β-Hydroxybutyrate Oxidation Promotes the Accumulation of Immunometabolites in Activated Microglia Cells
Abstract
1. Introduction
2. Results
2.1. Microglia Cells Oxidise β-Hydroxybutyrate in the TCA Cycle
2.2. β-Hydroxybutyrate Modulates the LPS-Induced Glycolytic Response
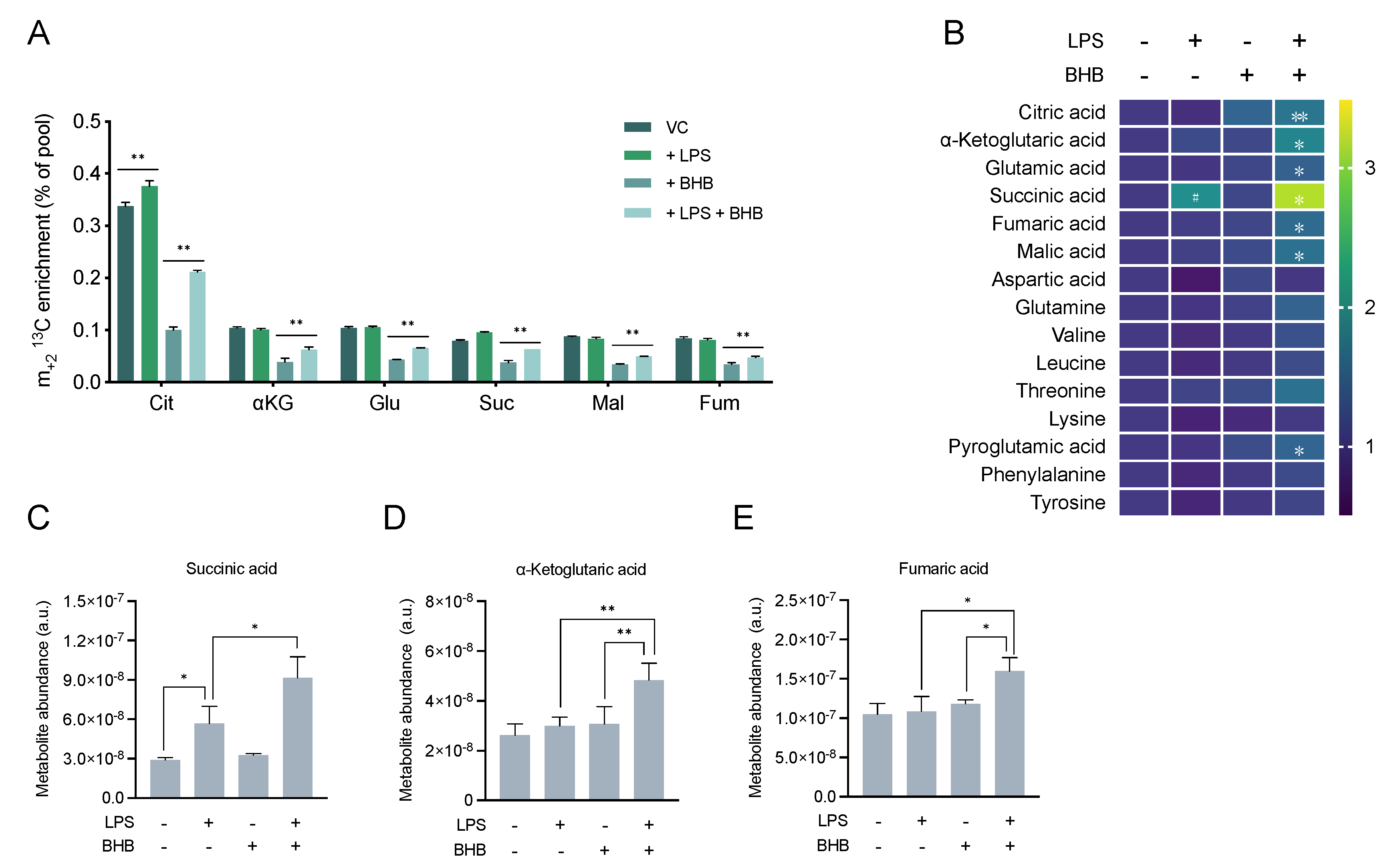
2.3. β-Hydroxybutyrate Promotes Mitochondrial Metabolism and Accumulation of TCA Cycle Intermediates
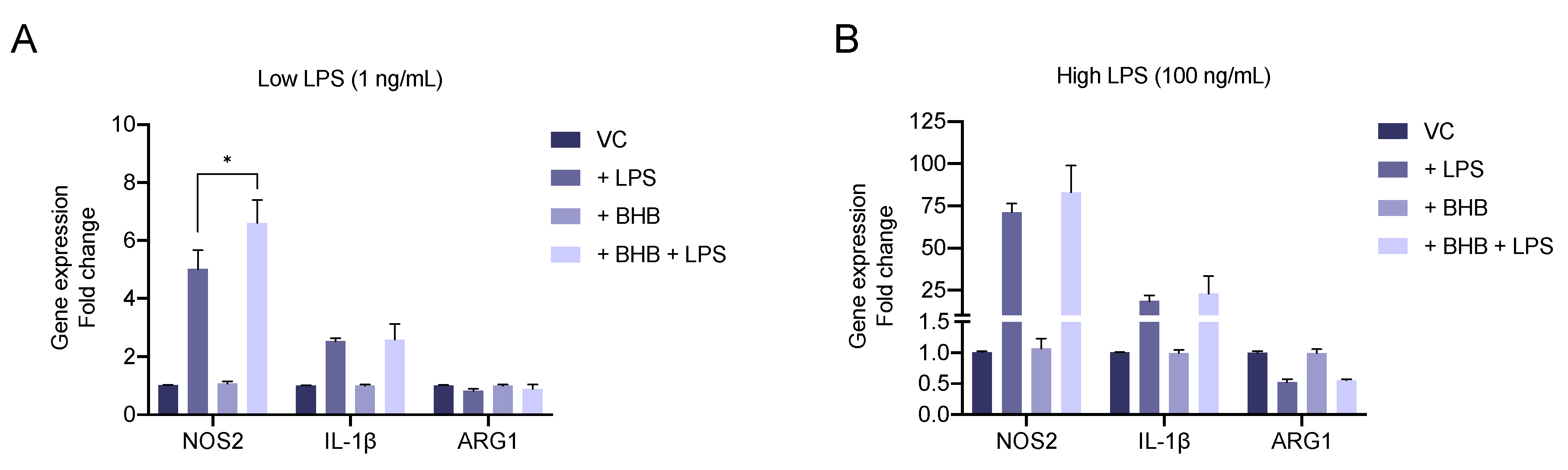
2.4. β-Hydroxybutyrate Enhances LPS-Induced Upregulation of the Pro-Inflammatory Marker NOS2
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Proliferation Assay
4.3. Gene Expression Assay
4.4. Stable-Isotope (13C) Tracing and Metabolic Profiling
4.5. 1H Nuclear Magnetic Resonance (NMR) Spectroscopy
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Salter, M.W.; Stevens, B. Microglia emerge as central players in brain disease. Nat. Med. 2017, 23, 1018–1027. [Google Scholar] [CrossRef]
- Jackson, L.; Dumanli, S.; Johnson, M.H.; Fagan, S.C.; Ergul, A. Microglia knockdown reduces inflammation and preserves cognition in diabetic animals after experimental stroke. J. Neuroinflamm. 2020, 17, 137. [Google Scholar] [CrossRef]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef]
- Ransohoff, R.M. A polarizing question: Do M1 and M2 microglia exist? Nat. Neurosci. 2016, 19, 987–991. [Google Scholar] [CrossRef]
- Lauro, C.; Limatola, C. Metabolic Reprograming of Microglia in the Regulation of the Innate Inflammatory Response. Front. Immunol. 2020, 11, 493. [Google Scholar] [CrossRef]
- Ghosh, S.; Castillo, E.; Frias, E.S.; Swanson, R.A. Bioenergetic regulation of microglia. Glia 2018, 66, 1200–1212. [Google Scholar] [CrossRef]
- Borst, K.; Schwabenland, M.; Prinz, M. Microglia metabolism in health and disease. Neurochem. Int. 2018, 130, 104331. [Google Scholar] [CrossRef]
- Baik, S.H.; Kang, S.; Lee, W.; Choi, H.; Chung, S.; Kim, J.-I.; Mook-Jung, I. A Breakdown in Metabolic Reprogramming Causes Microglia Dysfunction in Alzheimer’s Disease. Cell Metab. 2019, 30, 493–507. [Google Scholar] [CrossRef]
- Seim, G.L.; Britt, E.C.; John, S.V.; Yeo, F.J.; Johnson, A.R.; Eisenstein, R.S.; Pagliarini, D.J.; Fan, J. Two-stage metabolic remodelling in macrophages in response to lipopolysaccharide and interferon-γ stimulation. Nat. Metab. 2019, 1, 731–742. [Google Scholar] [CrossRef]
- Pearce, E.L.; Pearce, E.J. Metabolic pathways in immune cell activation and quiescence. Immunity 2013, 38, 633–643. [Google Scholar] [CrossRef]
- Geric, I.; Schoors, S.; Claes, C.; Gressens, P.; Verderio, C.; Verfaillie, C.M.; Veldhoven, P.P.V.; Carmeliet, P.; Baes, M. Metabolic Reprogramming during Microglia Activation. Immunometabolism 2019, 1. [Google Scholar] [CrossRef]
- Orihuela, R.; McPherson, C.A.; Harry, G.J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 2015, 173, 649–665. [Google Scholar] [CrossRef]
- Caputa, G.; Castoldi, A.; Pearce, E.J. Metabolic adaptations of tissue-resident immune cells. Nat. Immunol. 2019, 20, 793–801. [Google Scholar] [CrossRef]
- Qiu, J.; Villa, M.; Sanin, D.E.; Buck, M.D.; O’Sullivan, D.; Ching, R.; Matsushita, M.; Grzes, K.M.; Winkler, F.; Chang, C.-H.; et al. Acetate Promotes T Cell Effector Function during Glucose Restriction. Cell Rep. 2019, 27, 2063–2074.e5. [Google Scholar] [CrossRef]
- Davies, L.C.; Rice, C.M.; Palmieri, E.M.; Taylor, P.R.; Kuhns, D.B.; McVicar, D.W. Peritoneal tissue-resident macrophages are metabolically poised to engage microbes using tissue-niche fuels. Nat. Commun. 2017, 8, 2074. [Google Scholar] [CrossRef]
- Angela, M.; Endo, Y.; Asou, H.K.; Yamamoto, T.; Tumes, D.J.; Tokuyama, H.; Yokote, K.; Nakayama, T. Fatty acid metabolic reprogramming via mTOR-mediated inductions of PPARγ directs early activation of T cells. Nat. Commun. 2016, 7, 13683. [Google Scholar] [CrossRef]
- Bernier, L.; York, E.; Kamyabi, A.; Choi, H.; Weilinger, N.; MacVicar, B. Microglial metabolic flexibility supports immune surveillance of the brain parenchyma. Nat. Commun. 2020, 11, 1559. [Google Scholar] [CrossRef]
- Nagy, A.M.; Fekete, R.; Horvath, G.; Koncsos, G.; Kriston, C.; Sebestyen, A.; Giricz, Z.; Kornyei, Z.; Madarasz, E.; Tretter, L. Versatility of microglial bioenergetic machinery under starving conditions. Biochim. Biophys. Acta 2018, 1859, 201–214. [Google Scholar] [CrossRef]
- De Goede, K.E.; Harber, K.J.; den Bossche, J.V. Let’s Enter the Wonderful World of Immunometabolites. Trends Endocrinol. Metab. 2019, 30, 329–331. [Google Scholar] [CrossRef]
- Zasłona, Z.; O’Neill, L.A.J. Cytokine-like Roles for Metabolites in Immunity. Mol. Cell 2020. [Google Scholar] [CrossRef]
- Choi, I.Y.; Piccio, L.; Childress, P.; Bollman, B.; Ghosh, A.; Brandhorst, S.; Suarez, J.; Michalsen, A.; Cross, A.H.; Morgan, T.E.; et al. A Diet Mimicking Fasting Promotes Regeneration and Reduces Autoimmunity and Multiple Sclerosis Symptoms. Cell Rep. 2016, 15, 2136–2146. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, E.L.; Molony, R.D.; Kudo, E.; Sidorov, S.; Kong, Y.; Dixit, V.D.; Iwasaki, A. Ketogenic diet activates protective γδ T cell responses against influenza virus infection. Sci. Immunol. 2019, 4, eaav2026. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, E.L.; Shchukina, I.; Asher, J.L.; Sidorov, S.; Artyomov, M.N.; Dixit, V.D. Ketogenesis activates metabolically protective γδ T cells in visceral adipose tissue. Nat. Metab. 2020, 2, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Mattson, M.P. Fasting: Molecular mechanisms and clinical applications. Cell Metab. 2014, 19, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Orillion, A.R.; Damayanti, N.P.; Shen, L.; Adelaiye-Ogala, R.; Affronti, H.C.; Elbanna, M.; Chintala, S.; Ciesielski, M.J.; Fontana, L.; Kao, C.; et al. Dietary protein restriction reprograms tumor associated macrophages and enhances immunotherapy. Clin. Cancer Res. 2018, 24, 6383–6395. [Google Scholar] [CrossRef]
- Wu, Z.; Isik, M.; Moroz, N.; Steinbaugh, M.J.; Zhang, P.; Blackwell, T.K. Dietary Restriction Extends Lifespan through Metabolic Regulation of Innate Immunity. Cell Metab. 2019, 29, 1192–1205.e8. [Google Scholar] [CrossRef]
- Soldati, L.; Renzo, L.D.; Jirillo, E.; Ascierto, P.A.; Marincola, F.M.; Lorenzo, A.D. The influence of diet on anti-cancer immune responsiveness. J. Transl. Med. 2018, 16, 75. [Google Scholar] [CrossRef]
- Klement, R.J. Beneficial effects of ketogenic diets for cancer patients: A realist review with focus on evidence and confirmation. Med. Oncol. 2017, 34, 132. [Google Scholar] [CrossRef]
- Klement, R.J.; Brehm, N.; Sweeney, R.A. Ketogenic diets in medical oncology: A systematic review with focus on clinical outcomes. Med. Oncol. 2020, 37, 14. [Google Scholar] [CrossRef]
- Woolf, E.C.; Syed, N.; Scheck, A.C. Tumor Metabolism, the Ketogenic Diet and β-Hydroxybutyrate: Novel Approaches to Adjuvant Brain Tumor Therapy. Front. Mol. Neurosci. 2016, 9, 122. [Google Scholar] [CrossRef]
- Gasior, M.; Rogawski, M.A.; Hartman, A.L. Neuroprotective and disease-modifying effects of the ketogenic diet. Behav. Pharmacol. 2006, 17, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Lutas, A.; Yellen, G. The ketogenic diet: Metabolic influences on brain excitability and epilepsy. Trends Neurosci. 2013, 36, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Youm, Y.-H.; Nguyen, K.Y.; Grant, R.W.; Goldberg, E.L.; Bodogai, M.; Kim, D.; D’Agostino, D.; Planavsky, N.; Lupfer, C.; Kanneganti, T.D.; et al. The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat. Med. 2015, 21, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.A.; Kruger, S.L.; Broderick, C.; Amarlkhagva, T.; Agrawal, S.; Dodam, J.R.; Mrug, M.; Lyons, L.A.; Weimbs, T. Ketosis Ameliorates Renal Cyst Growth in Polycystic Kidney Disease. Cell Metab. 2019, 30, 1007–1023. [Google Scholar] [CrossRef]
- Newman, J.C.; Verdin, E. β-Hydroxybutyrate: A Signaling Metabolite. Annu. Rev. Nutr. 2017, 37, 51–76. [Google Scholar] [CrossRef]
- Trotta, M.C.; Maisto, R.; Guida, F.; Boccella, S.; Luongo, L.; Balta, C.; D’Amico, G.; Herman, H.; Hermenean, A.; Bucolo, C.; et al. The activation of retinal HCA2 receptors by systemic beta-hydroxybutyrate inhibits diabetic retinal damage through reduction of endoplasmic reticulum stress and the NLRP3 inflammasome. PLoS ONE 2019, 14, e0211005. [Google Scholar] [CrossRef]
- Han, Y.-M.; Bedarida, T.; Ding, Y.; Somba, B.K.; Lu, Q.; Wang, Q.; Song, P.; Zou, M.-H. β-Hydroxybutyrate Prevents Vascular Senescence through hnRNP A1-Mediated Upregulation of Oct4. Mol. Cell 2018, 71, 1064–1078. [Google Scholar] [CrossRef]
- Huang, C.; Wang, P.; Xu, X.; Zhang, Y.; Gong, Y.; Hu, W.; Gao, M.; Wu, Y.; Ling, Y.; Zhao, X.; et al. The ketone body metabolite β-hydroxybutyrate induces an antidepression-associated ramification of microglia via HDACs inhibition-triggered Akt-small RhoGTPase activation. Glia 2018, 66, 256–278. [Google Scholar] [CrossRef]
- Chen, Y.; Ouyang, X.; Hoque, R.; Garcia-Martinez, I.; Yousaf, M.N.; Tonack, S.; Offermanns, S.; Dubuquoy, L.; Louvet, A.; Mathurin, P.; et al. β-Hydroxybutyrate protects from alcohol-induced liver injury via a Hcar2-cAMP dependent pathway. J. Hepatol. 2018, 69, 687–696. [Google Scholar] [CrossRef]
- Taggart, A.K.P.; Kero, J.; Gan, X.; Cai, T.-Q.; Cheng, K.; Ippolito, M.; Ren, N.; Kaplan, R.; Wu, K.; Wu, T.-J.; et al. (D)-beta-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J. Biol. Chem. 2005, 280, 26649–26652. [Google Scholar] [CrossRef]
- Goldberg, E.L.; Asher, J.L.; Molony, R.D.; Shaw, A.C.; Zeiss, C.J.; Wang, C.; Morozova-Roche, L.A.; Herzog, R.I.; Iwasaki, A.; Dixit, V.D. β-Hydroxybutyrate Deactivates Neutrophil NLRP3 Inflammasome to Relieve Gout Flares. Cell Rep. 2017, 18, 2077–2087. [Google Scholar] [CrossRef]
- Stansley, B.; Post, J.; Hensley, K. A comparative review of cell culture systems for the study of microglial biology in Alzheimer’s disease. J. Neuroinflamm. 2012, 9, 115. [Google Scholar] [CrossRef]
- Russo, C.D.; Cappoli, N.; Coletta, I.; Mezzogori, D.; Paciello, F.; Pozzoli, G.; Navarra, P.; Battaglia, A. The human microglial HMC3 cell line: Where do we stand? A systematic literature review. J. Neuroinflamm. 2018, 15, 259. [Google Scholar] [CrossRef] [PubMed]
- Timmerman, R.; Burm, S.M.; Bajramovic, J.J. An Overview of in vitro Methods to Study Microglia. Front. Cell. Neurosci. 2018, 12, 242. [Google Scholar] [CrossRef] [PubMed]
- Cotter, D.G.; Schugar, R.C.; Crawford, P.A. Ketone body metabolism and cardiovascular disease. Am. J. Physiol.-Heart Circ. Physiol. 2013, 304, H1060–H1076. [Google Scholar] [CrossRef] [PubMed]
- Edmond, J.; Robbins, R.A.; Bergstrom, J.D.; Cole, R.A.; de Vellis, J. Capacity for substrate utilization in oxidative metabolism by neurons, astrocytes, and oligodendrocytes from developing brain in primary culture. J. Neurosci. Res. 1987, 18, 551–561. [Google Scholar] [CrossRef]
- Chechik, T.; Roeder, L.M.; Tildon, J.T.; Poduslo, S.E. Ketone body enzyme activities in purified neurons, astrocytes and oligodendroglia. Neurochem. Int. 1987, 10, 95–99. [Google Scholar] [CrossRef]
- Pan, J.W.; de Graaf, R.A.; Petersen, K.F.; Shulman, G.I.; Hetherington, H.P.; Rothman, D.L. [2,4-13C2]-beta-Hydroxybutyrate metabolism in human brain. J. Cereb. Blood Flow Metab. 2002, 22, 890–898. [Google Scholar] [CrossRef]
- Lopes-Cardozo, M.; Larsson, O.M.; Schousboe, A. Acetoacetate and Glucose as Lipid Precursors and Energy Substrates in Primary Cultures of Astrocytes and Neurons from Mouse Cerebral Cortex. J. Neurochem. 1986, 46, 773–778. [Google Scholar] [CrossRef]
- Jang, C.; Chen, L.; Rabinowitz, J.D. Metabolomics and Isotope Tracing. Cell 2018, 173, 822–837. [Google Scholar] [CrossRef]
- Shen, Y.; Kapfhamer, D.; Minnella, A.M.; Kim, J.-E.; Won, S.J.; Chen, Y.; Huang, Y.; Low, L.H.; Massa, S.M.; Swanson, R.A. Bioenergetic state regulates innate inflammatory responses through the transcriptional co-repressor CtBP. Nat. Commun. 2017, 8, 624. [Google Scholar] [CrossRef] [PubMed]
- Williamson, D.; Lund, P.; Krebs, H. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J. 1967, 103, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Chesser, A.S.; Grima, J.C.; Rappold, P.M.; Blum, D.; Przedborski, S.; Tieu, K. D-β-Hydroxybutyrate Is Protective in Mouse Models of Huntington’s Disease. PLoS ONE 2011, 6, e24620. [Google Scholar] [CrossRef] [PubMed]
- Kajitani, N.; Iwata, M.; Miura, A.; Tsunetomi, K.; Yamanashi, T.; Matsuo, R.; Nishiguchi, T.; Fukuda, S.; Nagata, M.; Shibushita, M.; et al. Prefrontal cortex infusion of beta-hydroxybutyrate, an endogenous NLRP3 inflammasome inhibitor, produces antidepressant-like effects in a rodent model of depression. Neuropsychopharmacol. Rep. 2020. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Muhammad, S.; Khan, M.A.; Chen, H.; Ridder, D.A.; Müller-Fielitz, H.; Pokorná, B.; Vollbrandt, T.; Stölting, I.; Nadrowitz, R.; et al. The β-hydroxybutyrate receptor HCA2 activates a neuroprotective subset of macrophages. Nat. Commun. 2014, 5, 3944. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.-P.; Wang, J.-F.; Xue, W.-J.; Liu, H.-M.; Liu, B.; Zeng, Y.-L.; Li, S.-N.; Huang, B.-X.; Lv, Q.-K.; Wang, W.; et al. Anti-inflammatory effects of BHBA in both in vivo and in vitro Parkinson’s disease models are mediated by GPR109A-dependent mechanisms. J. Neuroinflamm. 2015, 12, 9–14. [Google Scholar] [CrossRef]
- Deora, V.; Albornoz, E.A.; Zhu, K.; Woodruff, T.M.; Gordon, R. The Ketone Body β-Hydroxybutyrate Does Not Inhibit Synuclein Mediated Inflammasome Activation in Microglia. J. Neuroimmune Pharmacol. 2017, 12, 568–574. [Google Scholar] [CrossRef]
- Ryan, D.G.; Murphy, M.P.; Frezza, C.; Prag, H.A.; Chouchani, E.T.; O’Neill, L.A.; Mills, E.L. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat. Metab. 2019, 1, 16–33. [Google Scholar] [CrossRef]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef]
- Cordes, T.; Wallace, M.; Michelucci, A.; Divakaruni, A.S.; Sapcariu, S.C.; Sousa, C.; Koseki, H.; Cabrales, P.; Murphy, A.N.; Hiller, K.; et al. Immunoresponsive Gene 1 and Itaconate Inhibit Succinate Dehydrogenase to Modulate Intracellular Succinate Levels. J. Biol. Chem. 2016, 291, 14274–14284. [Google Scholar] [CrossRef]
- Lampropoulou, V.; Sergushichev, A.; Bambouskova, M.; Nair, S.; Vincent, E.E.; Loginicheva, E.; Cervantes-Barragan, L.; Ma, X.; Huang, S.C.-C.; Griss, T.; et al. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell Metab. 2016, 24, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Laffel, L. Ketone bodies: A review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes Metab. Res. Rev. 1999, 15, 412–426. [Google Scholar] [CrossRef]
- Owen, O.E.; Felig, P.; Morgan, A.P.; Wahren, J.; Cahill, G.F. Liver and kidney metabolism during prolonged starvation. J. Clin. Investig. 1969, 48, 574–583. [Google Scholar] [CrossRef]
- Cohen, C.W.; Fontaine, K.R.; Arend, R.C.; Gower, B.A. A Ketogenic Diet Is Acceptable in Women with Ovarian and Endometrial Cancer and Has No Adverse Effects on Blood Lipids: A Randomized, Controlled Trial. Nutr. Cancer 2019, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Klement, R.J.; Sweeney, R.A. Impact of a ketogenic diet intervention during radiotherapy on body composition: I. Initial clinical experience with six prospectively studied patients. BMC Res. Notes 2016, 9, 143. [Google Scholar] [CrossRef] [PubMed]
- Mergenthaler, P.; Lindauer, U.; Dienel, G.A.; Meisel, A. Sugar for the brain: The role of glucose in physiological and pathological brain function. Trends Neurosci. 2013, 36, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, S.V.; Nguyen, N.H.T.; Rise, F.; Hassel, B. Brain metabolism of exogenous pyruvate: Brain metabolism of pyruvate. J. Neurochem. 2005, 95, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Boumezbeur, F.; Petersen, K.F.; Cline, G.W.; Mason, G.F.; Behar, K.L.; Shulman, G.I.; Rothman, D.L. The contribution of blood lactate to brain energy metabolism in humans measured by dynamic 13C nuclear magnetic resonance spectroscopy. J. Neurosci. 2010, 30, 13983–13991. [Google Scholar] [CrossRef]
- Mächler, P.; Wyss, M.T.; Elsayed, M.; Stobart, J.; Gutierrez, R.; von Faber-Castell, A.; Kaelin, V.; Zuend, M.; Martín, A.S.; Romero-Gómez, I.; et al. In Vivo Evidence for a Lactate Gradient from Astrocytes to Neurons. Cell Metab. 2016, 23, 94–102. [Google Scholar] [CrossRef]
- Rae, C.; Fekete, A.D.; Kashem, M.A.; Nasrallah, F.A.; Bröer, S. Metabolism, Compartmentation, Transport and Production of Acetate in the Cortical Brain Tissue Slice. Neurochem. Res. 2012, 37, 2541–2553. [Google Scholar] [CrossRef]
- Pan, J.W.; Rothman, T.L.; Behar, K.L.; Stein, D.T.; Hetherington, H.P. Human brain beta-hydroxybutyrate and lactate increase in fasting-induced ketosis. J. Cereb. Blood Flow Metab. 2000, 20, 1502–1507. [Google Scholar] [CrossRef]
- Achanta, L.B.; Rowlands, B.D.; Thomas, D.S.; Housley, G.D.; Rae, C.D. β-Hydroxybutyrate Boosts Mitochondrial and Neuronal Metabolism but is not Preferred Over Glucose Under Activated Conditions. Neurochem. Res. 2017, 42, 1710–1723. [Google Scholar] [CrossRef]
- Halestrap, A.P. The SLC16 gene family-structure, role and regulation in health and disease. Mol. Asp. Med. 2013, 34, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Pellerin, L.; Bergersen, L.H.; Halestrap, A.P.; Pierre, K. Cellular and subcellular distribution of monocarboxylate transporters in cultured brain cells and in the adult brain. J. Neurosci. Res. 2005, 79, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Moreira, T.J.T.P.; Pierre, K.; Maekawa, F.; Repond, C.; Cebere, A.; Liljequist, S.; Pellerin, L. Enhanced cerebral expression of MCT1 and MCT2 in a rat ischemia model occurs in activated microglial cells. J. Cereb. Blood Flow Metab. 2009, 29, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Achanta, L.B.; Rae, C.D. β-Hydroxybutyrate in the Brain: One Molecule, Multiple Mechanisms. Neurochem. Res. 2016, 42, 35–49. [Google Scholar] [CrossRef]
- Bröer, S.; Bröer, A.; Schneider, H.-P.; Stegen, C.; Halestrap, A.P.; Deitmer, J.W. Characterization of the high-affinity monocarboxylate transporter MCT2 in Xenopus laevis oocytes. Biochem. J. 1999, 341, 529. [Google Scholar] [CrossRef]
- Fukao, T.; Lopaschuk, G.D.; Mitchell, G.A. Pathways and control of ketone body metabolism: On the fringe of lipid biochemistry. Prostaglandins Leukot Essent. Fat. Acids 2004, 70, 243–251. [Google Scholar] [CrossRef]
- Grabacka, M.; Pierzchalska, M.; Dean, M.; Reiss, K. Regulation of Ketone Body Metabolism and the Role of PPARα. Int. J. Mol. Sci. 2016, 17, 2093. [Google Scholar] [CrossRef]
- Huang, D.; Li, T.; Wang, L.; Zhang, L.; Yan, R.; Li, K.; Xing, S.; Wu, G.; Hu, L.; Jia, W.; et al. Hepatocellular carcinoma redirects to ketolysis for progression under nutrition deprivation stress. Cell Res. 2016, 26, 1112–1130. [Google Scholar] [CrossRef]
- Guo, K.; Lukacik, P.; Papagrigoriou, E.; Meier, M.; Lee, W.H.; Adamski, J.; Oppermann, U. Characterization of Human DHRS6, an Orphan Short Chain Dehydrogenase/Reductase Enzyme: A novel, cytosolic type 2 R-β-hydroxybutyrate dehydrogenase. J. Biol. Chem. 2005, 281, 10291–10297. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrovskii, Y.A. Antithrombin III, C1 inhibitor, methylglyoxal, and polymorphonuclear leukocytes in the development of vascular complications in diabetes mellitus. Thromb. Res. 1992, 67, 179–189. [Google Scholar] [CrossRef]
- Glew, R.H. You can get there from here: Acetone, anionic ketones and even-carbon fatty acids can provide substrates for gluconeogenesis. Niger. J. Physiol. Sci. Off. Publ. Physiol. Soc. Niger. 2010, 25, 2–4. [Google Scholar]
- Jagt, D.L.V.; Hassebrook, R.K.; Hunsaker, L.A.; Brown, W.M.; Royer, R.E. Metabolism of the 2-oxoaldehyde methylglyoxal by aldose reductase and by glyoxalase-I: Roles for glutathione in both enzymes and implications for diabetic complications. Chem. Biol. Interact. 2001, 130–132, 549–562. [Google Scholar] [CrossRef]
- Casazza, J.P.; Felver, M.E.; Veech, R.L. The metabolism of acetone in rat. J. Biol. Chem. 1984, 259, 231–236. [Google Scholar] [PubMed]
- Bhattacharyya, N.; Pal, A.; Patra, S.; Haldar, A.K.; Roy, S.; Ray, M. Activation of macrophages and lymphocytes by methylglyoxal against tumor cells in the host. Int. Immunopharmacol. 2008, 8, 1503–1512. [Google Scholar] [CrossRef]
- Dhananjayan, K.; Gunawardena, D.; Hearn, N.; Sonntag, T.; Moran, C.; Gyengesi, E.; Srikanth, V.; Münch, G. Activation of Macrophages and Microglia by Interferon-γ and Lipopolysaccharide Increases Methylglyoxal Production: A New Mechanism in the Development of Vascular Complications and Cognitive Decline in Type 2 Diabetes Mellitus? J. Alzheimers Dis. 2017, 59, 467–479. [Google Scholar] [CrossRef]
- Nair, S.; Sobotka, K.S.; Joshi, P.; Gressens, P.; Fleiss, B.; Thornton, C.; Mallard, C.; Hagberg, H. Lipopolysaccharide-induced alteration of mitochondrial morphology induces a metabolic shift in microglia modulating the inflammatory response in vitro and in vivo. Glia 2019, 91, 1345. [Google Scholar] [CrossRef]
- Gimeno-Bayón, J.; López-López, A.; Rodríguez, M.J.; Mahy, N. Glucose pathways adaptation supports acquisition of activated microglia phenotype. J. Neurosci. Res. 2014, 92, 723–731. [Google Scholar] [CrossRef]
- Berg, J.; Tymoczko, J.; Stryer, L. Section 16.1, Glycolysis Is an Energy-Conversion Pathway in Many Organisms. In Biochemistry; W H Freeman: New York, NY, USA, 2002. [Google Scholar]
- Hara, M.R.; Agrawal, N.; Kim, S.F.; Cascio, M.B.; Fujimuro, M.; Ozeki, Y.; Takahashi, M.; Cheah, J.H.; Tankou, S.K.; Hester, L.D.; et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 2005, 7, 665–674. [Google Scholar] [CrossRef]
- Mohr, S.; Stamler, J.S.; Brüne, B. Posttranslational Modification of Glyceraldehyde-3-phosphate Dehydrogenase by S -Nitrosylation and Subsequent NADH Attachment. J. Biol. Chem. 1996, 271, 4209–4214. [Google Scholar] [CrossRef] [PubMed]
- Souza, J.M.; Radi, R. Glyceraldehyde-3-Phosphate Dehydrogenase Inactivation by Peroxynitrite. Arch. Biochem. Biophys. 1998, 360, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Dimmeler, S.; Lottspeich, F.; Brüne, B. Nitric oxide causes ADP-ribosylation and inhibition of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 1992, 267, 16771–16774. [Google Scholar]
- Dimmeler, S.; Ankarcrona, M.; Nicotera, P.; Brüne, B. Exogenous nitric oxide (NO) generation or IL-1 beta-induced intracellular NO production stimulates inhibitory auto-ADP-ribosylation of glyceraldehyde-3-phosphate dehydrogenase in RINm5F cells. J. Immunol. 1993, 150, 2964–2971. [Google Scholar] [PubMed]
- Y Vedia, L.M.; McDonald, B.; Reep, B.; Brüne, B.; Silvio, M.D.; Billiar, T.R.; Lapetina, E.G. Nitric oxide-induced S-nitrosylation of glyceraldehyde-3-phosphate dehydrogenase inhibits enzymatic activity and increases endogenous ADP-ribosylation. J. Biol. Chem. 1992, 267, 24929–24932. [Google Scholar]
- Berg, J.; Tymoczko, J.; Stryer, L. Section 17.2, Entry to the Citric Acid Cycle and Metabolism through It Are Controlled. In Biochemistry; W H Freeman: New York, NY, USA, 2002. [Google Scholar]
- Mills, E.L.; Kelly, B.; Logan, A.; Costa, A.S.H.; Varma, M.; Bryant, C.E.; Tourlomousis, P.; Däbritz, J.H.M.; Gottlieb, E.; Latorre, I.; et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016, 167, 457–470.e13. [Google Scholar] [CrossRef]
- Michelucci, A.; Heurtaux, T.; Grandbarbe, L.; Morga, E.; Heuschling, P. Characterization of the microglial phenotype under specific pro-inflammatory and anti-inflammatory conditions: Effects of oligomeric and fibrillar amyloid-beta. J. Neuroimmunol. 2009, 210, 3–12. [Google Scholar] [CrossRef]
- Graff, E.C.; Fang, H.; Wanders, D.; Judd, R.L. Anti-inflammatory effects of the hydroxycarboxylic acid receptor 2. Metab. Clin. Exp. 2016, 65, 102–113. [Google Scholar] [CrossRef]
- Offermanns, S.; Schwaninger, M. Nutritional or pharmacological activation of HCA(2) ameliorates neuroinflammation. Trends Mol. Med. 2015, 21, 245–255. [Google Scholar] [CrossRef]
- Zandi-Nejad, K.; Takakura, A.; Jurewicz, M.; Chandraker, A.K.; Offermanns, S.; Mount, D.; Abdi, R. The role of HCA2 (GPR109A) in regulating macrophage function. FASEB J. 2013, 27, 4366–4374. [Google Scholar] [CrossRef]
- Shi, X.; Li, X.; Li, D.; Li, Y.; Song, Y.; Deng, Q.; Wang, J.; Zhang, Y.; Ding, H.; Yin, L.; et al. β-Hydroxybutyrate activates the NF-κB signaling pathway to promote the expression of pro-inflammatory factors in calf hepatocytes. Cell. Physiol. Biochem. 2014, 33, 920–932. [Google Scholar] [CrossRef]
- Henn, A.; Lund, S.; Hedtjärn, M.; Schrattenholz, A.; Pörzgen, P.; Leist, M. The suitability of BV2 cells as alternative model system for primary microglia cultures or for animal experiments examining brain inflammation. Altex 2009, 26, 83–94. [Google Scholar] [CrossRef]
- Carrillo-Jimenez, A.; Deniz, Ö.; Niklison-Chirou, M.V.; Ruiz, R.; Bezerra-Salomão, K.; Stratoulias, V.; Amouroux, R.; Yip, P.K.; Vilalta, A.; Cheray, M.; et al. TET2 Regulates the Neuroinflammatory Response in Microglia. Cell Rep. 2019, 29, 697–713.e8. [Google Scholar] [CrossRef] [PubMed]
- Burguillos, M.A.; Deierborg, T.; Kavanagh, E.; Persson, A.; Hajji, N.; Garcia-Quintanilla, A.; Cano, J.; Brundin, P.; Englund, E.; Venero, J.L.; et al. Caspase signalling controls microglia activation and neurotoxicity. Nature 2011, 472, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Kim, S.H.; Arifuzzaman, S.; Yoon, T.; Chai, J.C.; Lee, Y.S.; Park, K.S.; Jung, K.H.; Chai, Y.G. Transcriptome sequencing reveals that LPS-triggered transcriptional responses in established microglia BV2 cell lines are poorly representative of primary microglia. J. Neuroinflamm. 2016, 13, 182. [Google Scholar] [CrossRef]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 2013, 17, 131–143. [Google Scholar] [CrossRef]
- Abdelwahab, M.G.; Fenton, K.E.; Preul, M.C.; Rho, J.M.; Lynch, A.; Stafford, P.; Scheck, A.C. The ketogenic diet is an effective adjuvant to radiation therapy for the treatment of malignant glioma. PLoS ONE 2012, 7, e36197. [Google Scholar] [CrossRef] [PubMed]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Barnes, E.M.E.; Xu, Y.; Benito, A.; Herendi, L.; Siskos, A.P.; Aboagye, E.O.; Nijhuis, A.; Keun, H.C. Lactic acidosis induces resistance to the pan-Akt inhibitor uprosertib in colon cancer cells. Br. J. Cancer 2020, 122, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.-H.E.; Tredwell, G.D.; Ellis, J.K.; Lam, E.W.-F.; Keun, H.C. Metabolomic characterisation of the effects of oncogenic PIK3CA transformation in a breast epithelial cell line. Sci. Rep. 2017, 7, 46079. [Google Scholar] [CrossRef]
- Siderius, D. NIST Standard Reference Simulation Website. Available online: https://www.nist.gov/programs-projects/nist-standard-reference-simulation-website (accessed on 26 August 2020).
- Behrends, V.; Tredwell, G.D.; Bundy, J.G. A software complement to AMDIS for processing GC-MS metabolomic data. Anal. Biochem. 2011, 415, 206–208. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benito, A.; Hajji, N.; O’Neill, K.; Keun, H.C.; Syed, N. β-Hydroxybutyrate Oxidation Promotes the Accumulation of Immunometabolites in Activated Microglia Cells. Metabolites 2020, 10, 346. https://doi.org/10.3390/metabo10090346
Benito A, Hajji N, O’Neill K, Keun HC, Syed N. β-Hydroxybutyrate Oxidation Promotes the Accumulation of Immunometabolites in Activated Microglia Cells. Metabolites. 2020; 10(9):346. https://doi.org/10.3390/metabo10090346
Chicago/Turabian StyleBenito, Adrian, Nabil Hajji, Kevin O’Neill, Hector C. Keun, and Nelofer Syed. 2020. "β-Hydroxybutyrate Oxidation Promotes the Accumulation of Immunometabolites in Activated Microglia Cells" Metabolites 10, no. 9: 346. https://doi.org/10.3390/metabo10090346
APA StyleBenito, A., Hajji, N., O’Neill, K., Keun, H. C., & Syed, N. (2020). β-Hydroxybutyrate Oxidation Promotes the Accumulation of Immunometabolites in Activated Microglia Cells. Metabolites, 10(9), 346. https://doi.org/10.3390/metabo10090346





