SARS-CoV2 Infection Alters Tryptophan Catabolism and Phospholipid Metabolism
Abstract
:1. Introduction
2. Results
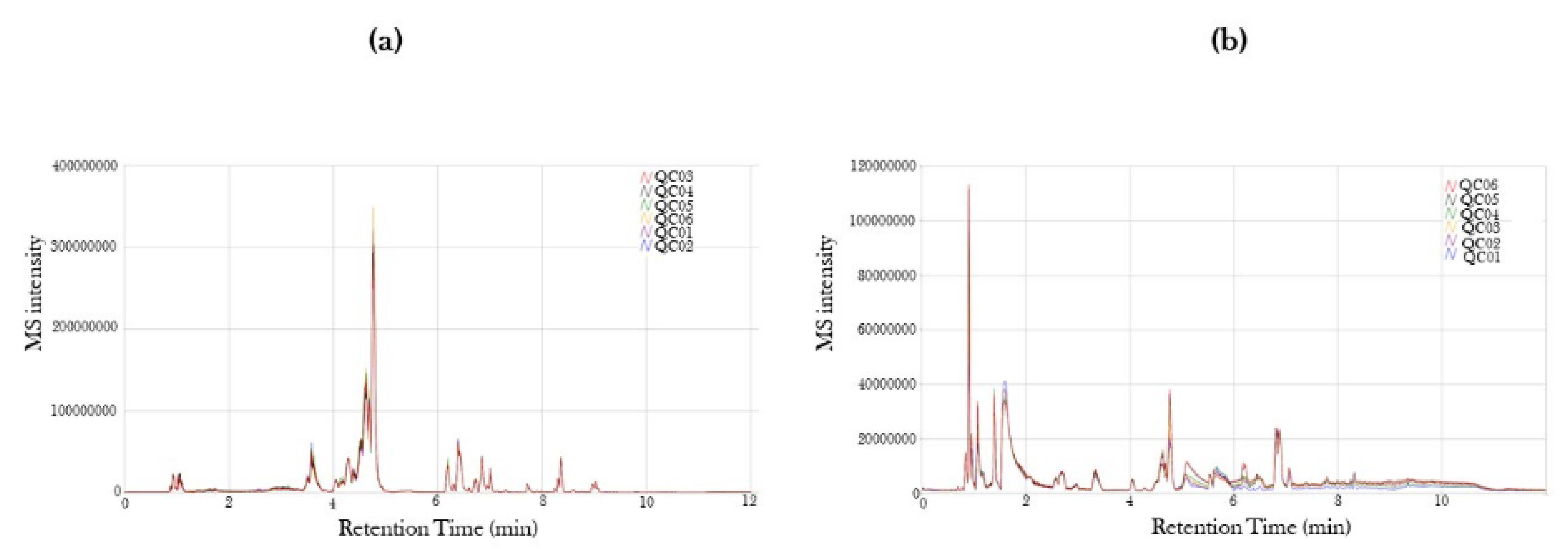
2.1. Mass Spectra in Positive and Negative Ion Mode of Serum from COVID-19-Positive and COVID-19-Recovered Subjects Analyzed by UPLC-MS
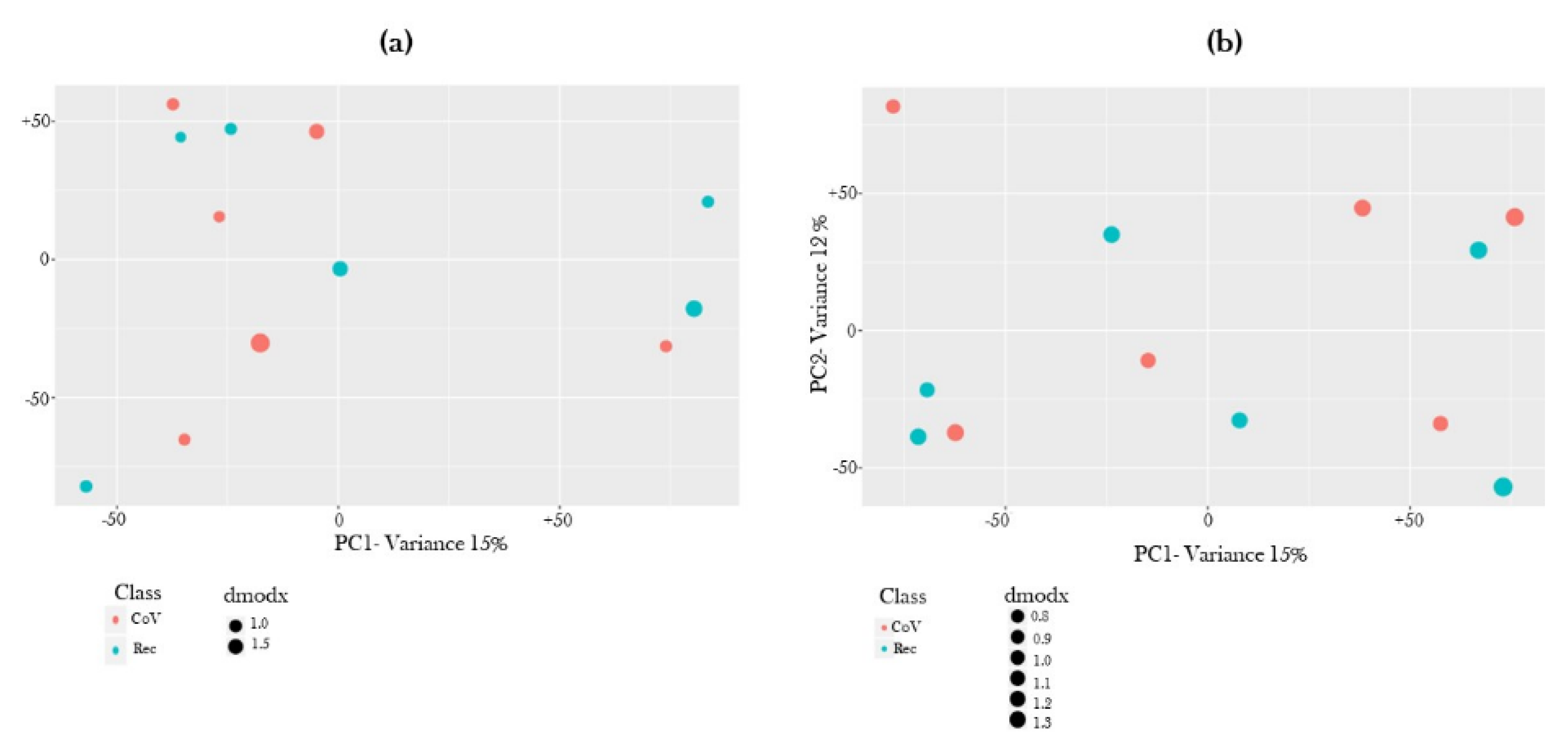
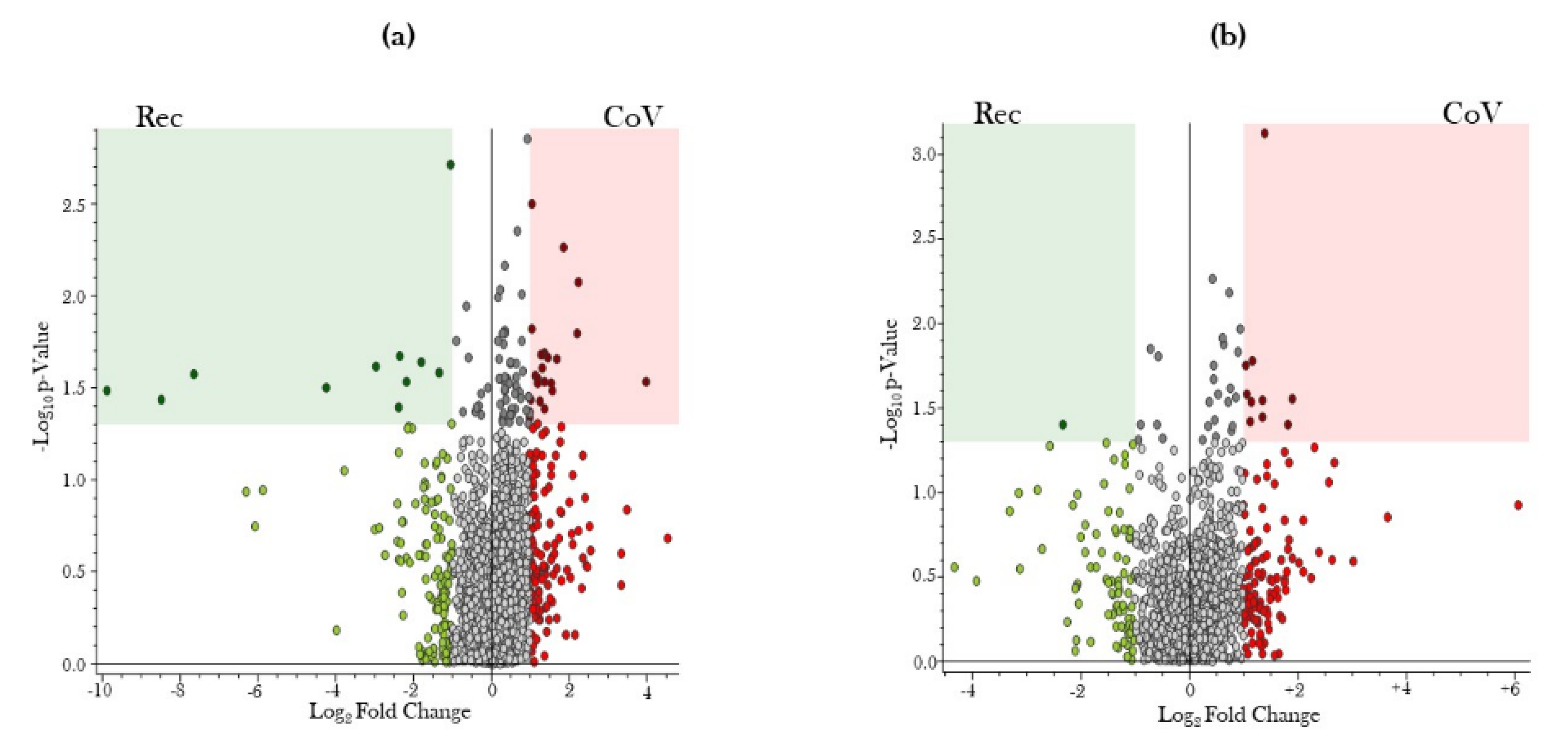
2.2. Global Metabolite Profiling Identified Significantly Dysregulated Metabolites in COVID-19-Positive Patients as Compared to COVID-19-Recovered Subjects
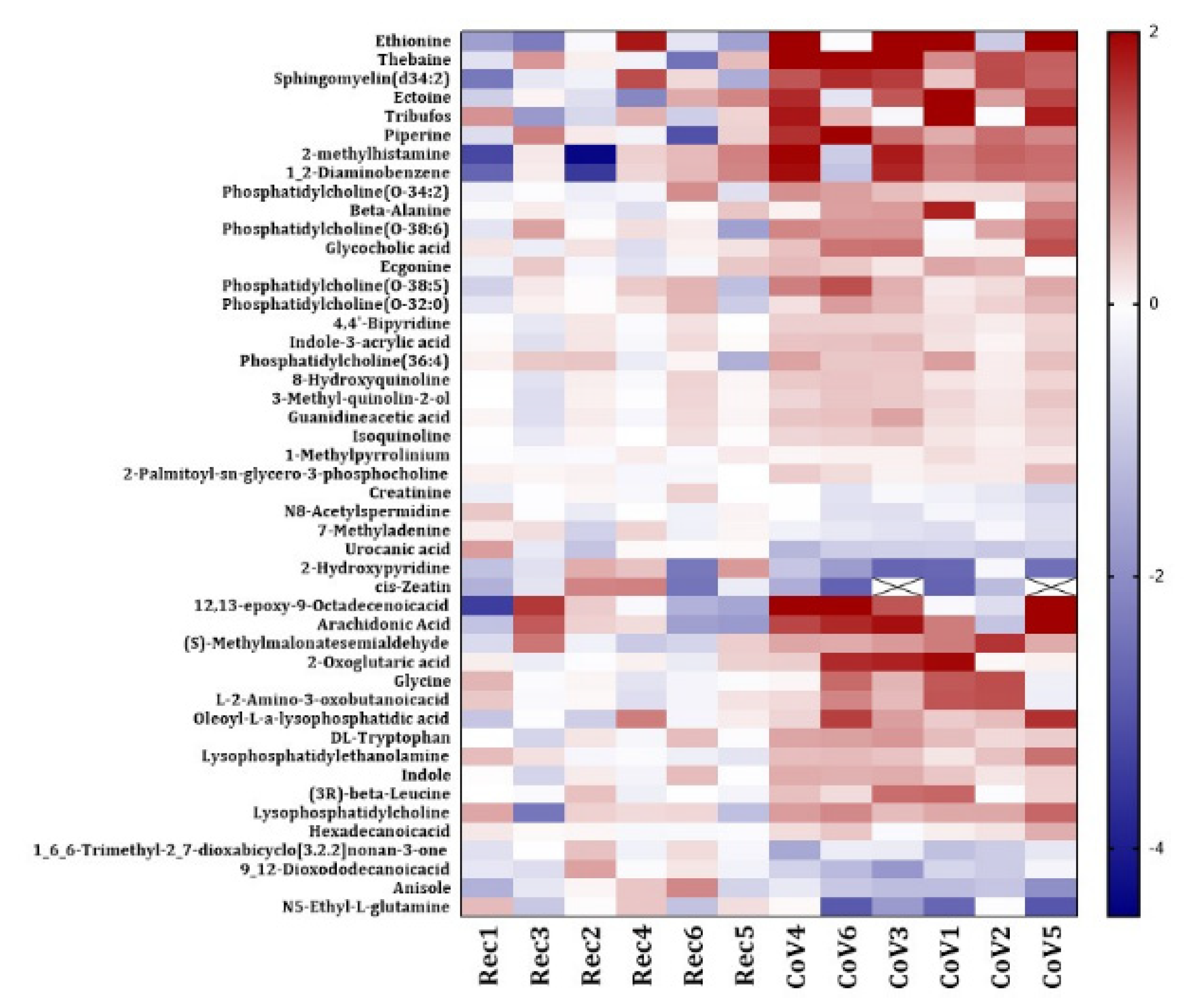
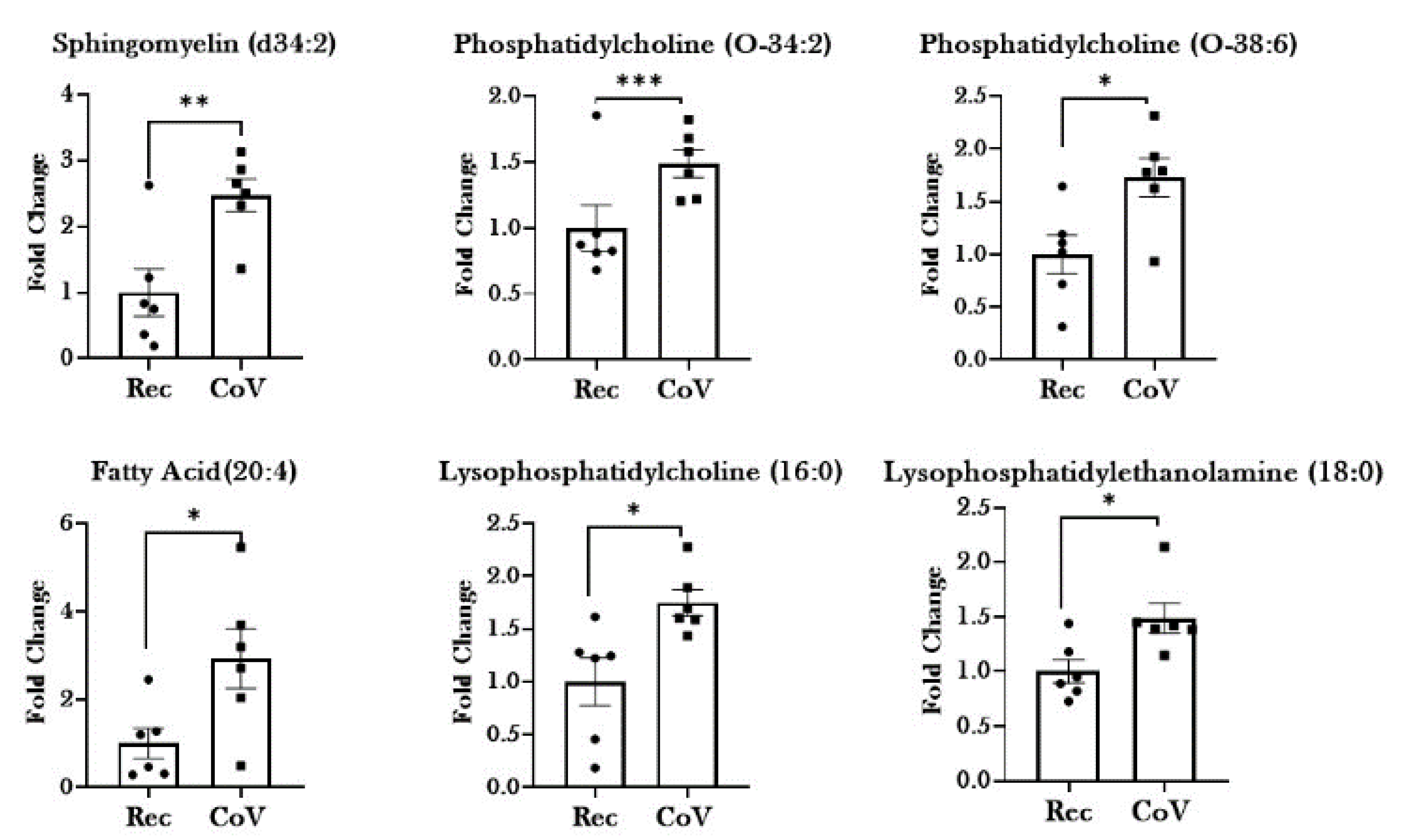
2.3. Dysregulated Lipid Metabolism in Patients with COVID-19
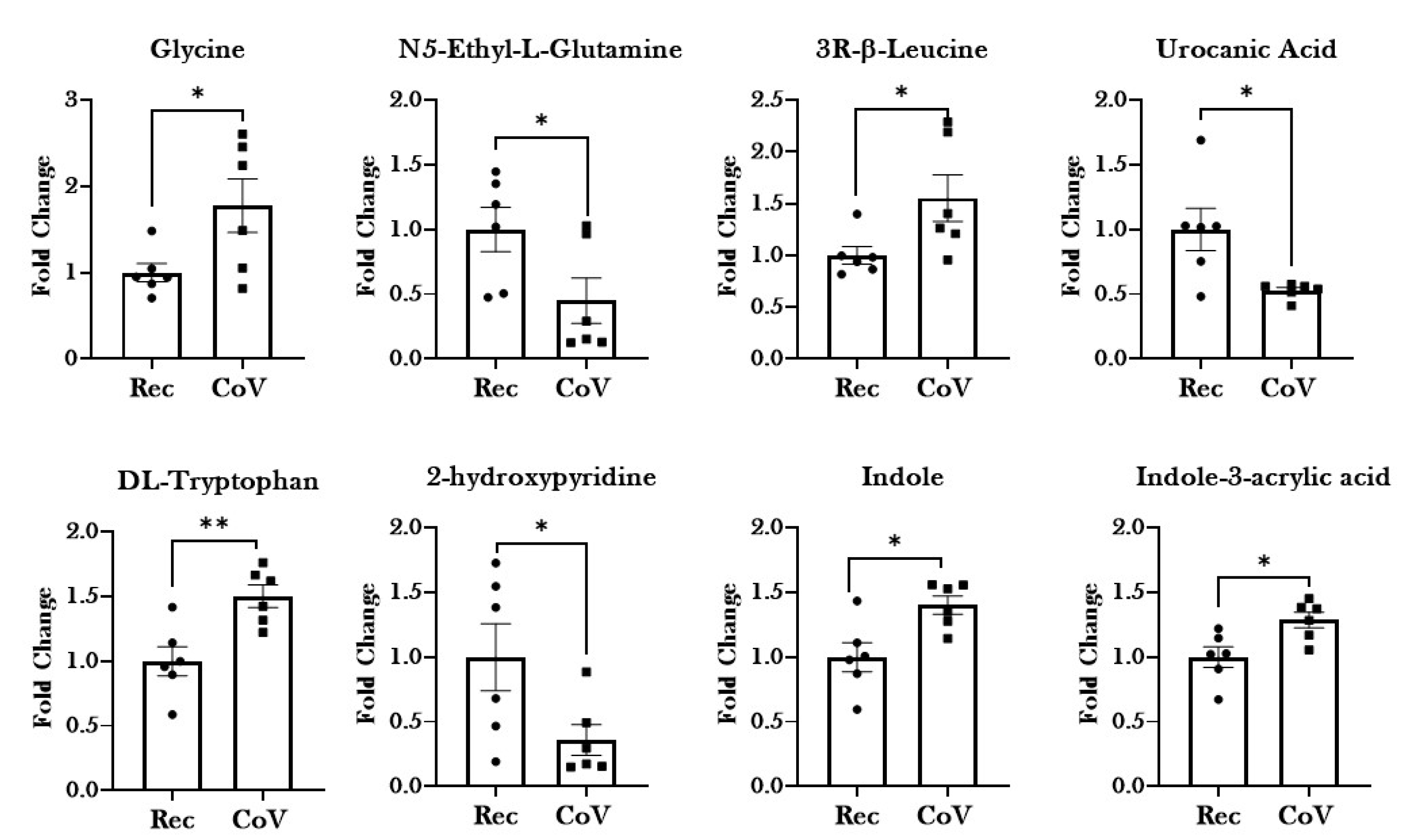
2.4. Significant Alterations in Products from Tryptophan Metabolism in Patients with COVID-19
2.5. Other Dysregulated Metabolites in the Serum from COVID-19-Positive Patients
3. Discussion
4. Methods
4.1. Ethics and Approvals
4.2. Human Subjects
4.3. Metabolomic Analyses
4.3.1. Nomenclature
4.3.2. Sample Preparation
4.3.3. LC/MS Data Acquisition
4.3.4. Data Analyses
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| COVID-19 | Coronavirus Disease-2019 |
| SARS-CoV2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| WHO | World Health Organization |
| UPLC-MS | Ultrahigh Performance Liquid Chromatography/tandem mass spectrometry |
| HIV | Human Immunodeficiency Virus |
| AhR | Aryl hydrocarbon Receptor |
| I3P | Indole-3-propionic acid |
| CD | Cluster of differentiation |
References
- Roth, M.D.; Connett, J.E.; D’Armiento, J.M.; Foronjy, R.F.; Friedman, P.J.; Goldin, J.G.; Louis, T.A.; Mao, J.T.; Muindi, J.R.; O’Connor, G.T.; et al. Feasibility of retinoids for the treatment of emphysema study. Chest 2006, 130, 1334–1345. [Google Scholar] [CrossRef] [PubMed]
- Blasco, H.; Bessy, C.; Plantier, L.; Lefevre, A.; Piver, E.; Bernard, L.; Marlet, J.; Stefic, K.; Benz-de Bretagne, I.; Cannet, P.; et al. The specific metabolome profiling of patients infected by SARS-COV-2 supports the key role of tryptophan-nicotinamide pathway and cytosine metabolism. Sci. Rep. 2020, 10, 16824. [Google Scholar] [CrossRef]
- Danlos, F.-X.; Grajeda-Iglesias, C.; Durand, S.; Sauvat, A.; Roumier, M.; Cantin, D.; Colomba, E.; Rohmer, J.; Pommeret, F.; Baciarello, G.; et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 2021, 12, 258. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Boasso, A.; Shearer, G.M. How does indoleamine 2,3-dioxygenase contribute to HIV-mediated immune dysregulation. Curr. Drug Metab. 2007, 8, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Severe Covid-19 GWAS Group. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef]
- D’Alessandro, A.; Thomas, T.; Dzieciatkowska, M.; Hill, R.C.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hod, E.A.; Spitalnik, S.L.; Hansen, K.C. Serum Proteomics in COVID-19 Patients: Altered Coagulation and Complement Status as a Function of IL-6 Level. J. Proteome Res. 2020, 19, 4417–4427. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Song, J.W.; Lam, S.M.; Fan, X.; Cao, W.J.; Wang, S.Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.P.; Xu, Z.; et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 2020, 32, 188–202.e5. [Google Scholar] [CrossRef]
- Li, S.; Ma, F.; Yokota, T.; Garcia, G., Jr.; Palermo, A.; Wang, Y.; Farrell, C.; Wang, Y.-C.; Wu, R.; Zhou, Z.; et al. Metabolic reprogramming and epigenetic changes of vital organs in SARS-CoV-2-induced systemic toxicity. JCI Insight 2021, 6, e145027. [Google Scholar] [CrossRef]
- Jia, H.; Liu, C.; Li, D.; Huang, Q.; Liu, D.; Zhang, Y.; Ye, C.; Zhou, D.; Wang, Y.; Tan, Y.; et al. Metabolomic analyses reveals new stage-specific features of the COVID-19. Eur. Respir. J. 2021, 2100284. [Google Scholar] [CrossRef]
- Migaud, M.; Gandotra, S.; Chand, H.S.; Gillespie, M.N.; Thannickal, V.J.; Langley, R.J. Metabolomics to Predict Antiviral Drug Efficacy in COVID-19. Am. J. Respir. Cell Mol. Biol. 2020, 63, 396–398. [Google Scholar] [CrossRef]
- Doğan, H.O.; Şenol, O.; Bolat, S.; Yıldız Ş, N.; Büyüktuna, S.A.; Sarıismailoğlu, R.; Doğan, K.; Hasbek, M.; Hekim, S.N. Understanding the pathophysiological changes via untargeted metabolomics in COVID-19 patients. J. Med. Virol. 2021, 93, 2340–2349. [Google Scholar] [CrossRef]
- Meoni, G.; Ghini, V.; Maggi, L.; Vignoli, A.; Mazzoni, A.; Salvati, L.; Capone, M.; Vanni, A.; Tenori, L.; Fontanari, P.; et al. Metabolomic/lipidomic profiling of COVID-19 and individual response to tocilizumab. PLOS Pathog. 2021, 17, e1009243. [Google Scholar] [CrossRef]
- Khovidhunkit, W.; Kim, M.S.; Memon, R.A.; Shigenaga, J.K.; Moser, A.H.; Feingold, K.R.; Grunfeld, C. Effects of infection and inflammation on lipid and lipoprotein metabolism: Mechanisms and consequences to the host. J Lipid Res. 2004, 45, 1169–1196. [Google Scholar] [CrossRef] [Green Version]
- Iida, M.; Harada, S.; Takebayashi, T. Application of Metabolomics to Epidemiological Studies of Atherosclerosis and Cardiovascular Disease. J. Atheroscler. Thromb. 2019, 26, 747–757. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.; Shah, S.H.; Corwin, E.J.; Fiehn, O.; Fitzgerald, R.L.; Gerszten, R.E.; Illig, T.; Rhee, E.P.; Srinivas, P.R.; Wang, T.J.; et al. Potential Impact and Study Considerations of Metabolomics in Cardiovascular Health and Disease: A Scientific Statement from the American Heart Association. Circ. Cardiovasc. Genet. 2017, 10, e000032. [Google Scholar] [CrossRef] [Green Version]
- Weiss, R.H.; Kim, K. Metabolomics in the study of kidney diseases. Nat. Rev. Nephrol. 2012, 8, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, S.; Baumgartner, T.; Meier, M.A.; Ottiger, M.; Voegeli, A.; Bernasconi, L.; Neyer, P.; Mueller, B.; Schuetz, P. The role of metabolomic markers for patients with infectious diseases: Implications for risk stratification and therapeutic modulation. Expert Rev. Anti-Infect. Ther. 2018, 16, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef] [PubMed]
- Arshad, H.; Alfonso, J.C.L.; Franke, R.; Michaelis, K.; Araujo, L.; Habib, A.; Zboromyrska, Y.; Lücke, E.; Strungaru, E.; Akmatov, M.K.; et al. Decreased plasma phospholipid concentrations and increased acid sphingomyelinase activity are accurate biomarkers for community-acquired pneumonia. J. Transl. Med. 2019, 17, 365. [Google Scholar] [CrossRef]
- Spadaro, F.; Cecchetti, S.; Fantuzzi, L. Macrophages and Phospholipases at the Intersection between Inflammation and the Pathogenesis of HIV-1 Infection. Int. J. Mol. Sci. 2017, 18, 1390. [Google Scholar] [CrossRef] [Green Version]
- Bryceson, Y.T.; Chiang, S.C.; Darmanin, S.; Fauriat, C.; Schlums, H.; Theorell, J.; Wood, S.M. Molecular mechanisms of natural killer cell activation. J. Innate Immun. 2011, 3, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Gallin, J.I.; Kaye, D.; O’Leary, W.M. Serum Lipids in Infection. N. Engl. J. Med. 1969, 281, 1081–1086. [Google Scholar] [CrossRef]
- Dissanayake, T.K.; Yan, B.; Ng, A.C.; Zhao, H.; Chan, G.; Yip, C.C.; Sze, K.H.; To, K.K. Differential role of sphingomyelin in influenza virus, rhinovirus and SARS-CoV-2 infection of Calu-3 cells. J. Gen. Virol. 2021, 102, 001593. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekharan, J.A.; Sharma-Walia, N. Arachidonic Acid Derived Lipid Mediators Influence Kaposi’s Sarcoma-Associated Herpesvirus Infection and Pathogenesis. Front. Microbiol. 2019, 10, 358. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. Genetic Variation, Diet, Inflammation, and the Risk for COVID-19. Lifestyle Genom. 2021, 14, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Barberis, E.; Timo, S.; Amede, E.; Vanella, V.V.; Puricelli, C.; Cappellano, G.; Raineri, D.; Cittone, M.G.; Rizzi, E.; Pedrinelli, A.R.; et al. Large-Scale Plasma Analysis Revealed New Mechanisms and Molecules Associated with the Host Response to SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 8623. [Google Scholar] [CrossRef]
- Das, U.N. Can Bioactive Lipids Inactivate Coronavirus (COVID-19)? Arch. Med Res. 2020, 51, 282–286. [Google Scholar] [CrossRef]
- Coperchini, F.; Chiovato, L.; Croce, L.; Magri, F.; Rotondi, M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020, 53, 25–32. [Google Scholar] [CrossRef]
- Shoieb, S.M.; El-Ghiaty, M.A.; El-Kadi, A.O.S. Targeting arachidonic acid-related metabolites in COVID-19 patients: Potential use of drug-loaded nanoparticles. Emergent Mater. 2020, 4, 265–277. [Google Scholar] [CrossRef]
- Kaur, G.; Yogeswaran, S.; Muthumalage, T.; Rahman, I. Persistently Increased Systemic ACE2 Activity Is Associated With an Increased Inflammatory Response in Smokers With COVID-19. Front. Physiol. 2021, 12, 653045. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yin, Y.-L.; Li, D.; Woo Kim, S.; Wu, G. Amino acids and immune function. Br. J. Nutr. 2007, 98, 237–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, W.; Rajendran, R.; Zhao, Y.; Tan, B.; Wu, G.; Bazer, F.W.; Zhu, G.; Peng, Y.; Huang, X.; Deng, J.; et al. Amino Acids As Mediators of Metabolic Cross Talk between Host and Pathogen. Front. Immunol. 2018, 9, 319. [Google Scholar] [CrossRef]
- Cheng, Z.-X.; Guo, C.; Chen, Z.-G.; Yang, T.-C.; Zhang, J.-Y.; Wang, J.; Zhu, J.-X.; Li, D.; Zhang, T.-T.; Li, H.; et al. Glycine, serine and threonine metabolism confounds efficacy of complement-mediated killing. Nat. Commun. 2019, 10, 3325. [Google Scholar] [CrossRef] [PubMed]
- Eroğlu, İ.; Eroğlu, B.Ç.; Güven, G.S. Altered tryptophan absorption and metabolism could underlie long-term symptoms in survivors of coronavirus disease 2019 (COVID-19). Nutrition 2021, 90, 111308. [Google Scholar] [CrossRef] [PubMed]
- Lionetto, L.; Ulivieri, M.; Capi, M.; De Bernardini, D.; Fazio, F.; Petrucca, A.; Pomes, L.M.; De Luca, O.; Gentile, G.; Casolla, B.; et al. Increased kynurenine-to-tryptophan ratio in the serum of patients infected with SARS-CoV2: An observational cohort study. Biochim. Et Biophys. Acta. Mol. Basis Dis. 2021, 1867, 166042. [Google Scholar] [CrossRef]
- Anderson, G.; Carbone, A.; Mazzoccoli, G. Tryptophan Metabolites and Aryl Hydrocarbon Receptor in Severe Acute Respiratory Syndrome, Coronavirus-2 (SARS-CoV-2) Pathophysiology. Int. J. Mol. Sci. 2021, 22, 1597. [Google Scholar] [CrossRef]
- Mehraj, V.; Routy, J.-P. Tryptophan Catabolism in Chronic Viral Infections: Handling Uninvited Guests. Int. J. Tryptophan Res. 2015, 8, 41–48. [Google Scholar] [CrossRef]
- Sorgdrager, F.J.H.; Naudé, P.J.W.; Kema, I.P.; Nollen, E.A.; Deyn, P.P.D. Tryptophan Metabolism in Inflammaging: From Biomarker to Therapeutic Target. Front. Immunol. 2019, 10, 2565. [Google Scholar] [CrossRef]
- Carlin, J.M.; Borden, E.C.; Byrne, G.I. Interferon-induced indoleamine 2,3-dioxygenase activity inhibits Chlamydia psittaci replication in human macrophages. J. Interferon Res. 1989, 9, 329–337. [Google Scholar] [CrossRef]
- Schmitz, J.L.; Carlin, J.M.; Borden, E.C.; Byrne, G.I. Beta interferon inhibits Toxoplasma gondii growth in human monocyte-derived macrophages. Infect. Immun. 1989, 57, 3254–3256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, S.V.; Schultze, J.L. New Insights into IDO Biology in Bacterial and Viral Infections. Front. Immunol. 2014, 5, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, O.; Besken, K.; Oberdörfer, C.; MacKenzie, C.R.; Rüssing, D.; Däubener, W. Inhibition of human herpes simplex virus type 2 by interferon gamma and tumor necrosis factor alpha is mediated by indoleamine 2,3-dioxygenase. Microbes Infect. 2004, 6, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Belladonna, M.L.; Orabona, C. Potential Benefits of Tryptophan Metabolism to the Efficacy of Tocilizumab in COVID-19. Front. Pharmacol. 2020, 11, 959. [Google Scholar] [CrossRef]
- Li, X.; Krysiak-Baltyn, K.; Richards, L.; Jarrold, A.; Stevens, G.W.; Bowser, T.; Speight, R.E.; Gras, S.L. High-Efficiency Biocatalytic Conversion of Thebaine to Codeine. ACS Omega 2020, 5, 9339–9347. [Google Scholar] [CrossRef]
- Ambre, J.J.; Ruo, T.-I.; Smith, G.L.; Backes, D.; Smith, C.M. Ecgonine Methyl Ester, A Major Metabolite of Cocaine. J. Anal. Toxicol. 1982, 6, 26–29. [Google Scholar] [CrossRef]
- Gordon, S.G.; Kittleson, M.D. Chapter 17—Drugs used in the management of heart disease and cardiac arrhythmias. In Small Animal Clinical Pharmacology, 2nd ed.; Maddison, J.E., Page, S.W., Church, D.B., Eds.; W.B. Saunders: Edinburgh, UK, 2008; pp. 380–457. [Google Scholar]
- Pippi, B.; Joaquim, A.R.; Merkel, S.; Zanette, R.A.; Nunes, M.E.M.; da Costa Silva, D.G.; Schimith, L.E.; Teixeira, M.L.; Franco, J.L.; Fernandes de Andrade, S.; et al. Antifungal activity and toxicological parameters of 8-hydroxyquinoline-5-sulfonamides using alternative animal models. J. Appl. Microbiol. 2021, 130, 1925–1934. [Google Scholar] [CrossRef]
- Wang, Q.; Ji, X.; Rahman, I. Dysregulated Metabolites Serve as Novel Biomarkers for Metabolic Diseases Caused by E-Cigarette Vaping and Cigarette Smoking. Metabolites 2021, 11, 345. [Google Scholar] [CrossRef]
- Huang, D.; Gaul, D.A.; Nan, H.; Kim, J.; Fernández, F.M. Deep Metabolomics of a High-Grade Serous Ovarian Cancer Triple-Knockout Mouse Model. J. Proteome Res. 2019, 18, 3184–3194. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. JMS 2010, 45, 703–714. [Google Scholar] [CrossRef]
| Name | Molecular Weight | Log2 Fold Change: (Co)/(Re) | p-Value: (Co)/(Re) |
|---|---|---|---|
| Lipid and Lipid metabolites | |||
| SM(d34:2) | 700.55 | 1.70 | 0.02 |
| PC(O-34:2) | 743.58 | 0.82 | 0.03 |
| PC(O-38:6) | 791.58 | 0.74 | 0.05 |
| PC(O-32:0) | 719.58 | 0.39 | 0.06 |
| PC(O-38:5) | 793.60 | 0.53 | 0.04 |
| PC(O-36:4) | 767.58 | 0.96 | 0.08 |
| PC(16:0 > PC(0:0/16:0) | 495.33 | 0.19 | 0.01 |
| FA(20:4) | 304.24 | 1.82 | 0.04 |
| Oleoyl-L-α-lysophosphatidic acid | 436.26 | 0.76 | 0.02 |
| Hexadecanoicacid | 256.24 | 0.25 | 0.05 |
| 1_6_6-Trimethyl-2_7-dioxabicyclo[3.2.2]nonan-3-one | 184.11 | −0.56 | 0.02 |
| 9_12-Dioxododecanoicacid | 228.14 | −0.70 | 0.01 |
| LPC(16:0) > LPC(16:0/0:0)_and_LPC(0:0/16:0) 2M + H2CO2 | 541.34 | 0.42 | 0.08 |
| LPE(18:0) > LPE(18:0/0:0)_and_LPE(0:0/18:0) | 481.32 | 0.61 | 0.01 |
| Amino Acid and Amino acid metabolites | |||
| DL-Tryptophan | 204.09 | 0.64 | 0.01 |
| Indole;1-Benzazole | 117.06 | 0.53 | 0.03 |
| 2-Hydroxypyridine | 95.04 | −2.13 | 0.05 |
| Indole-3-acrylic acid | 187.06 | 0.37 | 0.03 |
| Glycine | 75.03 | 0.98 | 0.05 |
| Guanidineacetic acid | 117.06 | 0.33 | 0.02 |
| L-2-Amino-3-oxobutanoicacid | 117.04 | 0.79 | 0.04 |
| (3R)-beta-Leucine | 131.09 | 0.47 | 0.04 |
| (S)-Methylmalonatesemialdehyde | 102.03 | 1.17 | 0.02 |
| N5-Ethyl-L-glutamine | 174.10 | −2.32 | 0.04 |
| Urocanic acid | 138.04 | −0.89 | 0.02 |
| Drug Metabolites | |||
| Thebaine | 311.15 | 1.87 | 0.01 |
| 1_2-Diaminobenzene | 108.07 | 0.89 | 0.10 |
| Ecgonine | 185.11 | 0.64 | 0.04 |
| 4,4’-Bipyridine | 174.08 | 0.35 | 0.02 |
| 8-Hydroxyquinoline | 145.05 | 0.36 | 0.03 |
| 1-Methylpyrrolinium | 83.07 | 0.21 | 0.03 |
| 3-Methyl-quinolin-2-ol | 159.07 | 0.35 | 0.03 |
| Isoquinoline | 129.06 | 0.31 | 0.02 |
| Others | |||
| Ectoine | 142.07 | 1.59 | 0.03 |
| cis-Zeatin | 219.11 | −2.10 | 0.05 |
| 2-methylhistamine | 125.10 | 0.91 | 0.10 |
| Glycocholic acid | 465.32 | 0.66 | 0.02 |
| Creatinine | 113.06 | −0.31 | 0.04 |
| 2-Oxoglutaric acid | 146.02 | 1.13 | 0.04 |
| Characteristics | COVID-19-Recovered | COVID-19-Positive | p-Value * |
|---|---|---|---|
| N | 6 | 6 | |
| Age (mean + SD) | 36.33 ± 9.4 | 47 ± 14.4 | 0.1926 |
| Male:Female | 3:3 | 4:2 | 0.6667 |
| Smokers, n (%) | 2 (33.33) | 2 (33.33) | 0.3333 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaur, G.; Ji, X.; Rahman, I. SARS-CoV2 Infection Alters Tryptophan Catabolism and Phospholipid Metabolism. Metabolites 2021, 11, 659. https://doi.org/10.3390/metabo11100659
Kaur G, Ji X, Rahman I. SARS-CoV2 Infection Alters Tryptophan Catabolism and Phospholipid Metabolism. Metabolites. 2021; 11(10):659. https://doi.org/10.3390/metabo11100659
Chicago/Turabian StyleKaur, Gagandeep, Xiangming Ji, and Irfan Rahman. 2021. "SARS-CoV2 Infection Alters Tryptophan Catabolism and Phospholipid Metabolism" Metabolites 11, no. 10: 659. https://doi.org/10.3390/metabo11100659
APA StyleKaur, G., Ji, X., & Rahman, I. (2021). SARS-CoV2 Infection Alters Tryptophan Catabolism and Phospholipid Metabolism. Metabolites, 11(10), 659. https://doi.org/10.3390/metabo11100659








