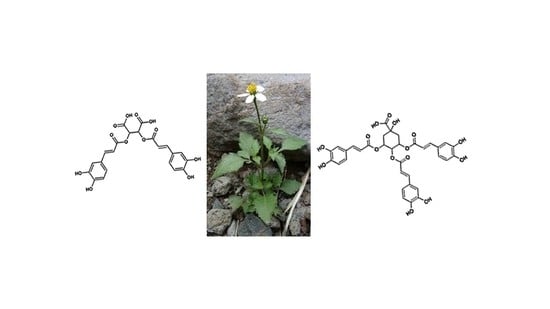
Profiling of Chlorogenic Acids from Bidens pilosa and Differentiation of Closely Related Positional Isomers with the Aid of UHPLC-QTOF-MS/MS-Based In-Source Collision-Induced Dissociation
Abstract
1. Introduction
2. Results
2.1. Profiling of Chlorogenic Acid Derivatives in Tissues and Cell Cultures of B. pilosa
2.2. Multivariate Data Analysis to Reveal Tissue-Specific and Cell Line-Specific Differences Within Tissues and Cell Cultures of B. pilosa
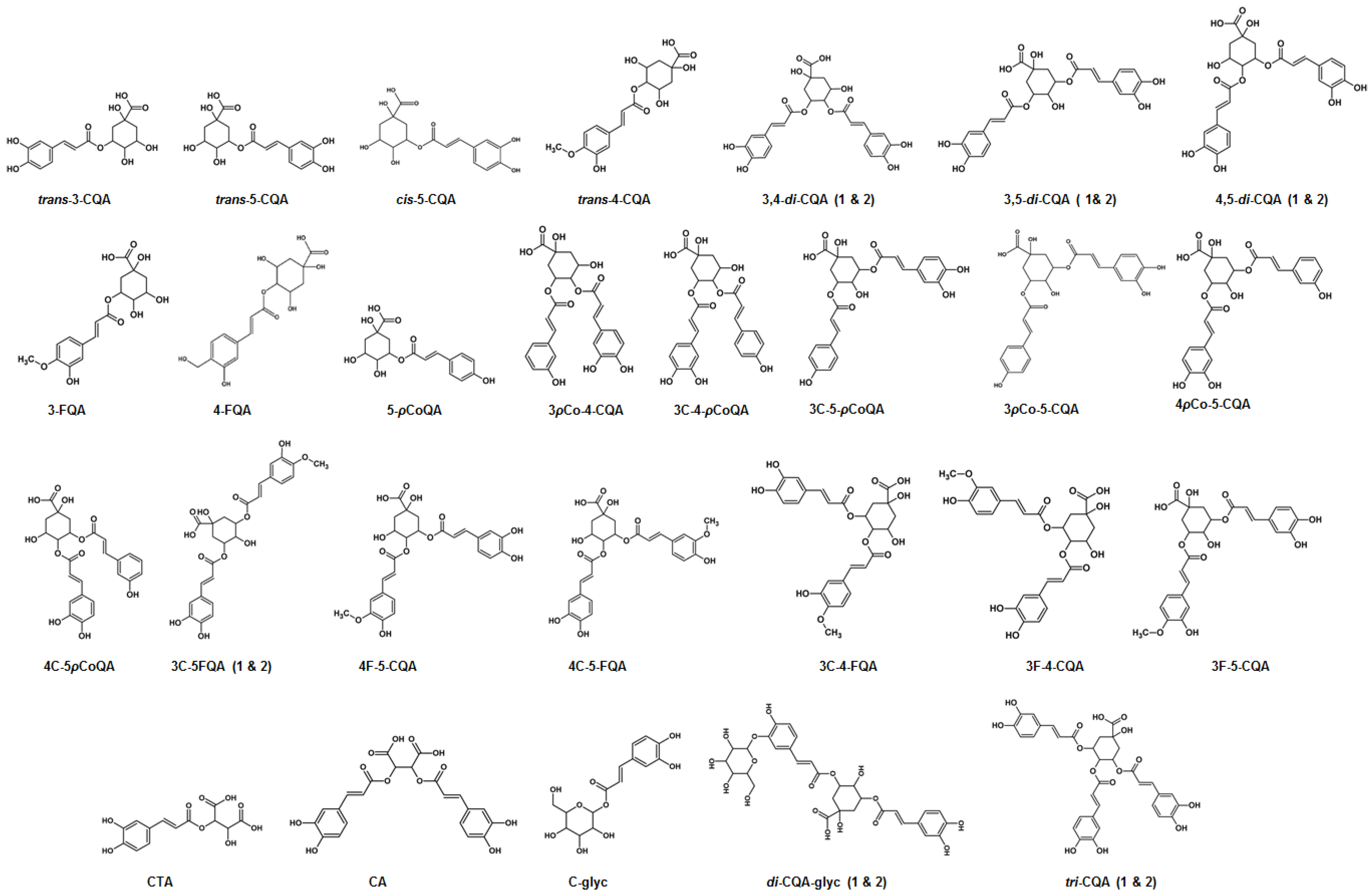
2.3. Metabolite Annotations
3. Discussion
3.1. Characterization of Mono-Acyl Chlorogenic Acids (CGAs)
3.2. Characterization of Caffeoylglycoside
3.3. Characterization of Hydroxycinnamoyl-Tartaric Acid Esters
3.4. Characterization of Di-Caffeoylquinic Acids (Di-CQAs)
3.5. Characterization of Tri-Caffeoylquinic Acids (Tri-CQAs) and Di-Caffeoylquinic Acid Glycosides
3.6. Characterization of p-Coumaroyl-Caffeoylquinic Acids (pCo-CQAs)
3.7. Characterization of Feruloyl-Caffeoylquinic Acids (F-CQAs)
3.8. Distribution of HCA Derivatives in Tissues and Cell Cultures of B. pilosa
4. Materials and Methods
4.1. Plant Cultivation Tissues and Undifferentiated Cells
4.2. Callus Initiation and Cell Suspension Cultures
4.3. Metabolite Extraction
4.4. Ultra High-Performance Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry (UHPLC-QTOF-MS/MS)
4.5. Data Processing, Multivariate Data Analysis (MVDA), and Metabolite Annotations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data Availability
References
- Arthur, G.D. Bidens pilosa L.: Agricultural and pharmaceutical importance. J. Med. Plants Res. 2012, 6, 3282–3287. [Google Scholar] [CrossRef]
- Bartolome, A.P.; Villaseñor, I.M.; Yang, W.-C. Bidens pilosa L. (Asteraceae): Botanical Properties, Traditional Uses, Phytochemistry, and Pharmacology. Evidence-Based Complement. Altern. Med. 2013, 2013, 1–51. [Google Scholar] [CrossRef] [PubMed]
- Xuan, T.D.; Khanh, T.D. Chemistry and pharmacology of Bidens pilosa: An overview. J. Pharm. Investig. 2016, 46, 91–132. [Google Scholar] [CrossRef] [PubMed]
- Bilanda, D.C.; Djomeni, P.D.D.; Kouakep, L.; Aboubakar, B.F.O.; Tedong, L.; Kamtchouing, P.; Dimo, T. Bidens pilosa Ethylene acetate extract can protect against L-NAME-induced hypertension on rats. BMC Complement. Altern. Med. 2017, 17, 479. [Google Scholar] [CrossRef] [PubMed]
- Rojas, D.F.C.; Chagas-Paula, D.; Da Costa, F.B.; Souza, C.R.F.; Oliveira, W. Bioactive compounds in Bidens pilosa L. populations: A key step in the standardization of phytopharmaceutical preparations. Rev. Bras. Farm. 2013, 23, 28–35. [Google Scholar] [CrossRef]
- Chien, S.C.; Young, P.H.; Hsu, Y.; Chen, C.H.; Tien, Y.J.; Shiu, S.Y.; Li, T.H.; Yang, C.W.; Marimuthu, P.; Tsai, L.F.; et al. Anti-diabetic properties of three common Bidens pilosa variants in Taiwan. Phytochemistry 2009, 70, 1246–1254. [Google Scholar] [CrossRef]
- Liang, Y.-C.; Yang, M.-T.; Lin, C.-J.; Chang, C.L.-T.; Yang, W.-C. Bidens pilosa and its active compound inhibit adipogenesis and lipid accumulation via down-modulation of the C/EBP and PPARγ pathways. Sci. Rep. 2016, 6, 24285. [Google Scholar] [CrossRef]
- De Wet, H.; Nzama, V.; Van Vuuren, S. Medicinal plants used for the treatment of sexually transmitted infections by lay people in northern Maputaland, KwaZulu–Natal Province, South Africa. South Afr. J. Bot. 2012, 78, 12–20. [Google Scholar] [CrossRef]
- Chang, S.-L.; Chiang, Y.-M.; Chang, C.L.-T.; Yeh, H.-H.; Shyur, L.-F.; Kuo, Y.-H.; Wu, T.-K.; Yang, W.-C. Flavonoids, centaurein and centaureidin, from Bidens pilosa, stimulate IFN-γ expression. J. Ethnopharmacol. 2007, 112, 232–236. [Google Scholar] [CrossRef]
- Owoyemi, O.O.; Oladunmoye, M.K. Phytochemical Screening and Antibacterial Activities of Bidens pilosa L. and Tridax procumbens L. on Skin Pathogens. Int. J. Mod. Biol. Med. 2017, 8, 24–46. [Google Scholar]
- Rao, S.R.; A Ravishankar, G. Plant cell cultures: Chemical factories of secondary metabolites. Biotechnol. Adv. 2002, 20, 101–153. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, R.; Rana, C.S. Plant secondary metabolites: A review. Int. J. Eng. Res. Gen. Sci. 2015, 3, 661–670. [Google Scholar]
- Ling, A.P.K.; Ong, S.L.; Sobri, H. Strategies in enhancing secondary metabolites production in plant cell cultures. Med. Aromat. Plant Sci. Biotechnol. 2011, 5, 94–101. [Google Scholar]
- Ochoa-Villarreal, M.; Howat, S.; Hong, S.; Jang, M.O.; Jin, Y.-W.; Lee, E.-K.; Loake, G.J. Plant cell culture strategies for the production of natural products. BMB Rep. 2016, 49, 149–158. [Google Scholar] [CrossRef]
- Chen, S.; Lin, J.; Liu, H.; Gong, Z.; Wang, X.; Li, M.; Aharoni, A.; Yang, Z.; Yu, X. Insights into Tissue-specific Specialized Metabolism in Tieguanyin Tea Cultivar by Untargeted Metabolomics. Molecules 2018, 23, 1817. [Google Scholar] [CrossRef]
- Hellwig, S.; Drossard, J.; Twyman, R.; Fischer, R. Plant cell cultures for the production of recombinant proteins. Nat. Biotechnol. 2004, 22, 1415–1422. [Google Scholar] [CrossRef]
- James, J.T.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Metabolomic Analysis of Methyl Jasmonate-Induced Triterpenoid Production in the Medicinal Herb Centella asiatica (L.) Urban. Molecules 2013, 18, 4267–4281. [Google Scholar] [CrossRef]
- Dorenenburg, H.; Knorr, D. Strategies to improve secondary metabolites in plant cultures. Enzyme Microb. Technol. 1995, 17, 674–684. [Google Scholar] [CrossRef]
- Laura, C.R.; Mauricio, R.L.; Aura, U.; Lucia, A.G. Effect of nutrients starvation in the cell cycle synchronization of cell suspension cultures of Jatropha curcas L. Curr. Plant Boil. 2018, 13, 23–31. [Google Scholar] [CrossRef]
- Clifford, M.N. Review Chlorogenic acids and other cinnamates–nature, occurrence and dietary burden. J. Sci. Food Agric. 1999, 79, 362–372. [Google Scholar] [CrossRef]
- Clifford, M.; Knight, S.; Kuhnert, N. Discriminating between the Six Isomers of Dicaffeoylquinic Acid by LC-MSn. J. Agric. Food Chem. 2005, 53, 3821–3832. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, R.; Sovdat, T.; Vivan, F.; Kuhnert, N. Profiling and characterization by LC-MSn of the chlorogenic acids and hydroxycinnamoylshikimate esters in maté (Ilex paraguariensis). J. Agric. Food Chem. 2010, 58, 5471–5484. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Renslow, R.S.; Makola, M.M.; Webb, I.K.; Deng, L.; Thomas, D.G.; Govind, N.; Ibrahim, Y.M.; Kabanda, M.M.; Dubery, I.A.; et al. Structural Elucidation of cis/trans Dicaffeoylquinic Acid Photoisomerization Using Ion Mobility Spectrometry-Mass Spectrometry. J. Phys. Chem. Lett. 2017, 8, 1381–1388. [Google Scholar] [CrossRef] [PubMed]
- Wianowska, D.; Gil, M. Recent advances in extraction and analysis procedures of natural chlorogenic acids. Phytochem. Rev. 2018, 18, 273–302. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Metabolomic fingerprinting of primed tobacco cells provide the first evidence for the biological origin of cis-chlorogenic acid. Biotechnol. Lett. 2014, 37, 205–209. [Google Scholar] [CrossRef]
- Lallemand, L.A.; Zubieta, C.; Lee, S.G.; Wang, Y.; Acajjaoui, S.; Timmins, J.; McSweeney, S.; Jez, J.M.; McCarthy, J.G.; McCarthy, A.A. A structural basis for the biosynthesis of the major chlorogenic acids found in coffee. Plant Physiol. 2012, 160, 249–260. [Google Scholar] [CrossRef]
- Taofiq, O.; González-Paramás, A.M.; Barreiro, M.F.; Ferreira, I.C.F.R. Hydroxycinnamic Acids and Their Derivatives: Cosmeceutical Significance, Challenges and Future Perspectives, a Review. Molecules 2017, 22, 281. [Google Scholar] [CrossRef]
- Kundu, A.; Vadassery, J. Chlorogenic acid-mediated chemical defence of plants against insect herbivores. Plant Boil. 2019, 21, 185–189. [Google Scholar] [CrossRef]
- Masike, K.; Dubery, I.; Steenkamp, P.; Smit, E.; Madala, N.E. Revising Reverse-Phase Chromatographic Behavior for Efficient Differentiation of Both Positional and Geometrical Isomers of Dicaffeoylquinic Acids. J. Anal. Methods Chem. 2018, 2018, 1–11. [Google Scholar] [CrossRef]
- Makola, M.M.; Steenkamp, P.; Dubery, I.A.; Kabanda, M.M.; Madala, N.E. Preferential alkali metal adduct formation by cis geometrical isomers of dicaffeoylquinic acids allows for efficient discrimination from their trans isomers during ultra-high-performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2016, 30, 1011–1018. [Google Scholar] [CrossRef]
- Clifford, M.; Johnston, K.L.; Knight, S.; Kuhnert, N. Hierarchical Scheme for LC-MSn Identification of Chlorogenic Acids. J. Agric. Food Chem. 2003, 51, 2900–2911. [Google Scholar] [CrossRef]
- Clifford, M.; Jaganath, I.B.; Ludwig, I.A.; Crozier, A. Chlorogenic acids and the acyl-quinic acids: Discovery, biosynthesis, bioavailability and bioactivity. Nat. Prod. Rep. 2017, 34, 1391–1421. [Google Scholar] [CrossRef] [PubMed]
- Asamenew, G.; Kim, H.-W.; Lee, M.-K.; Lee, S.-H.; Lee, S.; Cha, Y.-S.; Lee, S.H.; Yoo, S.M.; Kim, J.B. Comprehensive characterization of hydroxycinnamoyl derivatives in green and roasted coffee beans: A new group of methyl hydroxycinnamoyl quinate. Food Chem. X 2019, 2, 100033. [Google Scholar] [CrossRef] [PubMed]
- Madala, N.E.; Tugizimana, F.; Steenkamp, P.A. Development and Optimization of an UPLC-QTOF-MS/MS Method Based on an In-Source Collision Induced Dissociation Approach for Comprehensive Discrimination of Chlorogenic Acids Isomers from Momordica Plant Species. J. Anal. Methods Chem. 2014, 2014, 1–7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ncube, E.; I Mhlongo, M.; Piater, L.; Steenkamp, P.; Dubery, I.A.; Madala, N.E. Analyses of chlorogenic acids and related cinnamic acid derivatives from Nicotiana tabacum tissues with the aid of UPLC-QTOF-MS/MS based on the in-source collision-induced dissociation method. Chem. Central J. 2014, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Abrankó, L. Profiling of hydroxycinnamoylquinic acids in plant extracts using in-source CID fragmentation. J. Mass Spectrom. 2016, 51, 1130–1145. [Google Scholar] [CrossRef]
- Viacava, G.E.; Roura, S.I.; Berrueta, L.A.; Iriondo, C.; Gallo, B.; Alonso-Salces, R.M. Characterization of phenolic compounds in green and red oak-leaf lettuce cultivars by UHPLC-DAD-ESI-QTOF/MS using MSE scan mode. J. Mass Spectrom. 2017, 52, 873–902. [Google Scholar] [CrossRef]
- Chen, J.; Shiyanov, P.; Schlager, J.J.; Green, K.B. A Pseudo MS3 Approach for Identification of Disulfide-Bonded Proteins: Uncommon Product Ions and Database Search. J. Am. Soc. Mass Spectrom. 2011, 23, 225–243. [Google Scholar] [CrossRef]
- Achakzai, A.K.K.; Achakzai, P.; Masood, A.; Kayani, S.A.; Tareen, R.B. Response of plant parts and age on the distribution of secondary metabolites on plants found in Quetta. Pakistan J. Bot. 2009, 41, 2129–2135. [Google Scholar]
- Wiklund, S.; Johansson, E.; Sjöström, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-Based Metabolomics Data for Identification of Biochemically Interesting Compounds Using OPLS Class Models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.; Piater, L.; Dubery, I.A. A Conversation on Data Mining Strategies in LC-MS Untargeted Metabolomics: Pre-Processing and Pre-Treatment Steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Godzien, J.; Ciborowski, M.; Parreño, S.A.-D.; Barbas, C. From numbers to a biological sense: How the strategy chosen for metabolomics data treatment may affect final results. A practical example based on urine fingerprints obtained by LC-MS. Electrophoresis 2013, 34, 2812–2826. [Google Scholar] [CrossRef]
- Rodriguez, M.Z.; Comin, C.H.; Casanova, D.; Bruno, O.M.; Amancio, D.R.; Costa, L.D.F.; Rodrigues, F.A. Clustering algorithms: A comparative approach. PLOS ONE 2019, 14, e0210236. [Google Scholar] [CrossRef]
- Granato, D.; Santos, J.S.; Escher, G.B.; Ferreira, B.L.; Maggio, R.M. Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical perspective. Trends Food Sci. Technol. 2018, 72, 83–90. [Google Scholar] [CrossRef]
- Xie, C.; Yu, K.; Zhong, D.; Yuan, T.; Ye, F.; Jarrell, J.A.; Millar, A.; Chen, X. Investigation of Isomeric Transformations of Chlorogenic Acid in Buffers and Biological Matrixes by Ultra-performance Liquid Chromatography Coupled with Hybrid Quadrupole/Ion Mobility/Orthogonal Acceleration Time-of-Flight Mass Spectrometry. J. Agric. Food Chem. 2011, 59, 11078–11087. [Google Scholar] [CrossRef] [PubMed]
- Clifford, M.; Kirkpatrick, J.; Kuhnert, N.; Roozendaal, H.; Salgado, P.R. LC–MSn analysis of the cis isomers of chlorogenic acids. Food Chem. 2008, 106, 379–385. [Google Scholar] [CrossRef]
- Masike, K.; Mhlongo, M.I.; Mudau, S.P.; Nobela, O.; Ncube, E.; Tugizimana, F.; George, M.J.; Madala, N.E. Highlighting mass spectrometric fragmentation differences and similarities between hydroxycinnamoyl-quinic acids and hydroxycinnamoyl-isocitric acids. Chem. Central J. 2017, 11, 29. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Zheng, B.; Guan, Y.; Wang, L.; Chen, L.; Cai, W. Rapid characterization of chlorogenic acids in Duhaldea nervosa based on ultra-high-performance liquid chromatography-linear trap quadropole-Orbitrap-mass spectrometry and mass spectral trees similarity filter technique. J. Sep. Sci. 2018, 41, 1764–1774. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; A Barrett, D.; Beale, M.H.; Beger, R.D.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Linić, I.; Šamec, D.; Gruz, J.; Bok, V.V.; Strnad, M.; Sondi, B.S. Involvement of Phenolic Acids in Short-Term Adaptation to Salinity Stress is Species-Specific among Brassicaceae. Plants 2019, 8, 155. [Google Scholar] [CrossRef]
- Healy, E.F.; Sanders, J.; King, P.J.; Robinson, W.E. A docking study of l-chicoric acid with HIV-1 integrase. J. Mol. Graph. Model. 2009, 27, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Kuban-Jankowska, A.; Sahu, K.K.; Gorska, M.; Tuszynski, J.A.; Woźniak, M. Chicoric acid binds to two sites and decreases the activity of the YopH bacterial virulence factor. Oncotarget 2016, 7, 2229–2238. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Scagel, C. Chicoric acid levels in commercial basil (Ocimum basilicum) and Echinacea purpurea products. J. Funct. Foods 2010, 2, 77–84. [Google Scholar] [CrossRef]
- Khoza, B.; Gbashi, S.; Steenkamp, P.; Njobeh, P.B.; Madala, N. Identification of hydroxylcinnamoyl tartaric acid esters in Bidens pilosa by UPLC-tandem mass spectrometry. South Afr. J. Bot. 2016, 103, 95–100. [Google Scholar] [CrossRef]
- Jaiswal, R.; Müller, H.; Müller, A.; Gamaleldin, M.; Karar, E.; Kuhnert, N. Identification and characterization of chlorogenic acids, chlorogenic acid glycosides and flavonoids from Lonicera henryi L. (Caprifoliaceae) leaves by LC – MSn. Phytochemistry 2014, 108, 252–263. [Google Scholar] [CrossRef]
- Clifford, M.; Wu, W.; Kirkpatrick, J.; Kuhnert, N. Profiling the Chlorogenic Acids and Other Caffeic Acid Derivatives of Herbal Chrysanthemum by LC−MSn. J. Agric. Food Chem. 2007, 55, 929–936. [Google Scholar] [CrossRef]
- Clifford, M.N.; Marks, S.; Knight, S.; Kuhnert, N. Characterization by LC-MSn of four new classes of p-coumaric acid-containing diacyl chlorogenic acids in green coffee beans. J. Agric. Food Chem. 2006, 54, 4095–4101. [Google Scholar] [CrossRef]
- Mondolot, L.; La Fisca, P.; Buatois, B.; Talansier, E.; De Kochko, A.; Campa, C. Evolution in Caffeoylquinic Acid Content and Histolocalization During Coffea canephora Leaf Development. Ann. Bot. 2006, 98, 33–40. [Google Scholar] [CrossRef]
- Chen, Z.; Tang, N.; You, Y.; Lan, J.; Liu, Y.; Li, Z. Transcriptome Analysis Reveals the Mechanism Underlying the Production of a High Quantity of Chlorogenic Acid in Young Leaves of Lonicera macranthoides Hand.-Mazz. PLOS ONE 2015, 10, e0137212. [Google Scholar] [CrossRef]
- Lin, S.-D.; Sung, J.-M.; Chen, C.-L. Effect of drying and storage conditions on caffeic acid derivatives and total phenolics of Echinacea Ppurpurea grown in Taiwan. Food Chem. 2011, 125, 226–231. [Google Scholar] [CrossRef]
- Zhang, X.; Ding, X.; Ji, Y.; Wang, S.; Chen, Y.; Luo, J.; Shen, Y.; Peng, L. Measurement of metabolite variations and analysis of related gene expression in Chinese liquorice (Glycyrrhiza uralensis) plants under UV-B irradiation. Sci. Rep. 2018, 8, 6144. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M. Perennial peanut (Arachis glabrata Benth.) leaves contain hydroxycinnamoyl-CoA:tartaric acid hydroxycinnamoyl transferase activity and accumulate hydroxycinnamoyl-tartaric acid esters. Planta 2014, 239, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Zhang, W.; Yu, X. A combination of elicitation and precursor feeding leads to increased anthocyanin synthesis in cell suspension cultures of Vitis vinifera. Plant Cell Tissue Organ Cult. (PCTOC) 2011, 107, 261–269. [Google Scholar] [CrossRef]
- Nomura, T.; Ogita, S.; Kato, Y. Rational metabolic-flow switching for the production of exogenous secondary metabolites in bamboo suspension cells. Sci. Rep. 2018, 8, 13203. [Google Scholar] [CrossRef] [PubMed]
- Abouzid, S.F. Yield improvement strategies for the production of secondary metabolites in plant tissue culture: Silymarin from Silybum marianumtissue culture. Nat. Prod. Res. 2014, 28, 2102–2110. [Google Scholar] [CrossRef]
- Hussain, S.; Fareed, S.; Ansari, S.; Rahman, A.; Ahmad, I.Z.; Saeed, M. Current approaches toward production of secondary plant metabolites. J. Pharm. Bioallied Sci. 2012, 4, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qian, J.; Yao, L.; Lu, Y. Enhanced production of flavonoids by methyl jasmonate elicitation in cell suspension culture of Hypericum perforatum. Bioresour. Bioprocess. 2015, 2, 975. [Google Scholar] [CrossRef]
- Murthy, H.N.; Lee, E.-J.; Paek, K.Y. Production of secondary metabolites from cell and organ cultures: Strategies and approaches for biomass improvement and metabolite accumulation. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 118, 1–16. [Google Scholar] [CrossRef]
- Jamil, S.Z.M.R.; Rohani, E.R.; Baharum, S.N.; Noor, N.M. Metabolite profiles of callus and cell suspension cultures of mangosteen. 3 Biotech 2018, 8, 322. [Google Scholar] [CrossRef]
- Efferth, T. Biotechnology Applications of Plant Callus Cultures. Engineering 2019, 5, 50–59. [Google Scholar] [CrossRef]
- Sello, S.; Moscatiello, R.; La Rocca, N.; Baldan, B.; Navazio, L. A Rapid and Efficient Method to Obtain Photosynthetic Cell Suspension Cultures of Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Schütz, W.; Rave, G. The effect of cold stratification and light on the seed germination of temperate sedges (Carex) from various habitats and implications for regenerative strategies. Plant Ecol. 1999, 144, 215–230. [Google Scholar] [CrossRef]
- Maier, T.S.; Kuhn, J.; Müller, C. Proposal for field sampling of plants and processing in the lab for environmental metabolic fingerprinting. Plant Methods 2010, 6, 6. [Google Scholar] [CrossRef]
- Santos, R.B.; Abranches, R.; Fischer, R.; Sack, M.; Holland, T. Putting the Spotlight Back on Plant Suspension Cultures. Front. Plant Sci. 2016, 7, 121. [Google Scholar] [CrossRef]
- Gong, Z.-G.; Hu, J.; Wu, X.; Xu, Y.-J. The Recent Developments in Sample Preparation for Mass Spectrometry-Based Metabolomics. Crit. Rev. Anal. Chem. 2017, 47, 325–331. [Google Scholar] [CrossRef]
- Bartel, J.; Krumsiek, J.; Theis, F.J. Statistical methods for the analysis of high-throughput metabolomics data. Comput. Struct. Biotechnol. J. 2013, 4, e201301009. [Google Scholar] [CrossRef] [PubMed]
- Tugizimana, F.; Piater, L.; Dubery, I.A. Plant metabolomics: A new frontier in phytochemical analysis. South Afr. J. Sci. 2013, 109, 1–11. [Google Scholar] [CrossRef]
- Clifford, M.; Madala, N.E. Surrogate Standards: A Cost-Effective Strategy for Identification of Phytochemicals. J. Agric. Food Chem. 2017, 65, 3589–3590. [Google Scholar] [CrossRef] [PubMed]





| No | m/z | Mass Error (mDa) | Rt (min) | Fragment ions | Molecular Formula | Metabolite | Abbreviation | L | S | R | C-l | C-s | S-l | S-s |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 311.0392 | 8.8 | 4.51 | 179, 149, 135 | C13H12O9 | Caftaric acid | CTA | • | • | |||||
| 2 | 337.0822 | 5.4 | 9.45 | 191 | C16H18O8 | 5-Coumaroylquinic acid | 5-ρCoQA | • | • | • | • | • | ||
| 3 | 341.0829 | 2.4 | 7.33 | 179, 135 | C15H18O9 | Caffeoylglycoside | CFA-glyc | • | • | • | ||||
| 4 | 353.084 | 4.9 | 3.31 | 191, 179, 135 | C16H18O9 | trans-3-Caffeoylquinic acid | trans-3-CQA | • | • | • | • | • | • | |
| 5 | 353.0821 | 5.4 | 6.39 | 191 | C16H18O9 | trans-5-Caffeoylquinic acid | trans-5-CQA | • | • | |||||
| 6 | 353.0835 | 5.7 | 7.12 | 191, 179, 173, 135 | C16H18O9 | trans-4-Caffeoylquinic acid | trans-4-CQA | • | • | • | • | • | • | • |
| 7 | 353.0884 | 2.6 | 9.16 | 191 | C16H18O9 | cis-5-Caffeoylquinic acid | cis-5-CQA | • | • | • | • | • | • | |
| 8 | 367.1003 | 5.5 | 6.52 | 193 | C17H20O9 | 3-Feruloylqunic acid | 3-FQA | • | • | • | • | • | • | • |
| 9 | 367.0986 | 3.7 | 10.91 | 191, 173 | C17H20O9 | 4-Feruloylquinic acid | 4-FQA | • | • | • | • | |||
| 10 | 473.0673 | 2.7 | 14.89 | 311, 179, 149, 135 | C22H18O9 | Chicoric acid | CA | • | ||||||
| 11 | 499.1211 | 1.3 | 15.68 | 353, 337, 335, 191, 173, 163 | C25H24O11 | 3-Coumaroyl-4-caffeoylquinic acid | 3ρCo-4-CQA | • | • | • | ||||
| 12 | 499.1183 | 4.5 | 15.84 | 337, 335, 173, 164 | C25H24O11 | 3-Caffeoyl-4-coumaroylquinic acid | 3C-4-ρCoQA | • | • | • | ||||
| 13 | 499.1217 | 0.6 | 15.99 | 337, 163 | C25H24O11 | 3-Coumaroyl-5-caffeoylquinic acid | 3ρCo-5-CQA | • | • | |||||
| 14 | 499.12312 | 1.0 | 16.09 | 353, 337,191, 179 | C25H24O11 | 3-Caffeoyl-5-coumaroylquinic acid | 3C-5-ρCoQA | • | • | • | ||||
| 15 | 499.1352 | 4.2 | 16.65 | 337, 173, 163 | C25H24O11 | 4-Coumaroyl-5-caffeoylquinic acid | 4ρCo-5-CQA | • | • | • | ||||
| 16 | 499.1227 | 1.2 | 16.75 | 353, 337,191, 179, 173 | C25H24O11 | 4-Caffeoyl-5-coumaroylquinic acid | 4C-5-ρCoQA | • | • | • | ||||
| 17 | 515.1182 | 1.0 | 14.65 | 353, 335, 191, 179, 135 | C25H24O12 | 3,4-di-Caffeoylquinic acid | 3,4-di-CQA-1 | • | • | • | • | • | • | • |
| 18 | 515.1163 | 1.7 | 14.69 | 353, 335, 191, 179, 173, 135 | C25H24O12 | 3,4-di-Caffeoylquinic acid | 3,4-di-CQA-2 | • | ||||||
| 19 | 515.1210 | 6.2 | 14.93 | 353, 191, 179, 135 | C25H24O12 | 3,5-di-Caffeoylquinic acid | 3,5-di-CQA-1 | • | • | • | • | • | • | • |
| 20 | 515.1170 | 2.1 | 15.03 | 353, 191, 179, 135 | C25H24O12 | 3,5-di-Caffeoylqiunic acid | 3,5-di-CQA-2 | • | ||||||
| 21 | 515.1292 | 2.2 | 15.67 | 353, 335, 191, 179, 173, 135 | C25H24O12 | 4,5-di-Caffeoylquinic acid | 4,5-di-CQA-1 | • | • | • | • | • | • | • |
| 22 | 515.1122 | 2.0 | 16.89 | 353, 191, 179, 173 | C25H24O12 | 4,5-di-Caffeoylquinic acid | 4,5-di-CQA-2 | • | ||||||
| 23 | 529.1315 | 1.2 | 15.92 | 367, 353, 335, 193, 179, 173, 134 | C26H26O12 | 3-Feruloyl-4-caffeoylquinic acid | 3F-4-CQA | • | • | • | • | • | • | |
| 24 | 529.1381 | 1.9 | 16.11 | 367, 335, 193, 173 | C26H26O12 | 3-Caffeoyl-4-feruloylquinic acid | 3C-4-FQA | • | • | • | • | • | • | |
| 25 | 529.1296 | 3.5 | 16.37 | 367, 193, 134 | C26H26O12 | 3-Feruloyl-5-cafffeoylquinic acid | 3F-5-CQA | • | • | • | • | • | • | |
| 26 | 529.1422 | 0.8 | 16.49 | 367, 353, 191, 179 | C26H26O12 | 3-Caffeoyl-5-feruloylquinic acid | 3C-5-FQA-1 | • | • | • | • | • | • | |
| 27 | 529.1463 | 1.2 | 16.86 | 367, 193, 173 | C26H26O12 | 4-Feruloyl-5-caffeoylquinic acid | 4F-5-CQA | • | • | • | • | • | • | |
| 28 | 529.142 | 4.6 | 16.97 | 367, 353, 191, 179, 173, 135 | C26H26O12 | 4-Caffeoyl-5-feruloylquinic acid | 4C-5-FQA | • | • | • | • | • | • | |
| 29 | 529.1395 | 7.5 | 17.09 | 353, 191, 179 | C26H26O12 | 3-Caffeoyl-5-feruloylquinic acid | 3C-5FQA-2 | • | • | |||||
| 30 | 677.14 | 2.3 | 12.99 | 515, 353, 341, 353, 179, 173 | C31H33O17 | di-Caffeoylquinic acid glycoside | di-CQA-glc-1 | • | • | |||||
| 31 | 677.16 | 0.8 | 14.07 | 515, 353, 341 | C31H33O17 | di-Caffeoylquinic acid glycoside | di-CQA-glc-2 | • | • | |||||
| 32 | 677.1436 | 0.3 | 17.76 | 515, 353, 335, 191, 179, 173 | C34H30O15 | tri-Caffeoylquinic acid | tri-CQA-1 | • | • | |||||
| 33 | 677.15 | 3.3 | 18.21 | 515, 353,179, 173 | C34H30O15 | tri-Caffeoylquinic acid | tri-CQA-2 | • | • |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramabulana, A.-T.; Steenkamp, P.; Madala, N.; Dubery, I.A. Profiling of Chlorogenic Acids from Bidens pilosa and Differentiation of Closely Related Positional Isomers with the Aid of UHPLC-QTOF-MS/MS-Based In-Source Collision-Induced Dissociation. Metabolites 2020, 10, 178. https://doi.org/10.3390/metabo10050178
Ramabulana A-T, Steenkamp P, Madala N, Dubery IA. Profiling of Chlorogenic Acids from Bidens pilosa and Differentiation of Closely Related Positional Isomers with the Aid of UHPLC-QTOF-MS/MS-Based In-Source Collision-Induced Dissociation. Metabolites. 2020; 10(5):178. https://doi.org/10.3390/metabo10050178
Chicago/Turabian StyleRamabulana, Anza-Tshilidzi, Paul Steenkamp, Ntakadzeni Madala, and Ian A. Dubery. 2020. "Profiling of Chlorogenic Acids from Bidens pilosa and Differentiation of Closely Related Positional Isomers with the Aid of UHPLC-QTOF-MS/MS-Based In-Source Collision-Induced Dissociation" Metabolites 10, no. 5: 178. https://doi.org/10.3390/metabo10050178
APA StyleRamabulana, A.-T., Steenkamp, P., Madala, N., & Dubery, I. A. (2020). Profiling of Chlorogenic Acids from Bidens pilosa and Differentiation of Closely Related Positional Isomers with the Aid of UHPLC-QTOF-MS/MS-Based In-Source Collision-Induced Dissociation. Metabolites, 10(5), 178. https://doi.org/10.3390/metabo10050178