Dependence of the Staphylococcal Volatilome Composition on Microbial Nutrition
Abstract
1. Introduction
2. Results and Discussion
2.1. Staphylococcal Volatile Metabolites Produced in Rich Media
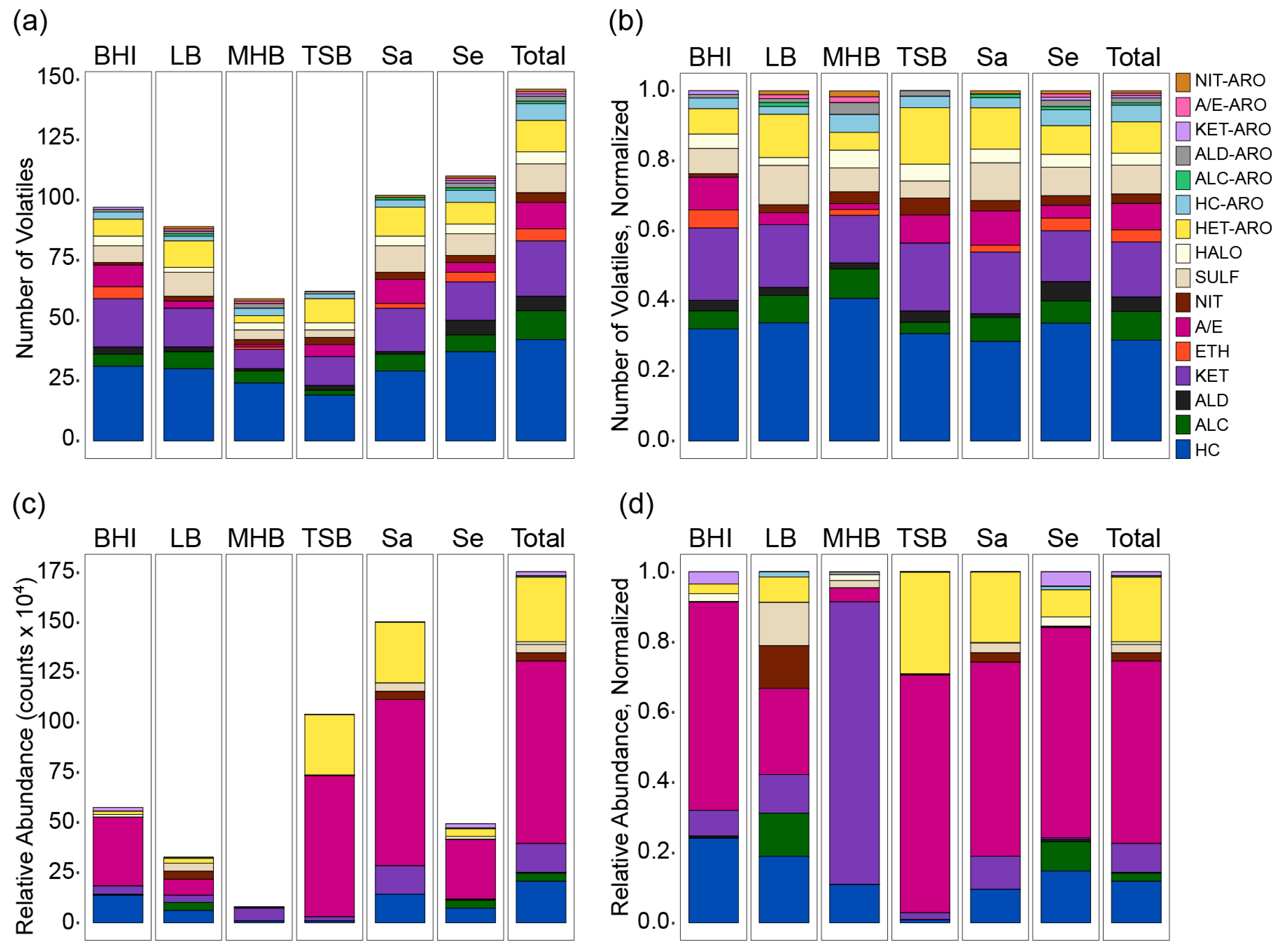
2.2. Chemical Composition of the S. aureus and S. epidermidis Volatilomes
2.3. Influence of Medium and Nutrition on the Chemical Composition of the Volatilome
2.4. Species-Specific Responses to Medium Nutrition
3. Materials and Methods
3.1. Bacterial Strains, Culture Conditions, and Metabolomics Analysis Sample Preparation
3.2. Analysis by HS-SPME-GC×GC-TOFMS
3.3. Statistical Analyses
3.4. Compound Identification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Somerville, G.A.; Proctor, R.A. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol. Mol. Biol. Rev. 2009, 73, 233–248. [Google Scholar] [CrossRef]
- Seidl, K.; Müller, S.; François, P.; Kriebitzsch, C.; Schrenzel, J.; Engelmann, S.; Bischoff, M.; Berger-Bächi, B. Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 2009, 9, 95. [Google Scholar] [CrossRef]
- Liebeke, M.; Dörries, K.; Zühlke, D.; Bernhardt, J.; Fuchs, S.; Pane-Farre, J.; Engelmann, S.; Völker, U.; Bode, R.; Dandekar, T. A metabolomics and proteomics study of the adaptation of Staphylococcus aureus to glucose starvation. Mol. Biosyst. 2011, 7, 1241–1253. [Google Scholar] [CrossRef]
- Richardson, A.R. Virulence and metabolism. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Schurig-Briccio, L.A.; Parraga Solorzano, P.K.; Lencina, A.M.; Radin, J.N.; Chen, G.Y.; Sauer, J.D.; Kehl-Fie, T.E.; Gennis, R.B. Role of respiratory NADH oxidation in the regulation of Staphylococcus aureus virulence. EMBO Rep. 2020, e45832. [Google Scholar] [CrossRef]
- Haag, A.F.; Bagnoli, F. The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. In Staphylococcus aureus; Springer: Berlin/Heidelberg, Germany, 2015; pp. 145–198. [Google Scholar]
- Scott, W. Water relations of Staphylococcus aureus at 30 °C. Aust. J. Biol. Sci. 1953, 6, 549–564. [Google Scholar] [CrossRef]
- Graham, J.E.; Wilkinson, B.J. Staphylococcus aureus osmoregulation: Roles for choline, glycine betaine, proline, and taurine. J. Bacteriol. 1992, 174, 2711–2716. [Google Scholar] [CrossRef]
- Chan, P.F.; Foster, S.J. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiol. SGM 1998, 144, 2469–2479. [Google Scholar] [CrossRef]
- Clements, M.O.; Foster, S.J. Stress resistance in Staphylococcus aureus. Trends Microbiol. 1999, 7, 458–462. [Google Scholar] [CrossRef]
- Cebrián, G.; Sagarzazu, N.; Pagán, R.; Condón, S.; Mañas, P. Development of stress resistance in Staphylococcus aureus after exposure to sublethal environmental conditions. Int. J. Food Microbiol. 2010, 140, 26–33. [Google Scholar] [CrossRef]
- Kehl-Fie, T.E.; Chitayat, S.; Hood, M.I.; Damo, S.; Restrepo, N.; Garcia, C.; Munro, K.A.; Chazin, W.J.; Skaar, E.P. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 2011, 10, 158–164. [Google Scholar] [CrossRef]
- Liebeke, M.; Lalk, M. Staphylococcus aureus metabolic response to changing environmental conditions—A metabolomics perspective. Int. J. Med. Microbiol. 2014, 304, 222–229. [Google Scholar] [CrossRef]
- Alreshidi, M.M.; Dunstan, R.H.; Macdonald, M.M.; Smith, N.D.; Gottfries, J.; Roberts, T.K. Amino acids and proteomic acclimation of Staphylococcus aureus when incubated in a defined minimal medium supplemented with 5% sodium chloride. MicrobiologyOpen 2019, 8, e00772. [Google Scholar] [CrossRef]
- Solórzano, P.K.P.; Yao, J.; Rock, C.O.; Kehl-Fie, T.E. Disruption of glycolysis by nutritional immunity activates a two-component system that coordinates a metabolic and antihost response by Staphylococcus aureus. Mbio 2019, 10, e01321-19. [Google Scholar]
- Collins, F.; Lascelles, J. The effect of growth conditions on oxidative and dehydrogenase activity in Staphylococcus aureus. J. Gen. Microbiol. 1962, 29, 531–535. [Google Scholar] [CrossRef]
- Strasters, K.; Winkler, K. Carbohydrate metabolism of Staphylococcus aureus. J. Gen. Microbiol. 1963, 33, 213–229. [Google Scholar] [CrossRef]
- Somerville, G.A.; Cockayne, A.; Dürr, M.; Peschel, A.; Otto, M.; Musser, J.M. Synthesis and deformylation of Staphylococcus aureus δ-toxin are linked to tricarboxylic acid cycle activity. J. Bacteriol. 2003, 185, 6686–6694. [Google Scholar] [CrossRef]
- Kaiser, J.C.; King, A.N.; Grigg, J.C.; Sheldon, J.R.; Edgell, D.R.; Murphy, M.E.; Brinsmade, S.R.; Heinrichs, D.E. Repression of branched-chain amino acid synthesis in Staphylococcus aureus is mediated by isoleucine via CodY, and by a leucine-rich attenuator peptide. PLoS Genet. 2018, 14, e1007159. [Google Scholar] [CrossRef]
- Ramsey, H. Endogenous respiration of Staphylococcus aureus. J. Bacteriol. 1962, 83, 507–514. [Google Scholar] [CrossRef]
- Krzemiński, Z.; Mikucki, J.; Szarapińska-Kwaszewska, J. Endogenous metabolism of Staphylococcus aureus. Folia Microbiol. 1972, 17, 46–54. [Google Scholar] [CrossRef]
- Lakshmi, H.P.; Prasad, U.V.; Yeswanth, S.; Swarupa, V.; Prasad, O.H.; Narasu, M.L.; Sarma, P.V.G.K. Molecular characterization of α-amylase from Staphylococcus aureus. Bioinformation 2013, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Rayner, M.H.; Sadler, P.J.; Scawen, M.D. NMR studies of a bacterial cell culture medium (LB broth): Cyclic nucleosides in yeast extracts. FEMS Microbiol. Lett. 1990, 68, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef] [PubMed]
- Wijesinghe, G.; Dilhari, A.; Gayani, B.; Kottegoda, N.; Samaranayake, L.; Weerasekera, M. Influence of laboratory culture media on in vitro growth, adhesion, and biofilm formation of Pseudomonas aeruginosa and Staphylococcus aureus. Med. Princ. Pract. 2019, 28, 28–35. [Google Scholar] [CrossRef]
- Macleod, P.; Jensen, R.; Gander, G.; Sampugna, J. Quantity and fatty acid composition of lipid extracted from cells of Streptococcus lactis. J. Bacteriol. 1962, 83, 806–810. [Google Scholar] [CrossRef]
- Huston, C.K.; Albro, P.W. Lipids of Sarcina lutea I.: Fatty acid composition of the extractable lipids. J. Bacteriol. 1964, 88, 425–432. [Google Scholar] [CrossRef]
- Hiron, A.; Borezée-Durant, E.; Piard, J.-C.; Juillard, V. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 2007, 189, 5119–5129. [Google Scholar] [CrossRef]
- Zhang, Y.-M.; Rock, C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008, 6, 222–233. [Google Scholar] [CrossRef]
- Sen, S.; Sirobhushanam, S.; Johnson, S.R.; Song, Y.; Tefft, R.; Gatto, C.; Wilkinson, B.J. Growth-environment dependent modulation of Staphylococcus aureus branched-chain to straight-chain fatty acid ratio and incorporation of unsaturated fatty acids. PLoS ONE 2016, 11, e0165300. [Google Scholar] [CrossRef]
- Tiwari, K.B.; Gatto, C.; Wilkinson, B.J. Plasticity of coagulase-negative staphylococcal membrane fatty acid composition and implications for responses to antimicrobial agents. Antibiotics 2020, 9, 214. [Google Scholar] [CrossRef]
- Schulz, S.; Dickschat, J.S. Bacterial volatiles: The smell of small organisms. Nat. Prod. Rep. 2007, 24, 814–842. [Google Scholar] [CrossRef]
- Citron, C.A.; Rabe, P.; Dickschat, J.S. The scent of bacteria: Headspace analysis for the discovery of natural products. J. Nat. Prod. 2012, 75, 1765–1776. [Google Scholar] [CrossRef]
- Veselova, M.; Plyuta, V.; Khmel, I. Volatile compounds of bacterial origin: Structure, biosynthesis, and biological activity. Microbiology 2019, 88, 261–274. [Google Scholar] [CrossRef]
- Dryahina, K.; Sovova, K.; Nemec, A.; Spanel, P. Differentiation of pulmonary bacterial pathogens in cystic fibrosis by volatile metabolites emitted by their in vitro cultures: Pseudomonas aeruginosa, Staphylococcus aureus, Stenotrophomonas maltophilia and the Burkholderia cepacia complex. J. Breath Res. 2016, 10, 037102. [Google Scholar] [CrossRef]
- Rees, C.A.; Nordick, K.V.; Franchina, F.A.; Lewis, A.E.; Hirsch, E.B.; Hill, J.E. Volatile metabolic diversity of Klebsiella pneumoniae in nutrient-replete conditions. Metabolomics 2017, 13, 18. [Google Scholar] [CrossRef]
- Wolfe, A.J. The acetate switch. Microbiol. Mol. Biol. Rev. 2005, 69, 12–50. [Google Scholar] [CrossRef]
- Baptista, I.; Santos, M.; Rudnitskaya, A.; Saraiva, J.A.; Almeida, A.; Rocha, S.M. A comprehensive look into the volatile exometabolome of enteroxic and non-enterotoxic Staphylococcus aureus strains. Int. J. Biochem. Cell Biol. 2019, 108, 40–50. [Google Scholar] [CrossRef]
- Berrou, K.; Dunyach-Remy, C.; Lavigne, J.-P.; Roig, B.; Cadiere, A. Multiple stir bar sorptive extraction combined with gas chromatography-mass spectrometry analysis for a tentative identification of bacterial volatile and/or semi-volatile metabolites. Talanta 2019, 195, 245–250. [Google Scholar] [CrossRef]
- Jia, B.; Sohnlein, B.; Mortelmans, K.; Coggiola, M.; Oser, H. Distinguishing methicillin-resistant and sensitive Staphylococcus aureus using volatile headspace metabolites. IEEE Sens. J. 2010, 10, 71–75. [Google Scholar] [CrossRef]
- Preti, G.; Thaler, E.; Hanson, C.W.; Troy, M.; Eades, J.; Gelperin, A. Volatile compounds characteristic of sinus-related bacteria and infected sinus mucus: Analysis by solid-phase microextraction and gas chromatography-mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009, 877, 2011–2018. [Google Scholar] [CrossRef]
- Timm, C.M.; Lloyd, E.P.; Egan, A.; Mariner, R.; Karig, D. Direct growth of bacteria in headspace vials allows for screening of volatiles by gas chromatography mass spectrometry. Front. Microbiol. 2018, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Rees, C.A.; Shen, A.; Hill, J.E. Characterization of the Clostridium difficile volatile metabolome using comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016, 1039, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.J.; DeMilo, A.B.; Moreno, D.S.; Martinez, A.J. Analysis of the volatile components of a bacterial fermentation attractive to the Mexican fruit fly (Anastrepha ludens). J. Agric. Food Chem. 1995, 43, 1348–1351. [Google Scholar] [CrossRef]
- Hettinga, K.A.; van Valenberg, H.J.; Lam, T.J.; van Hooijdonk, A.C. The origin of the volatile metabolites found in mastitis milk. Vet. Microbiol. 2009, 137, 384–387. [Google Scholar] [CrossRef]
- Filipiak, W.; Sponring, A.; Filipiak, A.; Baur, M.; Ager, C.; Wiesenhofer, H.; Margesin, R.; Nagl, M.; Troppmair, J.; Amann, A. Volatile organic compounds (VOCs) released by pathogenic microorganisms in vitro: Potential breath biomarkers. In Volatile Biomarkers; Elsevier: Amsterdam, The Netherlands, 2013; p. 463. [Google Scholar]
- Filipiak, W.; Beer, R.; Sponring, A.; Filipiak, A.; Ager, C.; Schiefecker, A.; Lanthaler, S.; Helbok, R.; Nagl, M.; Troppmair, J. Breath analysis for in vivo detection of pathogens related to ventilator-associated pneumonia in intensive care patients: A prospective pilot study. J. Breath Res. 2015, 9, 016004. [Google Scholar] [CrossRef]
- Pizzini, A.; Filipiak, W.; Wille, J.; Ager, C.; Wiesenhofer, H.; Kubinec, R.; Blaško, J.; Tschurtschenthaler, C.; Mayhew, C.A.; Weiss, G. Analysis of volatile organic compounds in the breath of patients with stable or acute exacerbation of chronic obstructive pulmonary disease. J. Breath Res. 2018, 12, 036002. [Google Scholar] [CrossRef]
- Lemfack, M.C.; Ravella, S.R.; Lorenz, N.; Kai, M.; Jung, K.; Schulz, S.; Piechulla, B. Novel volatiles of skin-borne bacteria inhibit the growth of Gram-positive bacteria and affect quorum-sensing controlled phenotypes of Gram-negative bacteria. Syst. Appl. Microbiol. 2016, 39, 503–515. [Google Scholar] [CrossRef]
- Chen, J.; Tang, J.; Shi, H.; Tang, C.; Zhang, R. Characteristics of volatile organic compounds produced from five pathogenic bacteria by headspace-solid phase micro-extraction/gas chromatography-mass spectrometry. J. Basic Microbiol. 2017, 57, 228–237. [Google Scholar] [CrossRef]
- Bean, H.D.; Dimandja, J.M.; Hill, J.E. Bacterial volatile discovery using solid phase microextraction and comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2012, 901, 41–46. [Google Scholar] [CrossRef]
- Vergnais, L.; Masson, F.; Montel, M.C.; Berdague, J.L.; Talon, R. Evaluation of solid-phase microextraction for analysis of volatile metabolites produced by staphylococci. J. Agric. Food Chem. 1998, 46, 228–234. [Google Scholar] [CrossRef]
- Rees, C.A.; Smolinska, A.; Hill, J.E. The volatile metabolome of Klebsiella pneumoniae in human blood. J. Breath Res. 2016, 10, 027101. [Google Scholar] [CrossRef] [PubMed]
- Hang, Y.; Chingin, K.; Liang, J.; Wang, X.; Hu, L. Fast detection of volatile organic compounds from staphylococcal blood cultures by CDI-MS. RSC Adv. 2017, 7, 24789–24794. [Google Scholar] [CrossRef]
- Tait, E.; Perry, J.D.; Stanforth, S.P.; Dean, J.R. Identification of volatile organic compounds produced by bacteria using HS-SPME-GC-MS. J. Chromatogr. Sci. 2014, 52, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Storer, M.K.; Hibbard-Melles, K.; Davis, B.; Scotter, J. Detection of volatile compounds produced by microbial growth in urine by selected ion flow tube mass spectrometry (SIFT-MS). J. Microbiol. Methods 2011, 87, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Thorn, R.M.; Reynolds, D.M.; Greenman, J. Multivariate analysis of bacterial volatile compound profiles for discrimination between selected species and strains in vitro. J. Microbiol. Methods 2011, 84, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Allardyce, R.A.; Langford, V.S.; Hill, A.L.; Murdoch, D.R. Detection of volatile metabolites produced by bacterial growth in blood culture media by selected ion flow tube mass spectrometry (SIFT-MS). J. Microbiol. Methods 2006, 65, 361–365. [Google Scholar] [CrossRef]
- Verhulst, N.O.; Andriessen, R.; Groenhagen, U.; Bukovinszkine Kiss, G.; Schulz, S.; Takken, W.; van Loon, J.J.; Schraa, G.; Smallegange, R.C. Differential attraction of malaria mosquitoes to volatile blends produced by human skin bacteria. PLoS ONE 2010, 5, e15829. [Google Scholar] [CrossRef]
- Nizio, K.D.; Perrault, K.A.; Troobnikoff, A.N.; Ueland, M.; Shoma, S.; Iredell, J.R.; Middleton, P.G.; Forbes, S.L. In vitro volatile organic compound profiling using GC×GC-TOFMS to differentiate bacteria associated with lung infections: A proof-of-concept study. J. Breath Res. 2016, 10, 026008. [Google Scholar] [CrossRef]
- Elgaali, H.; Hamilton-Kemp, T.R.; Newman, M.C.; Collins, R.W.; Yu, K.; Archbold, D.D. Comparison of long-chain alcohols and other volatile compounds emitted from food-borne and related Gram positive and Gram negative bacteria. J. Basic Microbiol. 2002, 42, 373–380. [Google Scholar] [CrossRef]
- Robacker, D.C.; Bartelt, R.J. Chemicals attractive to Mexican fruit fly from Klebsiella pneumoniae and Citrobacter freundii. Cultures sampled by solid-phase microextraction. J. Chem. Ecol. 1997, 23, 2897–2915. [Google Scholar] [CrossRef]
- Nasir, M.; Bean, H.D.; Smolinska, A.; Rees, C.A.; Zemanick, E.T.; Hill, J.E. Volatile molecules from bronchoalveolar lavage fluid can ‘rule-in’ Pseudomonas aeruginosa and ‘rule-out’ Staphylococcus aureus infections in cystic fibrosis patients. Sci. Rep. 2018, 8, 826. [Google Scholar] [CrossRef] [PubMed]
- Filipiak, W.; Sponring, A.; Baur, M.M.; Filipiak, A.; Ager, C.; Wiesenhofer, H.; Nagl, M.; Troppmair, J.; Amann, A. Molecular analysis of volatile metabolites released specifically by Staphylococcus aureus and Pseudomonas aeruginosa. BMC Microbiol. 2012, 12, 113. [Google Scholar] [CrossRef] [PubMed]
- Bean, H.D.; Rees, C.A.; Hill, J.E. Comparative analysis of the volatile metabolomes of Pseudomonas aeruginosa clinical isolates. J. Breath Res. 2016, 10, 047102. [Google Scholar] [CrossRef]
- Smart, A.; de Costello, B.L.; White, P.; Avison, M.; Batty, C.; Turner, C.; Persad, R.; Ratcliffe, N. Sniffing out resistance-rapid identification of urinary tract infection-causing bacteria and their antibiotic susceptibility using volatile metabolite profiles. J. Pharm. Biomed. Anal. 2019, 167, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Lawal, O.; Muhamadali, H.; Ahmed, W.M.; White, I.R.; Nijsen, T.M.E.; Goodacre, R.; Fowler, S.J. Headspace volatile organic compounds from bacteria implicated in ventilator-associated pneumonia analysed by TD-GC/MS. J. Breath Res. 2018, 12, 026002. [Google Scholar] [CrossRef] [PubMed]
- Jünger, M.; Vautz, W.; Kuhns, M.; Hofmann, L.; Ulbricht, S.; Baumbach, J.I.; Quintel, M.; Perl, T. Ion mobility spectrometry for microbial volatile organic compounds: A new identification tool for human pathogenic bacteria. Appl. Microbiol. Biotechnol. 2012, 93, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Epsky, N.D.; Heath, R.R.; Dueben, B.D.; Lauzon, C.R.; Proveaux, A.T.; MacCollom, G.B. Attraction of 3-methyl-1-butanol and ammonia identified from Enterobacter agglomerans to Anastrepha suspensa. J. Chem. Ecol. 1998, 24, 1867–1880. [Google Scholar] [CrossRef]
- Franchina, F.A.; Purcaro, G.; Burklund, A.; Beccaria, M.; Hill, J.E. Evaluation of different adsorbent materials for the untargeted and targeted bacterial VOC analysis using GC×GC-MS. Anal. Chim. Acta 2019, 1066, 146–153. [Google Scholar] [CrossRef]
- Karami, N.; Mirzajani, F.; Rezadoost, H.; Karimi, A.; Fallah, F.; Ghassempour, A.; Aliahmadi, A. Initial study of three different pathogenic microorganisms by gas chromatography-mass spectrometry. F1000research 2017, 6. [Google Scholar] [CrossRef]
- Neerincx, A.H.; Geurts, B.P.; van Loon, J.; Tiemes, V.; Jansen, J.J.; Harren, F.J.; Kluijtmans, L.A.; Merkus, P.J.; Cristescu, S.M.; Buydens, L.M.; et al. Detection of Staphylococcus aureus in cystic fibrosis patients using breath VOC profiles. J. Breath Res. 2016, 10, 046014. [Google Scholar] [CrossRef]
- Rees, C.A.; Franchina, F.A.; Nordick, K.V.; Kim, P.J.; Hill, J.E. Expanding the Klebsiella pneumoniae volatile metabolome using advanced analytical instrumentation for the detection of novel metabolites. J. Appl. Microbiol. 2017, 122, 785–795. [Google Scholar] [CrossRef]
- Boots, A.W.; Smolinska, A.; van Berkel, J.J.; Fijten, R.R.; Stobberingh, E.E.; Boumans, M.L.; Moonen, E.J.; Wouters, E.F.; Dallinga, J.W.; Van Schooten, F.J. Identification of microorganisms based on headspace analysis of volatile organic compounds by gas chromatography-mass spectrometry. J. Breath Res. 2014, 8, 027106. [Google Scholar] [CrossRef] [PubMed]
- Rees, C.A.; Burklund, A.; Stefanuto, P.H.; Schwartzman, J.D.; Hill, J.E. Comprehensive volatile metabolic fingerprinting of bacterial and fungal pathogen groups. J. Breath Res. 2018, 12, 026001. [Google Scholar] [CrossRef] [PubMed]
- Phan, J.; Meinardi, S.; Barletta, B.; Blake, D.R.; Whiteson, K. Stable isotope profiles reveal active production of VOCs from human-associated microbes. J. Breath Res. 2017, 11, 017101. [Google Scholar] [CrossRef] [PubMed]
- Power, D.A.; Johnson, J.A. Difco™ & BBL™ Manual; Becton, Dickinson and Company: Sparks, MD, USA, 2009. [Google Scholar]
- Jenkins, C.L.; Bean, H.D. Influence of media on the differentiation of Staphylococcus spp. by volatile compounds. J. Breath Res. 2020, 14, 016007. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Nair, K.S. Metabolomics workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016, 44, D463–D470. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- Fiehn, O.; Kristal, B.; van Ommen, B.; Sumner, L.W.; Sansone, S.A.; Taylor, C.; Hardy, N.; Kaddurah-Daouk, R. Establishing reporting standards for metabolomic and metabonomic studies: A call for participation. OMICS 2006, 10, 158–163. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]


| Log10 Peak Relative Abundances | Compound Name | CAS # | References | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus | S. epidermidis | |||||||||
| BHI | LB | MHB | TSB | BHI | LB | MHB | TSB | |||
| ACIDS and ESTERS | ||||||||||
| 4.7 | 4.9 | 3.5 | 2-methylbutanoic acid | 116-53-0 | [41,42,43,44,45] | |||||
| 4.1 | 4.2 | 2-methylbutyl acetate | 624-41-9 | [46,47] | ||||||
| 4.3 | 4.6 | 3-methylbut-3-enyl acetate | 5205-07-2 | [38,48] | ||||||
| 2.3 | 5.8 | 5.3 | 2.8 | 5.0 | 3-methylbutanoic acid | 503-74-2 | [36,38,39,40,41,42,43,44,45,46,47,49,50,51,52] | |||
| 4.2 | 4.3 | acetic acid | 64-19-7 | [35,38,39,40,41,42,45,46,47,48,53,54,55,56,57,58] | ||||||
| 5.1 | butyl 2-methylbutanoate | 15706-73-7 | [51,59] | |||||||
| 0.6 | butyl 2-methylpropanoate | 97-87-0 | [59] | |||||||
| 0.6 | butyl acetate | 123-86-4 | [38,46,47,48,51,59,60] | |||||||
| 0.8 | butyl propanoate | 590-01-2 | ||||||||
| 0.6 | 0.6 | dec-5-en-1-ol acetic acid | [60] | |||||||
| AROMATIC ACIDS and ESTERS | ||||||||||
| 0.8 | methyl benzoate | 93-58-3 | [38,58] | |||||||
| ALCOHOLS | ||||||||||
| 1.8 | 2-butyloctan-1-ol | 3913-02-8 | ||||||||
| 4.6 | 2-methylbutan-1-ol | 137-32-6 | [40,43,61,62] | |||||||
| 0.4 | 2-methylbutan-2-ol | 75-85-4 | [60,63] | |||||||
| 0.5 | 2-methylpropan-2-ol | 75-65-0 | [36,52,64] | |||||||
| 0.4 | 3-methylbut-3-en-1-ol | 763-32-6 | [36,38,65] | |||||||
| 0.9 | 0.8 | 0.6 | 1.1 | 2.8 | 2.5 | 1.3 | 1.5 | 3-methylbutan-1-ol | 123-51-3 | [36,38,39,40,42,44,46,47,48,51,52,56,60,61,62,64,66,67,68,69,70] |
| 0.6 | 0.5 | 6-methylheptan-1-ol | 1653-40-3 | [38] | ||||||
| 0.4 | octan-1-ol | 111-87-5 | [38,43,50,55,61] | |||||||
| AROMATIC ALCOHOLS | ||||||||||
| 0.4 | 0.4 | 2-phenylpropan-2-ol | 617-94-7 | |||||||
| ALDEHYDES | ||||||||||
| 1.0 | 2-methylidenebutanal | 922-63-4 | [47] | |||||||
| 0.3 | 2-methylpropanal | 78-84-2 | [36,38,39,46,47,48,57] | |||||||
| 0.3 | 3-methylbutanal | 590-86-3 | [40,42,45,46,47,48,50,52,55,62,67,68] | |||||||
| 3.5 | oct-2-enal | 2363-89-5 | [64] | |||||||
| AROMATIC ALDEHYDES | ||||||||||
| 2.8 | benzaldehyde | 100-52-7 | [38,39,40,41,42,46,47,54,62,71] | |||||||
| 1.1 | 0.7 | 1.1 | 0.5 | 2-phenylacetaldehyde | 122-78-1 | [42,52,71] | ||||
| ETHERS | ||||||||||
| 0.4 | 2-butoxyethanol | 111-76-2 | ||||||||
| 0.4 | 2-methyl-2-propan-2-yloxypropane | 17348-59-3 | ||||||||
| HYDROCARBONS | ||||||||||
| 0.5 | 1-methyl-4-prop-1-en-2-ylcyclohexene | 138-86-3 | [38] | |||||||
| 1.2 | 1.8 | 0.9 | 2,4,6,8-tetramethylundec-1-ene | 59920-26-2 | ||||||
| 0.7 | 1.0 | 0.7 | 1.0 | 0.7 | 0.4 | 3,3-dimethyloctane | 4110-44-5 | [60] | ||
| 1.0 | 1.4 | 1.6 | 1.5 | 1.2 | 1.7 | 1.0 | 0.8 | 3,7-dimethyloct-1-ene | 4984-01-4 | |
| 0.4 | 4-methylheptane | 589-53-7 | [36,48] | |||||||
| 3.3 | 3.9 | 7-methyl-3-methylideneocta-1,6-diene | 123-35-3 | |||||||
| 1.4 | 1.2 | 1.5 | 1.8 | 1.8 | 1.0 | 0.6 | decane | 124-18-5 | [48,65] | |
| 1.3 | dodecane | 112-40-3 | ||||||||
| 0.7 | 0.9 | hexan-2-ylcyclopropane | 6976-28-9 | |||||||
| 1.0 | 1.3 | 2.3 | 1.2 | 1.2 | 1.5 | 1.4 | 0.4 | tridec-3-ene | ||
| 0.9 | 0.3 | undec-3-ene | 60669-40-1 | |||||||
| 0.7 | 0.6 | undecane | 1120-21-4 | [36,48,66,72] | ||||||
| AROMATIC HYDROCARBONS | ||||||||||
| 3.7 | 1,1,5,6-tetramethyl-2H-naphthalene | 220766-68-7 | ||||||||
| 0.4 | 0.3 | 1,2-xylene | 95-47-6 | |||||||
| 0.3 | 0.4 | 1,3-ditert-butylbenzene | 1014-60-4 | |||||||
| 0.4 | 0.3 | 1,4-xylene | 106-42-3 | |||||||
| 0.3 | 1-ethyl-2-methylbenzene | 611-14-3 | [43] | |||||||
| 0.4 | 0.3 | ethylbenzene | 100-41-4 | [36,48,73] | ||||||
| KETONES | ||||||||||
| 1.1 | 0.7 | 0.9 | 1.1 | 3-methylbutan-2-one | 563-80-4 | [36,60,63,64,65] | ||||
| 0.6 | 4,6-dimethylheptan-2-one | 19549-80-5 | [36] | |||||||
| 1.7 | 2.7 | 1.6 | 1.6 | 1.7 | 3.0 | 4-methylheptan-2-one | 6137-06-0 | [36,38,39] | ||
| 0.9 | 0.8 | 0.8 | 0.6 | 4-methylpentan-2-one | 108-10-1 | [38,46,65] | ||||
| 0.4 | 6-methylhept-5-en-2-one | 110-93-0 | [38,48,71] | |||||||
| 2.0 | 1.9 | 0.9 | 1.1 | 6-methylheptan-2-one | 928-68-7 | [36,38] | ||||
| 0.3 | 0.3 | but-3-en-2-one | 78-94-4 | [46,48,65,74] | ||||||
| 0.4 | butan-2-one | 78-93-3 | [35,36,38,42,46,55,56,62,63,64,65,66,69,72,75] | |||||||
| 0.5 | 1.6 | 0.5 | butane-2,3-dione | 431-03-8 | [36,46,47,48,53,55,60,62,64,74,75,76] | |||||
| 0.4 | cyclohexanone | 108-94-1 | [38,43,48] | |||||||
| 2.3 | 2.9 | 1.0 | 1.6 | decan-2-one | 693-54-9 | [38,65] | ||||
| 1.2 | 0.7 | dodecan-2-one | 6175-49-1 | [65] | ||||||
| 0.4 | 1.9 | 1.1 | 1.3 | 0.7 | heptane-2,3-dione | 96-04-8 | [38,64,68] | |||
| 1.6 | 1.4 | 0.7 | 1.6 | 0.6 | 1.0 | nonan-2-one | 821-55-6 | [35,36,38,42,43,44,46,48,49,50,55,60,61,64,65,68,73] | ||
| 2.9 | 2.8 | 2.1 | 2.7 | 1.6 | 1.6 | 0.8 | 0.5 | octan-2-one | 111-13-7 | [35,38,67,68,73] |
| 1.2 | 3.6 | 0.9 | 1.3 | 1.3 | 3.1 | octan-3-one | 106-68-3 | [38,46,48,65] | ||
| 0.7 | 0.6 | pentan-2-one | 107-87-9 | [35,38,39,42,45,46,47,48,52,53,56,62,64,65] | ||||||
| 0.5 | propan-2-one | 67-64-1 | [35,38,48,55,56,57,60,65,66,72,74] | |||||||
| 2.7 | 3.6 | 2.8 | 0.5 | tetradecan-2-one | 2345-27-9 | [38] | ||||
| 2.4 | 4.5 | 2.0 | 2.1 | tridecan-2-one | 593-08-8 | [38,61] | ||||
| 2.8 | 4.8 | 4.2 | undecan-2-one | 112-12-9 | [38,39,42,49,50,61,64,65,68] | |||||
| AROMATIC KETONES | ||||||||||
| 4.3 | 1-phenylpropan-2-one | 103-79-7 | [36,38,73] | |||||||
| NITROGENS | ||||||||||
| 2.5 | 0.7 | 0.5 | 3-methylbutanenitrile | 625-28-5 | ||||||
| AROMATIC NITROGENS | ||||||||||
| 0.4 | 0.4 | 0.5 | benzonitrile | 100-47-0 | [36,38,64,68,73] | |||||
| HETEROAROMATICS | ||||||||||
| 0.7 | 1.0 | 2.5 | 0.5 | 1.2 | 0.8 | 2,3,5-trimethylpyrazine | 14667-55-1 | [41,44,62,68] | ||
| 0.4 | 2,3-dimethylpyrazine | 5910-89-4 | [41,73] | |||||||
| 4.2 | 4.0 | 2,4-dimethyl-1H-pyrrole | 625-82-1 | |||||||
| 1.4 | 2.0 | 2.3 | 0.5 | 1.0 | 1.0 | 1.1 | 2,5-dimethylpyrazine | 123-32-0 | [43,44,53,62,71] | |
| 3.3 | 2.8 | 2-ethyl-6-methylpyrazine | 13925-03-6 | [66] | ||||||
| 1.5 | 1.2 | 1.2 | 0.7 | 1.3 | 2-methyl-5-propylpyrazine | 29461-03-8 | ||||
| 0.5 | 0.7 | 0.9 | 1.5 | 0.3 | 0.4 | 0.7 | 2-methylpyrazine | 109-08-0 | [36,41,44,62,64,68] | |
| 0.3 | 4.3 | 3.0 | 3,5-dimethyl-2-(3-methylbutyl)pyrazine | 111150-30-2 | [73] | |||||
| 0.5 | 1.7 | 3.2 | 3-butyl-2,5-dimethylpyrazine | 40790-29-2 | ||||||
| 2.7 | 3.8 | 4.1 | 3-ethyl-2,5-dimethylpyrazine | 13360-65-1 | ||||||
| SULFURS | ||||||||||
| 0.8 | 1.0 | 1.1 | 2-methylundecane-2-thiol | 10059-13-9 | [38,71] | |||||
| 0.4 | 0.7 | 0.3 | isothiocyanatocyclohexane | 1122-82-3 | ||||||
| 0.4 | 0.4 | (methyldisulfanyl)methane | 624-92-0 | [35,36,38,41,46,47,48,55,56,58,61,62,65,66,67,68,74] | ||||||
| 0.5 | 0.4 | 0.5 | 0.6 | 0.4 | (methyltrisulfanyl)methane | 3658-80-8 | [35,38,46,48,55,61,65,66,67,68] | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jenkins, C.L.; Bean, H.D. Dependence of the Staphylococcal Volatilome Composition on Microbial Nutrition. Metabolites 2020, 10, 347. https://doi.org/10.3390/metabo10090347
Jenkins CL, Bean HD. Dependence of the Staphylococcal Volatilome Composition on Microbial Nutrition. Metabolites. 2020; 10(9):347. https://doi.org/10.3390/metabo10090347
Chicago/Turabian StyleJenkins, Carrie L., and Heather D. Bean. 2020. "Dependence of the Staphylococcal Volatilome Composition on Microbial Nutrition" Metabolites 10, no. 9: 347. https://doi.org/10.3390/metabo10090347
APA StyleJenkins, C. L., & Bean, H. D. (2020). Dependence of the Staphylococcal Volatilome Composition on Microbial Nutrition. Metabolites, 10(9), 347. https://doi.org/10.3390/metabo10090347







