez, F.J. Challenges and opportunities of metabolomics. J. Cell. Physiol. 2012, 227, 2975–2981. [Google Scholar] [CrossRef]
- Johnson, C.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Simon, I.; Zhuo, S.; Corral, L.; Diamandis, E.; Sarno, M.J.; Wolfert, R.L.; Kim, N.W. B7-H4 Is a Novel Membrane-Bound Protein and a Candidate Serum and Tissue Biomarker for Ovarian Cancer. Cancer Res. 2006, 66, 1570–1575. [Google Scholar] [CrossRef]
- Cramer, D.W.; Bast, R.; Berg, C.D.; Diamandis, E.; Godwin, A.K.; Hartge, P.; Lokshin, A.E.; Lu, K.H.; McIntosh, M.W.; Mor, G.; et al. Ovarian Cancer Biomarker Performance in Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial Specimens. Cancer Prev. Res. 2011, 4, 365–374. [Google Scholar] [CrossRef]
- Xu, Y.; Shen, Z.; Wiper, D.W.; Wu, M.; Morton, R.E.; Elson, P.; Kennedy, A.W.; Belinson, J.; Markman, M.; Casey, G. Lysophosphatidic Acid as a Potential Biomarker for Ovarian and Other Gynecologic Cancers. JAMA 1998, 280, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.L. Plasma Lysophosphatidic Acid Concentration and Ovarian Cancer. JAMA 2002, 287, 3081–3082. [Google Scholar] [CrossRef]
- Stephan, C.; Wilkosz, J.; Różański, W.; Ecke, T.H.; Lein, M.; Bryś, M.; Krześlak, A.; Chwatko, G.; Jung, K. Urinary thiosulfate as failed prostate cancer biomarker—An exemplary multicenter re-evaluation study. Clin. Chem. Lab. Med. 2015, 53, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Massaro, A.; Negro, A.; Bragolusi, M.; Miano, B.; Tata, A.; Suman, M.; Piro, R. Oregano authentication by mid-level data fusion of chemical fingerprint signatures acquired by ambient mass spectrometry. Food Control 2021, 126, 108058. [Google Scholar] [CrossRef]
- Damiani, T.; Cavanna, D.; Serani, A.; Dall’Asta, C.; Suman, M. GC-IMS and FGC-Enose fingerprint as screening tools for revealing extra virgin olive oil blending with soft-refined olive oils: A feasibility study. Microchem. J. 2020, 159, 105374. [Google Scholar] [CrossRef]
- Kunz, M.; Ottaway, J.; Kalivas, J.H.; Georgiou, C.; Mousdis, G. Updating a Synchronous Fluorescence Spectroscopic Virgin Olive Oil Adulteration Calibration to a New Geographical Region. J. Agric. Food Chem. 2011, 59, 1051–1057. [Google Scholar] [CrossRef]
- Riedl, J.; Esslinger, S.; Fauhl-Hassek, C. Review of validation and reporting of non-targeted fingerprinting approaches for food authentication. Anal. Chim. Acta 2015, 885, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Schymanski, E.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.; Hollender, J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef] [PubMed]
- Nishi, A.; Ohbuchi, K.; Kaifuchi, N.; Shimobori, C.; Kushida, H.; Yamamoto, M.; Kita, Y.; Tokuoka, S.M.; Yachie, A.; Matsuoka, Y.; et al. Publisher Correction: LimeMap: A comprehensive map of lipid mediator metabolic pathways. NPJ Syst. Biol. Appl. 2021, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Nishi, A.; Ohbuchi, K.; Kaifuchi, N.; Shimobori, C.; Kushida, H.; Yamamoto, M.; Kita, Y.; Tokuoka, S.M.; Yachie, A.; Matsuoka, Y.; et al. LimeMap: A comprehensive map of lipid mediator metabolic pathways. NPJ Syst. Biol. Appl. 2021, 7, 6. [Google Scholar] [CrossRef]
- Johnson, C.; Ivanisevic, J.; Benton, H.P.; Siuzdak, G. Bioinformatics: The Next Frontier of Metabolomics. Anal. Chem. 2015, 87, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Thiele, I.; Swainston, N.; Fleming, R.M.; Hoppe, A.; Sahoo, S.; Aurich, M.K.; Haraldsdottir, H.; Mo, M.L.; Rolfsson, O.; Stobbe, M.D.; et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013, 31, 419–425. [Google Scholar] [CrossRef]
- Zamboni, N.; Saghatelian, A.; Patti, G.J. Defining the Metabolome: Size, Flux, and Regulation. Mol. Cell 2015, 58, 699–706. [Google Scholar] [CrossRef]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting Network Activity from High Throughput Metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef]
- Cottret, L.; Wildridge, D.; Vinson, F.; Barrett, M.; Charles, H.; Sagot, M.-F.; Jourdan, F. MetExplore: A web server to link metabolomic experiments and genome-scale metabolic networks. Nucleic Acids Res. 2010, 38, W132–W137. [Google Scholar] [CrossRef]
- Scalbert, A.; Brennan, L.; Manach, C.; Andres-Lacueva, C.; Dragsted, L.O.; Draper, J.; Rappaport, S.M.; van der Hooft, J.J.; Wishart, D.S. The food metabolome: A window over dietary exposure. Am. J. Clin. Nutr. 2014, 99, 1286–1308. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef]
- Margulis, K.; Zhou, Z.; Fang, Q.; Sievers, R.E.; Lee, R.J.; Zare, R.N. Combining Desorption Electrospray Ionization Mass Spectrometry Imaging and Machine Learning for Molecular Recognition of Myocardial Infarction. Anal. Chem. 2018, 90, 12198–12206. [Google Scholar] [CrossRef]
- Schaffer, S.W.; Jong, C.J.; Kc, R.; Azuma, J. Physiological roles of taurine in heart and muscle. J. Biomed. Sci. 2010, 17 (Suppl. 1), S2–S8. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, S.W.; Jong, C.J.; Ito, T.; Azuma, J. Effect of taurine on ischemia-reperfusion injury. Amino Acids 2014, 46, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Ardisson, L.P.; Rafacho, B.P.; Santos, P.P.; Assalin, H.; Gonçalves, A.F.; Azevedo, P.S.; Minicucci, M.F.; Polegato, B.F.; Okoshi, K.; Marchini, J.S.; et al. Taurine attenuates cardiac remodeling after myocardial infarction. Int. J. Cardiol. 2013, 168, 4925–4926. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Banerjee, S.; Zare, R.N.; Tibshirani, R.J.; Kunder, C.; Nolley, R.; Fan, R.; Brooks, J.D.; Sonn, G.A. Diagnosis of prostate cancer by desorption electrospray ionization mass spectrometric imaging of small metabolites and lipids. Proc. Natl. Acad. Sci. USA 2017, 114, 3334–3339. [Google Scholar] [CrossRef] [PubMed]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Liebisch, G.; Vizcaino, J.A.; Köfeler, H.; Trötzmüller, M.; Griffiths, W.; Schmitz, G.; Spener, F.; Wakelam, M. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
- Lipidomics Standards Initiative Consortium. Lipidomics needs more standardization. Nat. Metab. 2019, 1, 745–747. [Google Scholar] [CrossRef] [PubMed]
| Current Problems | Potential Solutions | |
|---|---|---|
Discovery | Poor design, conduct, and analysis | Methodological rigor |
Unaccounted multiplicity | Appropriate use of statistics | |
Small studies | Larger, collaborative studies | |
Extreme case selection | Proper case-control or cohort selection | |
Nonrigorous exploratory nature of studies | More rigorous training of scientists | |
Poor reporting | Use of reporting standards | |
Selective reporting | Preregistration | |
Spin in interpretation | Careful editorial and peer-review | |
Validation | Any and all problems seen in discovery studies | Similar solutions, as above |
Lack of replication efforts | Incentives for running replication studies | |
Inbred replications (same populations, same investigators) | More emphasis on external, independent validation | |
Incomplete, suboptimal validation | Careful consideration of independence | |
No systematic reviews | Good-quality systematic reviews | |
Inflation in early, small studies | Large validation studies, ideally from collaborations without bias | |
Spurious variability in measurements, methods, analyses across studies | Standardization and harmonization of processes, collaborative consortia | |
Transition to clinical translation | Inappropriate perusal of clinical translation | Rigorous systematic reviews |
Poor prioritization | Rigorous umbrella reviews | |
Sponsor bias driving translation urge | Independent assessment of the evidence | |
Inappropriate stagnation without clinical translation | Incentives to translate | |
Evaluation | Focus only on accuracy and process measures | Emphasize patient outcomes |
Few randomized trials of biomarkers | Promote randomized trials of biomarker use | |
Use for unclear informational purposes | Evaluate utility of information for the sake of information and potential collateral harms | |
Improper use for selection and stratified/subgroup analyses in trials | Validation of utility of stratified/subgroup analyses | |
Implementation and deimplementation | Poor understanding of the use of biomarkers in real-life settings | Implementation studies assessing use and outcomes in diverse settings |
Lack of rigorous guidelines | Standardized, nonconflicted guidelines | |
Discordant guidelines | Strengthening of regulation for biomarkers | |
Not well-defined regulatory landscape | Testing of utility of long-used biomarkers | |
Entrenched useless biomarkers | Overcoming resistance from conflicted stakeholders, higher barrier for reimbursement | |
Resistance to deimplementation even with convincing negative evidence |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katz, L.; Tata, A.; Woolman, M.; Zarrine-Afsar, A. Lipid Profiling in Cancer Diagnosis with Hand-Held Ambient Mass Spectrometry Probes: Addressing the Late-Stage Performance Concerns. Metabolites 2021, 11, 660. https://doi.org/10.3390/metabo11100660
Katz L, Tata A, Woolman M, Zarrine-Afsar A. Lipid Profiling in Cancer Diagnosis with Hand-Held Ambient Mass Spectrometry Probes: Addressing the Late-Stage Performance Concerns. Metabolites. 2021; 11(10):660. https://doi.org/10.3390/metabo11100660
Chicago/Turabian StyleKatz, Lauren, Alessandra Tata, Michael Woolman, and Arash Zarrine-Afsar. 2021. "Lipid Profiling in Cancer Diagnosis with Hand-Held Ambient Mass Spectrometry Probes: Addressing the Late-Stage Performance Concerns" Metabolites 11, no. 10: 660. https://doi.org/10.3390/metabo11100660
APA StyleKatz, L., Tata, A., Woolman, M., & Zarrine-Afsar, A. (2021). Lipid Profiling in Cancer Diagnosis with Hand-Held Ambient Mass Spectrometry Probes: Addressing the Late-Stage Performance Concerns. Metabolites, 11(10), 660. https://doi.org/10.3390/metabo11100660






