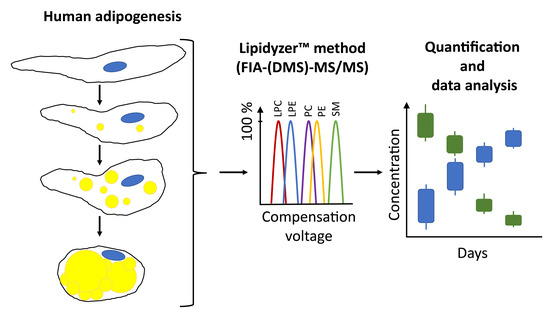
Lipidomic Phenotyping Reveals Extensive Lipid Remodeling during Adipogenesis in Human Adipocytes
Abstract
1. Introduction
2. Results
2.1. Analytical Method Validation
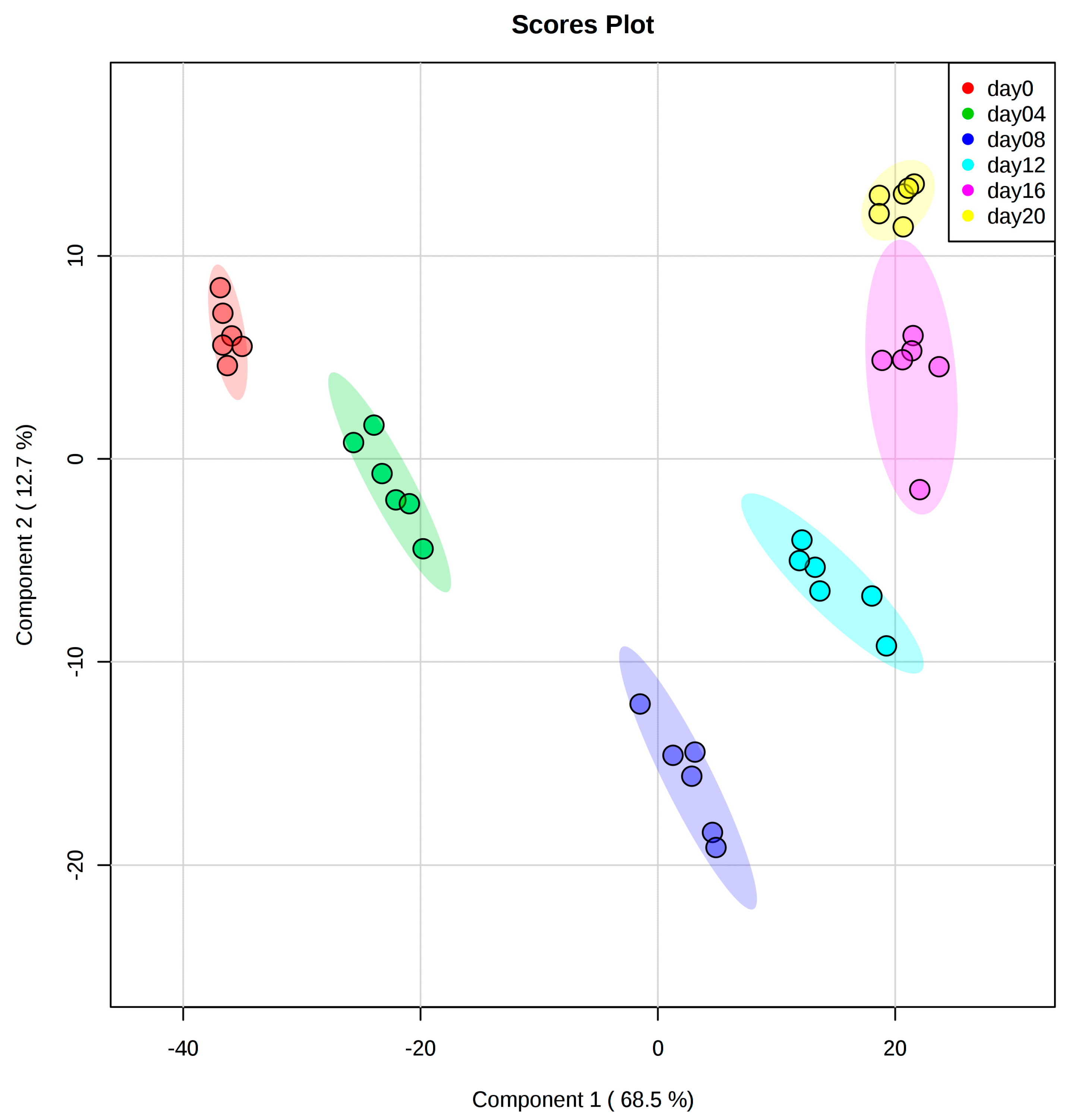
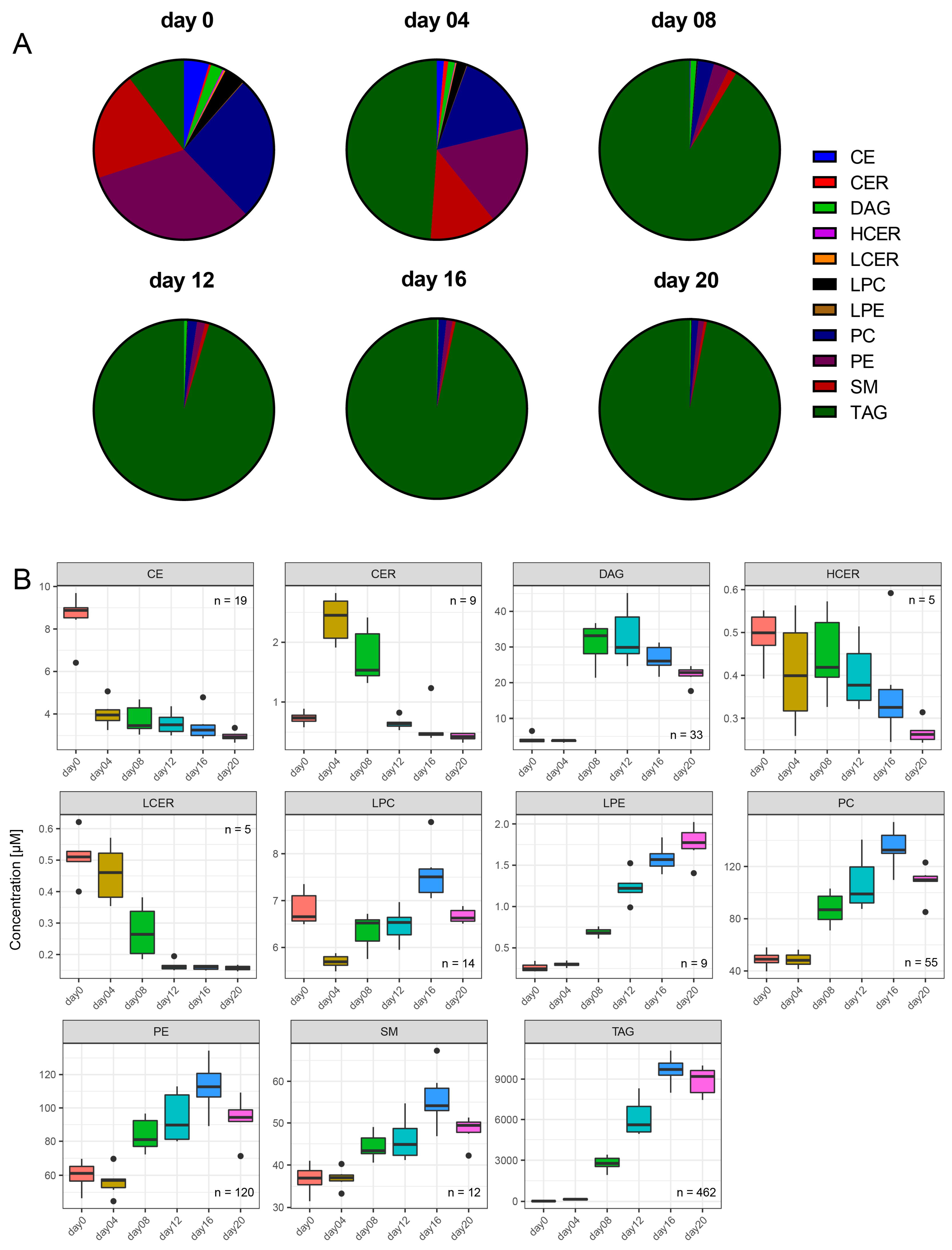
2.2. Cellular Lipid Composition Undergoes Remodeling During Adipogenesis to Mainly TAG
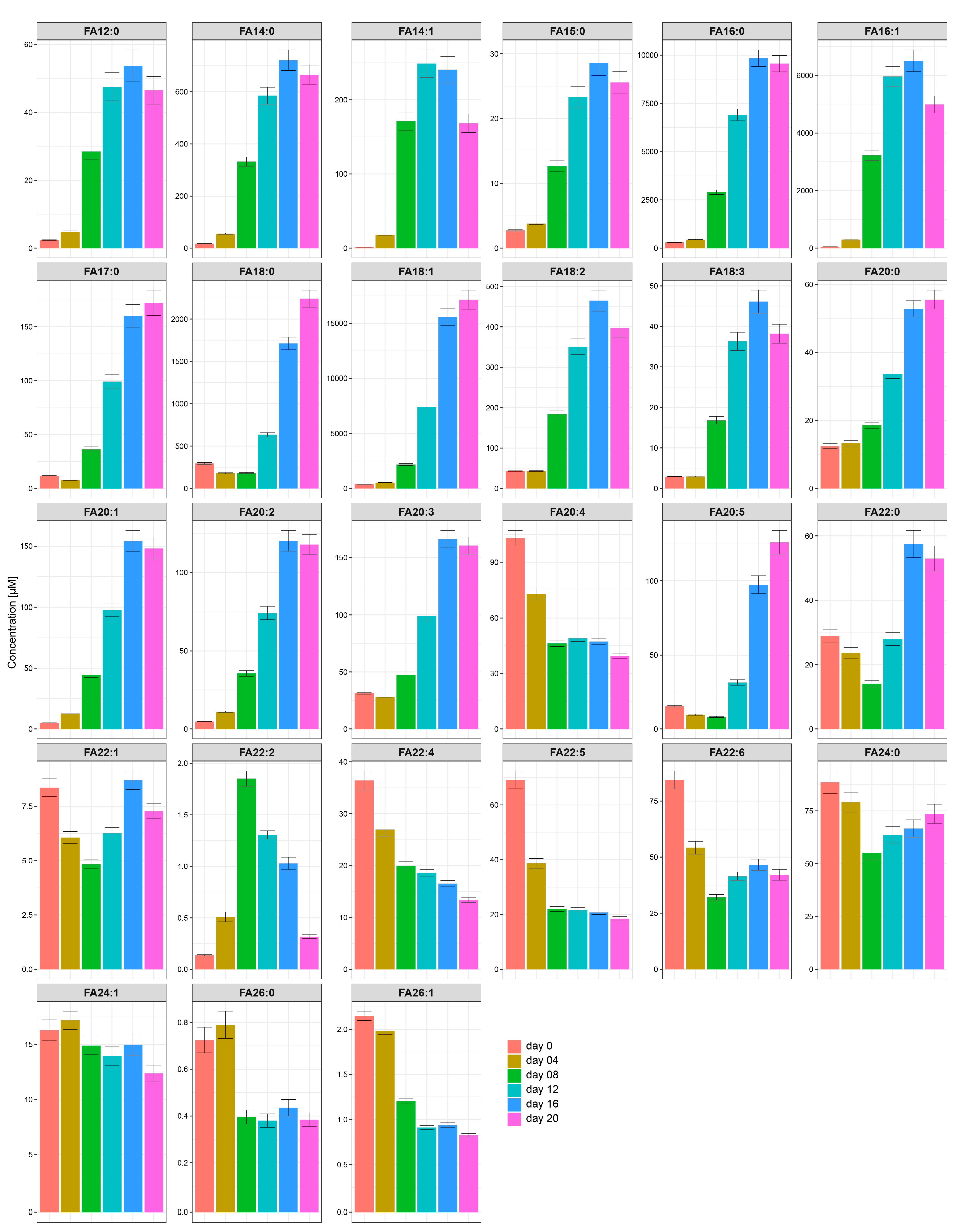
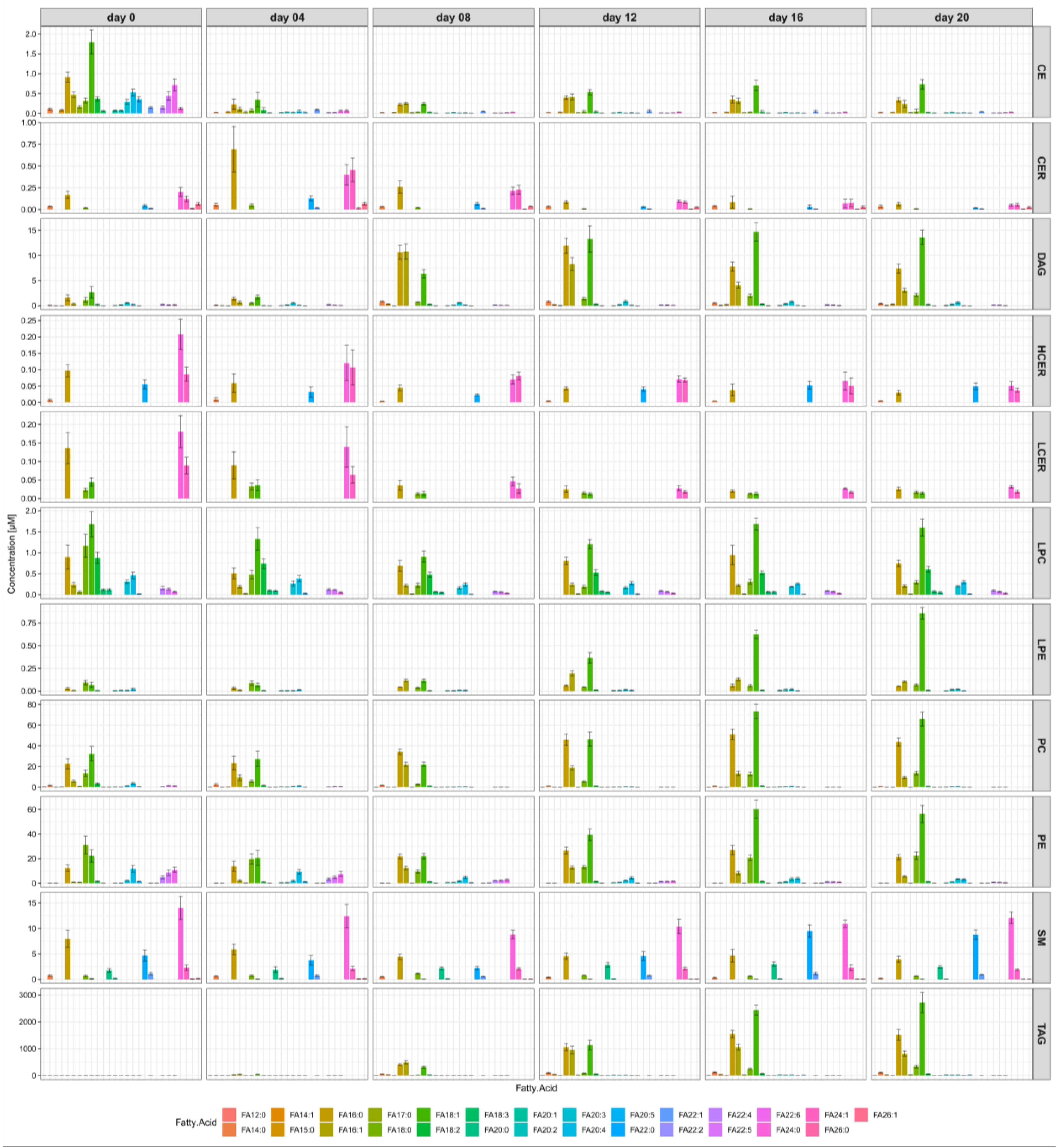
2.3. The Most Abundant Fatty Acids in Differentiated Human SGBS Cells Are C16:0, C16:1, C18:0, and C18:1
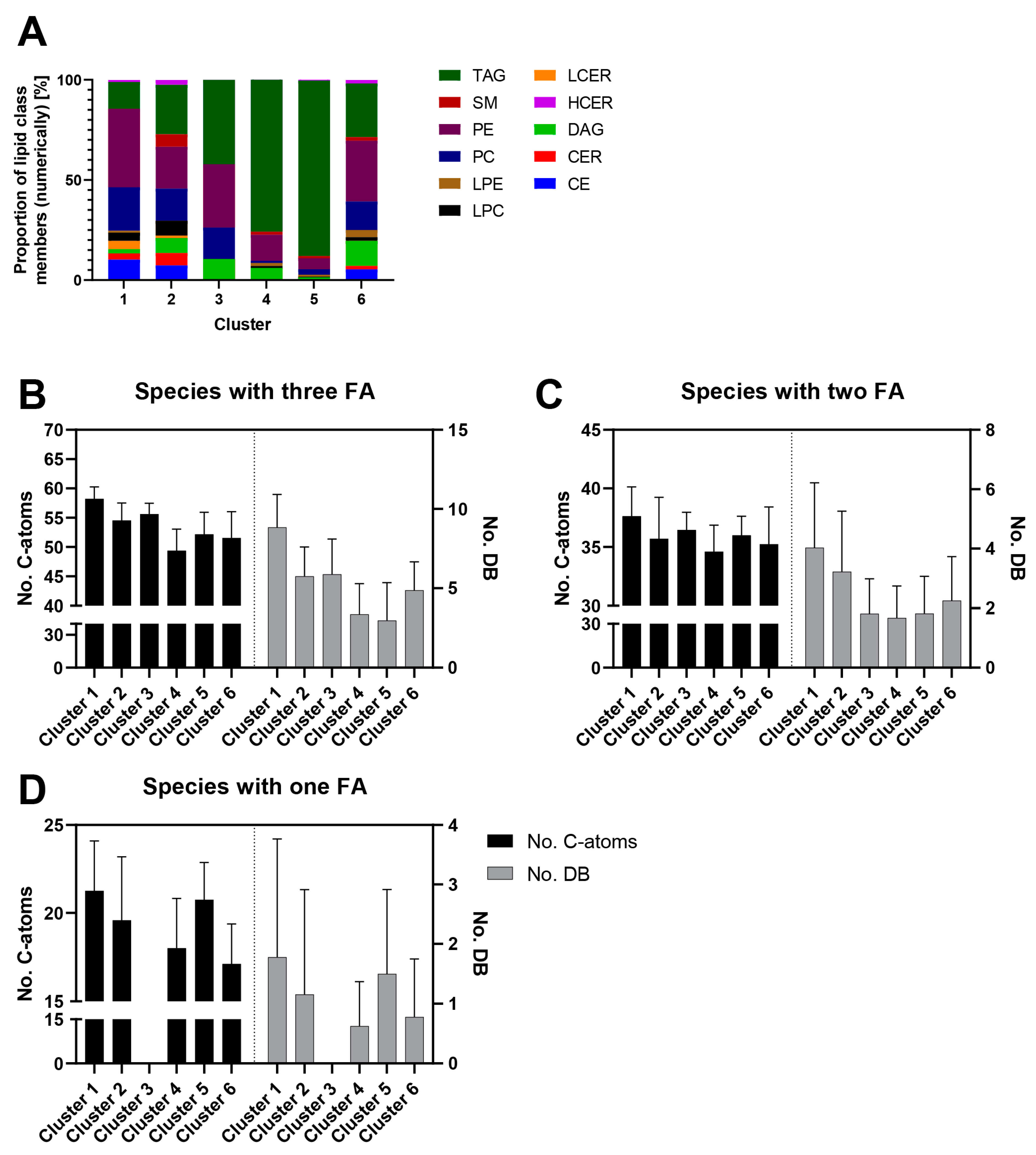
2.4. Correlations between Concentration Profiles of Lipid Species from Different Lipid Classes Reveal Extensive Lipid Remodeling during Adipogenesis
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Cell Harvesting and Homogenization
4.2. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
4.3. Hoechst Assay for DNA Quantification
4.4. Lipid Extraction and Targeted Lipidomics Analysis
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chooi, Y.C.; Ding, C.; Magkos, F. The epidemiology of obesity. Metabolism 2019, 92, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.M.; Danaei, G.; Farzadfar, F.; Stevens, G.A.; Woodward, M.; Wormser, D.; Kaptoge, S.; Whitlock, G.; Qiao, Q.; Lewington, S.; et al. The age-specific quantitative effects of metabolic risk factors on cardiovascular diseases and diabetes: A pooled analysis. PLoS ONE 2013, 8, e651740. [Google Scholar] [CrossRef] [PubMed]
- Czernichow, S.; Kengne, A.P.; Stamatakis, E.; Hamer, M.; Batty, G.D. Body mass index, waist circumference and waist-hip ratio: Which is the better discriminator of cardiovascular disease mortality risk? Evidence from an individual-participant meta-analysis of 82 864 participants from nine cohort studies. Obes. Rev. 2011, 12, 680–687. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, S.E.; Blackburn, O.A.; Marchildon, F.; Cohen, P. Insights into the Link Between Obesity and Cancer. Curr. Obes. Rep. 2017, 6, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.T.; Hochfeld, W.E.; Myburgh, R.; Pepper, M.S. Adipocyte and adipogenesis. Eur. J. Cell Biol. 2013, 92, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Lazar, M.A. New developments in adipogenesis. Trends Endocrinol. Metab. 2009, 20, 107–114. [Google Scholar] [CrossRef]
- Tontonoz, P.; Spiegelman, B.M. Fat and Beyond: The Diverse Biology of PPARγ. Annu. Rev. Biochem. 2008, 77, 289–312. [Google Scholar] [CrossRef]
- Otto, T.C.; Lane, M.D. Adipose Development: From Stem Cell to Adipocyte. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 229–242. [Google Scholar] [CrossRef]
- Roberts, L.D.; Virtue, S.; Vidal-Puig, A.; Nicholls, A.W.; Griffin, J.L. Metabolic phenotyping of a model of adipocyte differentiation. Physiol. Genom. 2009, 39, 109–119. [Google Scholar] [CrossRef]
- Guo, X.; Liao, K. Analysis of gene expression profile during 3T3-L1 preadipocyte differentiation. Gene 2000, 251, 45–53. [Google Scholar] [CrossRef]
- Ross, S.E.; Erickson, R.L.; Gerin, I.; DeRose, P.M.; Bajnok, L.; Longo, K.A.; Misek, D.E.; Kuick, R.; Hanash, S.M.; Atkins, K.B.; et al. Microarray analyses during adipogenesis: Understanding the effects of Wnt signaling on adipogenesis and the roles of liver X receptor alpha in adipocyte metabolism. Mol. Cell. Biol. 2002, 22, 5989–5999. [Google Scholar] [CrossRef] [PubMed]
- Green, C.R.; Wallace, M.; Divakaruni, A.S.; Phillips, S.A.; Murphy, A.N.; Ciaraldi, T.P.; Metallo, C.M. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nat. Chem. Biol. 2016, 12, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Molina, H.; Yang, Y.; Ruch, T.; Kim, J.-W.; Mortensen, P.; Otto, T.; Nalli, A.; Tang, Q.-Q.; Lane, M.D.; Chaerkady, R.; et al. Temporal profiling of the adipocyte proteome during differentiation using a five-plex SILAC based strategy. J. Proteome Res. 2009, 8, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Welsh, G.I.; Griffiths, M.R.; Webster, K.J.; Page, M.J.; Tavaré, J.M. Proteome analysis of adipogenesis. Proteom. Clin. Appl. 2004, 4, 1042–1051. [Google Scholar] [CrossRef]
- Ye, F.; Zhang, H.; Yang, Y.-X.; Hu, H.-D.; Sze, S.K.; Meng, W.; Qian, J.; Ren, H.; Yang, B.-L.; Luo, M.-Y.; et al. Comparative proteome analysis of 3T3-L1 adipocyte differentiation using iTRAQ-coupled 2D LC-MS/MS. J. Cell. Biochem. 2011, 112, 3002–3014. [Google Scholar] [CrossRef]
- Halama, A.; Horsch, M.; Kastenmuller, G.; Moller, G.; Kumar, P.; Prehn, C.; Laumen, H.; Hauner, H.; Hrabe de Angelis, M.; Beckers, J.; et al. Metabolic switch during adipogenesis: From branched chain amino acid catabolism to lipid synthesis. Arch. Biochem. Biophys. 2016, 589, 93–107. [Google Scholar] [CrossRef]
- Kirkwood, J.S.; Miranda, C.L.; Bobe, G.; Maier, C.S.; Stevens, J.F. 18O-Tracer Metabolomics Reveals Protein Turnover and CDP-Choline Cycle Activity in Differentiating 3T3-L1 Pre-Adipocytes. PLoS ONE 2016, 11, e0157118. [Google Scholar] [CrossRef]
- Liaw, L.; Prudovsky, I.; Koza, R.A.; Anunciado-Koza, R.V.; Siviski, M.E.; Lindner, V.; Friesel, R.E.; Rosen, C.J.; Baker, P.R.; Simons, B.; et al. Lipid Profiling of In Vitro Cell Models of Adipogenic Differentiation: Relationships With Mouse Adipose Tissues. J. Cell. Biochem. 2016, 117, 2182–2193. [Google Scholar] [CrossRef]
- Collins, J.M.; Neville, M.J.; Pinnick, K.E.; Hodson, L.; Ruyter, B.; van Dijk, T.H.; Reijngoud, D.-J.; Fielding, M.D.; Frayn, K.N. De novo lipogenesis in the differentiating human adipocyte can provide all fatty acids necessary for maturation. J. Lipid Res. 2011, 52, 1683–1692. [Google Scholar] [CrossRef]
- Ubhi, B.K. Direct Infusion-Tandem Mass Spectrometry (DI-MS/MS) Analysis of Complex Lipids in Human Plasma and Serum Using the Lipidyzer Platform. Methods Mol. Biol. 2018, 1730, 227–236. [Google Scholar] [CrossRef]
- Cao, Z.; Schmitt, T.C.; Varma, V.; Sloper, D.; Beger, R.D.; Sun, J. Evaluation of the Performance of Lipidyzer Platform and Its Application in the Lipidomics Analysis in Mouse Heart and Liver. J. Proteome Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Contrepois, K.; Mahmoudi, S.; Ubhi, B.K.; Papsdorf, K.; Hornburg, D.; Brunet, A.; Snyder, M. Cross-Platform Comparison of Untargeted and Targeted Lipidomics Approaches on Aging Mouse Plasma. Sci. Rep. 2018, 8, 17747. [Google Scholar] [CrossRef] [PubMed]
- Lepropre, S.; Kautbally, S.; Octave, M.; Ginion, A.; Onselaer, M.-B.; Steinberg, G.R.; Kemp, B.E.; Hego, A.; Wéra, O.; Brouns, S.; et al. AMPK-ACC signaling modulates platelet phospholipids and potentiates thrombus formation. Blood 2018, 132, 1180. [Google Scholar] [CrossRef] [PubMed]
- Franko, A.; Merkel, D.; Kovarova, M.; Hoene, M.; Jaghutriz, B.A.; Heni, M.; Konigsrainer, A.; Papan, C.; Lehr, S.; Haring, H.U.; et al. Dissociation of Fatty Liver and Insulin Resistance in I148M PNPLA3 Carriers: Differences in Diacylglycerol (DAG) FA18:1 Lipid Species as a Possible Explanation. Nutrients 2018, 10, 1314. [Google Scholar] [CrossRef]
- Alarcon-Barrera, J.C.; von Hegedus, J.H.; Brouwers, H.; Steenvoorden, E.; Ioan-Facsinay, A.; Mayboroda, O.A.; Ondo-Mendez, A.; Giera, M. Lipid metabolism of leukocytes in the unstimulated and activated states. Anal. Bioanal. Chem. 2020, 412, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Lintonen, T.P.; Baker, P.R.; Suoniemi, M.; Ubhi, B.K.; Koistinen, K.M.; Duchoslav, E.; Campbell, J.L.; Ekroos, K. Differential mobility spectrometry-driven shotgun lipidomics. Anal. Chem. 2014, 86, 9662–9669. [Google Scholar] [CrossRef]
- Yao, C.-H.; Liu, G.-Y.; Yang, K.; Gross, R.W.; Patti, G.J. Inaccurate quantitation of palmitate in metabolomics and isotope tracer studies due to plastics. Metabolomics 2016, 12, 143. [Google Scholar] [CrossRef]
- Bourbon, N.A.; Yun, J.; Kester, M. Ceramide Directly Activates Protein Kinase C ζ to Regulate a Stress-activated Protein Kinase Signaling Complex. J. Biol. Chem. 2000, 275, 35617–35623. [Google Scholar] [CrossRef]
- Reichert, M.; Eick, D. Analysis of cell cycle arrest in adipocyte differentiation. Oncogene 1999, 18, 459–466. [Google Scholar] [CrossRef]
- Ikonen, E. Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell Biol. 2008, 9, 125–138. [Google Scholar] [CrossRef]
- Prattes, S.; Horl, G.; Hammer, A.; Blaschitz, A.; Graier, W.F.; Sattler, W.; Zechner, R.; Steyrer, E. Intracellular distribution and mobilization of unesterified cholesterol in adipocytes: Triglyceride droplets are surrounded by cholesterol-rich ER-like surface layer structures. J. Cell Sci. 2000, 113, 2977. [Google Scholar] [PubMed]
- Wang, Y.; Castoreno, A.B.; Stockinger, W.; Nohturfft, A. Modulation of endosomal cholesteryl ester metabolism by membrane cholesterol. J. Biol. Chem. 2005, 280, 11876–11886. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.B.; Kennedy, E.P.; Kiyasu, J.Y. The Enzymatic Synthesis of Triglycerides. J. Biol. Chem. 1960, 235, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.A.; Mashek, D.G. Mammalian triacylglycerol metabolism: Synthesis, lipolysis, and signaling. Chem. Rev. 2011, 111, 6359–6386. [Google Scholar] [CrossRef] [PubMed]
- Fukami, K. Structure, Regulation, and Function of Phospholipase C Isozymes. J. Biochem. 2002, 131, 293–299. [Google Scholar] [CrossRef]
- Fong, W.-K.; Sánchez-Ferrer, A.; Rappolt, M.; Boyd, B.J.; Mezzenga, R. Structural Transformation in Vesicles upon Hydrolysis of Phosphatidylethanolamine and Phosphatidylcholine with Phospholipase C. Langmuir 2019, 35, 14949–14958. [Google Scholar] [CrossRef]
- Casares, D.; Escriba, P.V.; Rossello, C.A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef]
- Haczeyni, F.; Bell-Anderson, K.S.; Farrell, G.C. Causes and mechanisms of adipocyte enlargement and adipose expansion. Obes. Rev. 2018, 19, 406–420. [Google Scholar] [CrossRef]
- Kasturi, R.; Wakil, S.J. Increased synthesis and accumulation of phospholipids during differentiation of 3T3-L1 cells into adipocytes. J. Biol. Chem. 1983, 258, 3559–3564. [Google Scholar]
- Gauster, M.; Rechberger, G.; Sovic, A.; Horl, G.; Steyrer, E.; Sattler, W.; Frank, S. Endothelial lipase releases saturated and unsaturated fatty acids of high density lipoprotein phosphatidylcholine. J. Lipid Res. 2005, 46, 1517–1525. [Google Scholar] [CrossRef]
- Su, X.; Han, X.; Yang, J.; Mancuso, D.J.; Chen, J.; Bickel, P.E.; Gross, R.W. Sequential Ordered Fatty Acid α Oxidation and Δ9 Desaturation Are Major Determinants of Lipid Storage and Utilization in Differentiating Adipocytes. Biochemistry 2004, 43, 5033–5044. [Google Scholar] [CrossRef] [PubMed]
- Jeucken, A.; Brouwers, J.F. High-Throughput Screening of Lipidomic Adaptations in Cultured Cells. Biomolecules 2019, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Klusóczki, Á.; Veréb, Z.; Vámos, A.; Fischer-Posovszky, P.; Wabitsch, M.; Bacso, Z.; Fésüs, L.; Kristóf, E. Differentiating SGBS adipocytes respond to PPARγ stimulation, irisin and BMP7 by functional browning and beige characteristics. Sci. Rep. 2019, 9, 5823. [Google Scholar] [CrossRef]
- Yeo, C.R.; Agrawal, M.; Hoon, S.; Shabbir, A.; Shrivastava, M.K.; Huang, S.; Khoo, C.M.; Chhay, V.; Yassin, M.S.; Tai, E.S.; et al. SGBS cells as a model of human adipocyte browning: A comprehensive comparative study with primary human white subcutaneous adipocytes. Sci. Rep. 2017, 7, 4031. [Google Scholar] [CrossRef]
- Tews, D.; Fischer-Posovszky, P.; Fromme, T.; Klingenspor, M.; Fischer, J.; Rüther, U.; Marienfeld, R.; Barth, T.F.; Möller, P.; Debatin, K.M.; et al. FTO Deficiency Induces UCP-1 Expression and Mitochondrial Uncoupling in Adipocytes. Endocrinology 2013, 154, 3141–3151. [Google Scholar] [CrossRef] [PubMed]
- Tews, D.; Fromme, T.; Keuper, M.; Hofmann, S.M.; Debatin, K.M.; Klingenspor, M.; Wabitsch, M.; Fischer-Posovszky, P. Teneurin-2 (TENM2) deficiency induces UCP1 expression in differentiating human fat cells. Mol. Cell. Endocrinol. 2017, 443, 106–113. [Google Scholar] [CrossRef]
- Wabitsch, M.; Brenner, R.E.; Melzner, I.; Braun, M.; Möller, P.; Heinze, E.; Debatin, K.M.; Hauner, H. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int. J. Obes. 2001, 25, 8–15. [Google Scholar] [CrossRef]
- Shadid, S.; Koutsari, C.; Jensen, M.D. Direct Free Fatty Acid Uptake Into Human Adipocytes In Vivo Relation to Body Fat Distribution. Diabetes 2007, 56, 1369–1375. [Google Scholar] [CrossRef]
- Chamberlain, C.A.; Rubio, V.Y.; Garrett, T.J. Impact of matrix effects and ionization efficiency in non-quantitative untargeted metabolomics. Metabolomics 2019, 15, 135. [Google Scholar] [CrossRef]
- Triebl, A.; Trotzmuller, M.; Hartler, J.; Stojakovic, T.; Kofeler, H.C. Lipidomics by ultrahigh performance liquid chromatography-high resolution mass spectrometry and its application to complex biological samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1053, 72–80. [Google Scholar] [CrossRef]
- Koivusalo, M.; Haimi, P.; Heikinheimo, L.; Kostiainen, R.; Somerharju, P. Quantitative determination of phospholipid compositions by ESI-MS: Effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J. Lipid Res. 2001, 42, 663–672. [Google Scholar] [PubMed]
- Fischer-Posovszky, P.; Newell, F.S.; Wabitsch, M.; Tornqvist, H.E. Human SGBS cells—A unique tool for studies of human fat cell biology. Obes. Facts 2008, 1, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Muschet, C.; Moller, G.; Prehn, C.; de Angelis, M.H.; Adamski, J.; Tokarz, J. Removing the bottlenecks of cell culture metabolomics: Fast normalization procedure, correlation of metabolites to cell number, and impact of the cell harvesting method. Metabolomics 2016, 12, 151. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Liebisch, G.; Vizcaíno, J.A.; Köfeler, H.; Trötzmüller, M.; Griffiths, W.J.; Schmitz, G.; Spener, F.; Wakelam, M.J.O. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef]
- Vitello, D.J.; Ripper, R.M.; Fettiplace, M.R.; Weinberg, G.L.; Vitello, J.M. Blood Density Is Nearly Equal to Water Density: A Validation Study of the Gravimetric Method of Measuring Intraoperative Blood Loss. J. Vet. Med. 2015, 2015, 152730. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 24 April 2020).

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miehle, F.; Möller, G.; Cecil, A.; Lintelmann, J.; Wabitsch, M.; Tokarz, J.; Adamski, J.; Haid, M. Lipidomic Phenotyping Reveals Extensive Lipid Remodeling during Adipogenesis in Human Adipocytes. Metabolites 2020, 10, 217. https://doi.org/10.3390/metabo10060217
Miehle F, Möller G, Cecil A, Lintelmann J, Wabitsch M, Tokarz J, Adamski J, Haid M. Lipidomic Phenotyping Reveals Extensive Lipid Remodeling during Adipogenesis in Human Adipocytes. Metabolites. 2020; 10(6):217. https://doi.org/10.3390/metabo10060217
Chicago/Turabian StyleMiehle, Florian, Gabriele Möller, Alexander Cecil, Jutta Lintelmann, Martin Wabitsch, Janina Tokarz, Jerzy Adamski, and Mark Haid. 2020. "Lipidomic Phenotyping Reveals Extensive Lipid Remodeling during Adipogenesis in Human Adipocytes" Metabolites 10, no. 6: 217. https://doi.org/10.3390/metabo10060217
APA StyleMiehle, F., Möller, G., Cecil, A., Lintelmann, J., Wabitsch, M., Tokarz, J., Adamski, J., & Haid, M. (2020). Lipidomic Phenotyping Reveals Extensive Lipid Remodeling during Adipogenesis in Human Adipocytes. Metabolites, 10(6), 217. https://doi.org/10.3390/metabo10060217