Network Analysis Provides Insight into Tomato Lipid Metabolism
Abstract
1. Introduction
2. Results
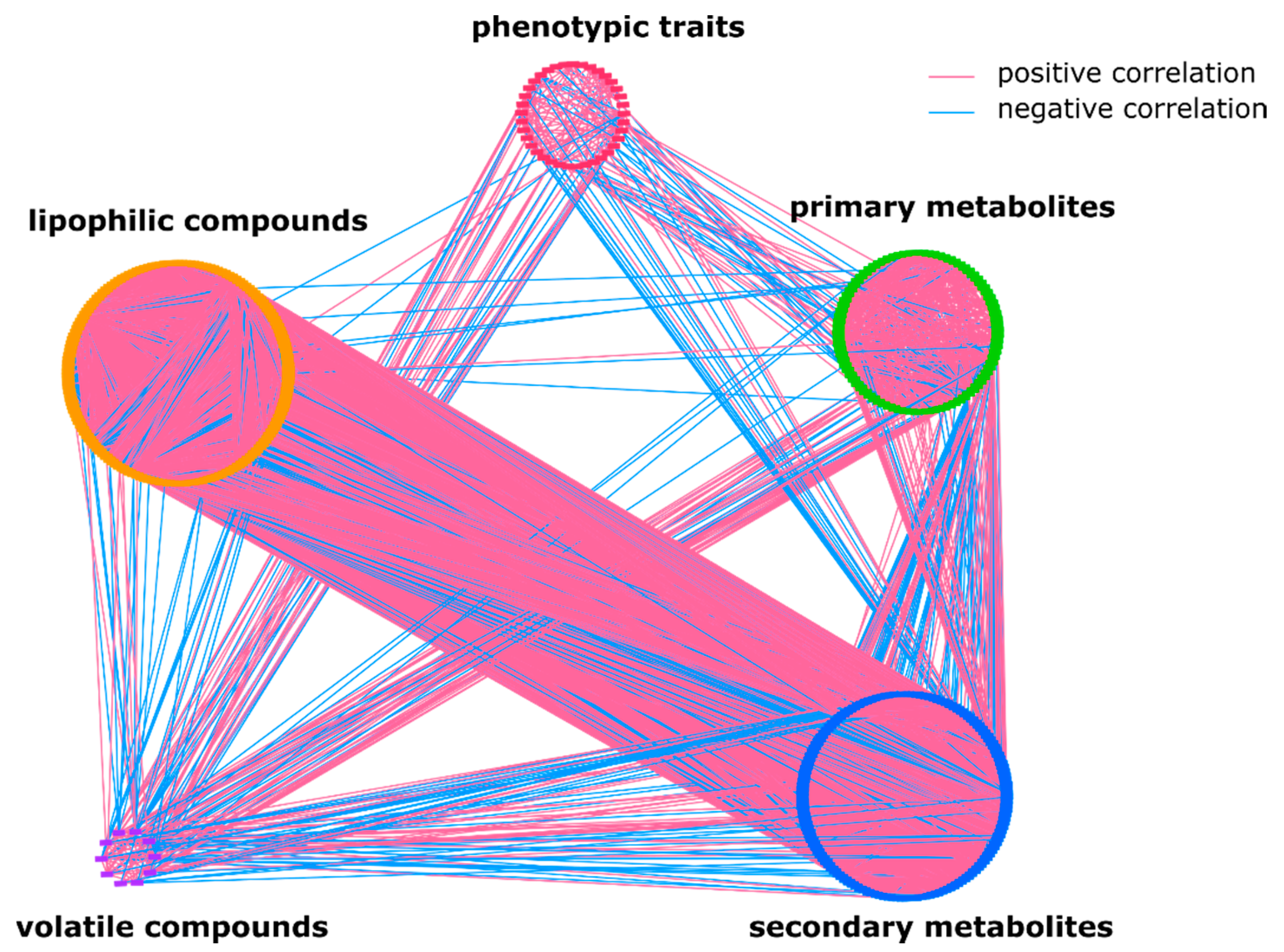
2.1. The Overall Metabolic Network in S. pennellii ILs
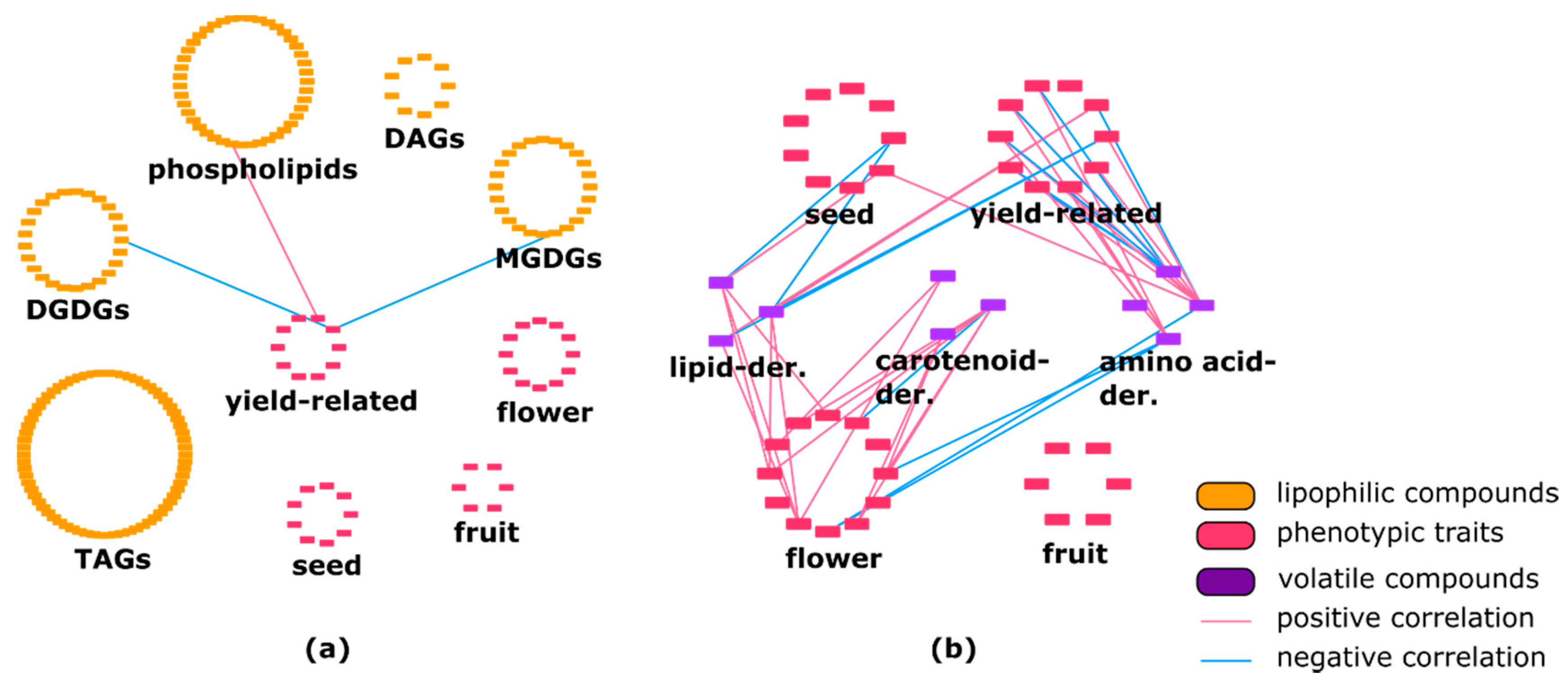
2.2. Lipophilic Compounds Expose Weak Correlation with Phenotypic Traits and Primary Metabolites
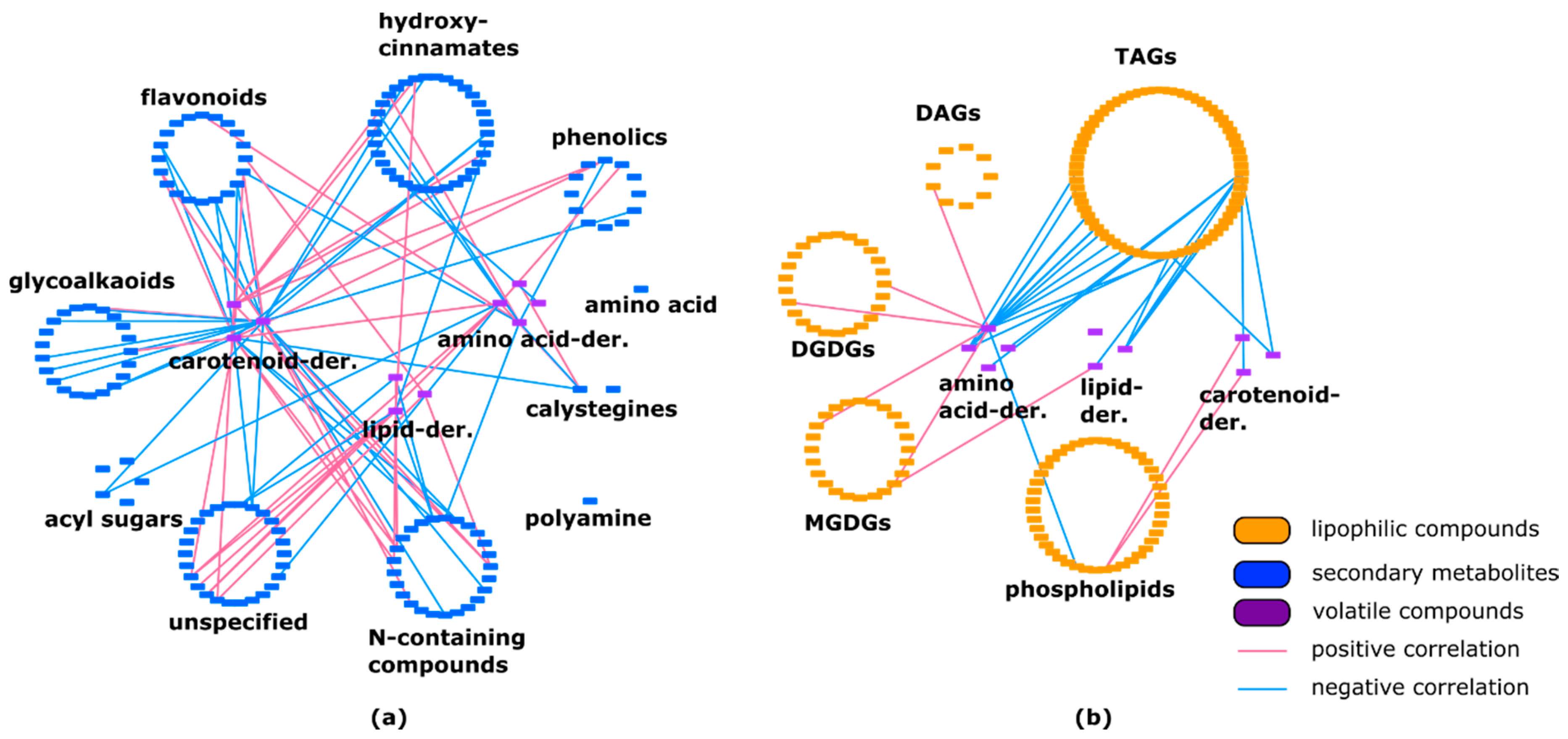
2.3. Lipids Are Highly Correlated with Specialized Metabolites and Show Some Negative Correlations with Volatiles
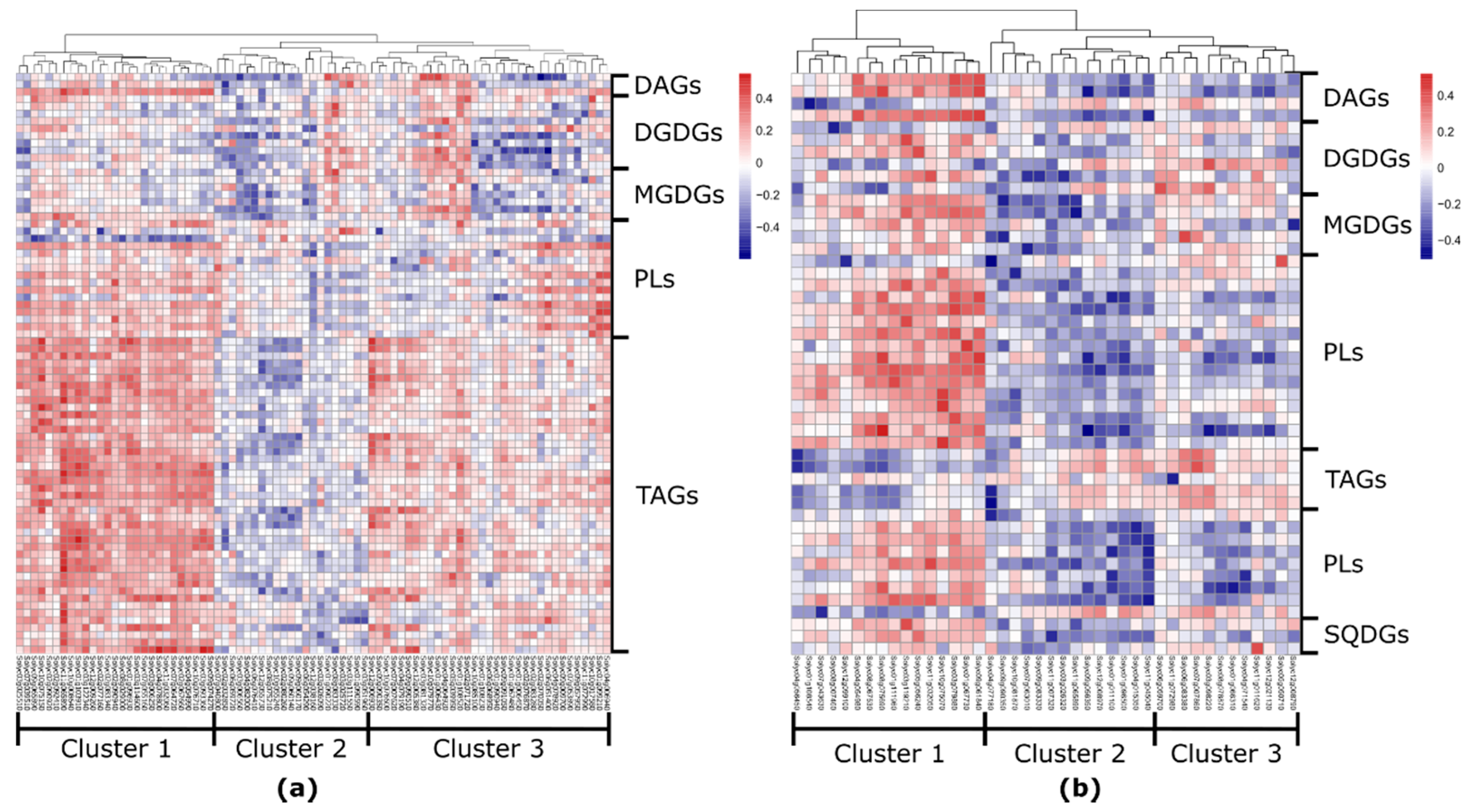
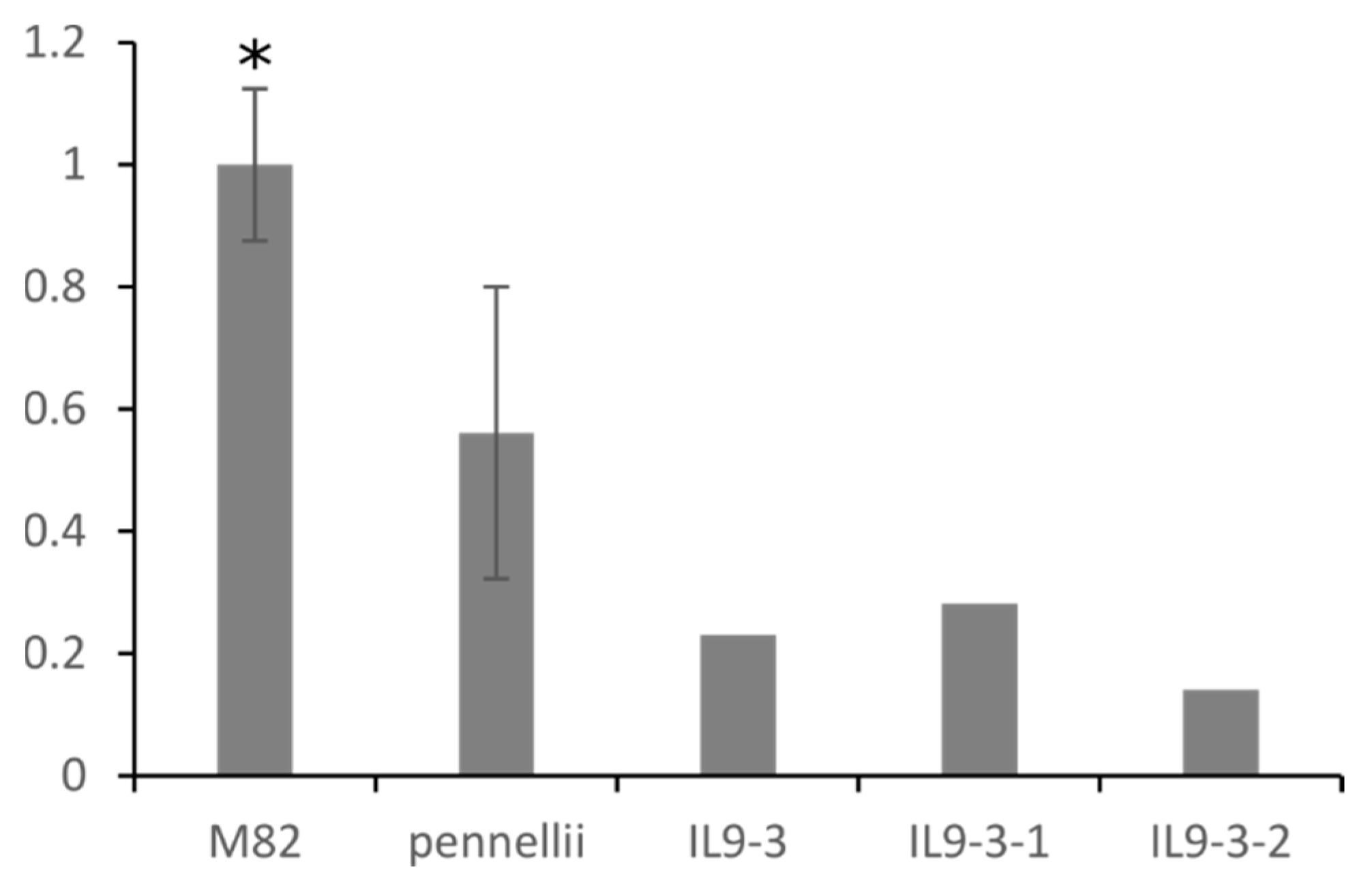
2.4. Fruit-Specific Lipid-Related Genes Show a Mainly Positive Pattern of Change with Lipophilic Compounds
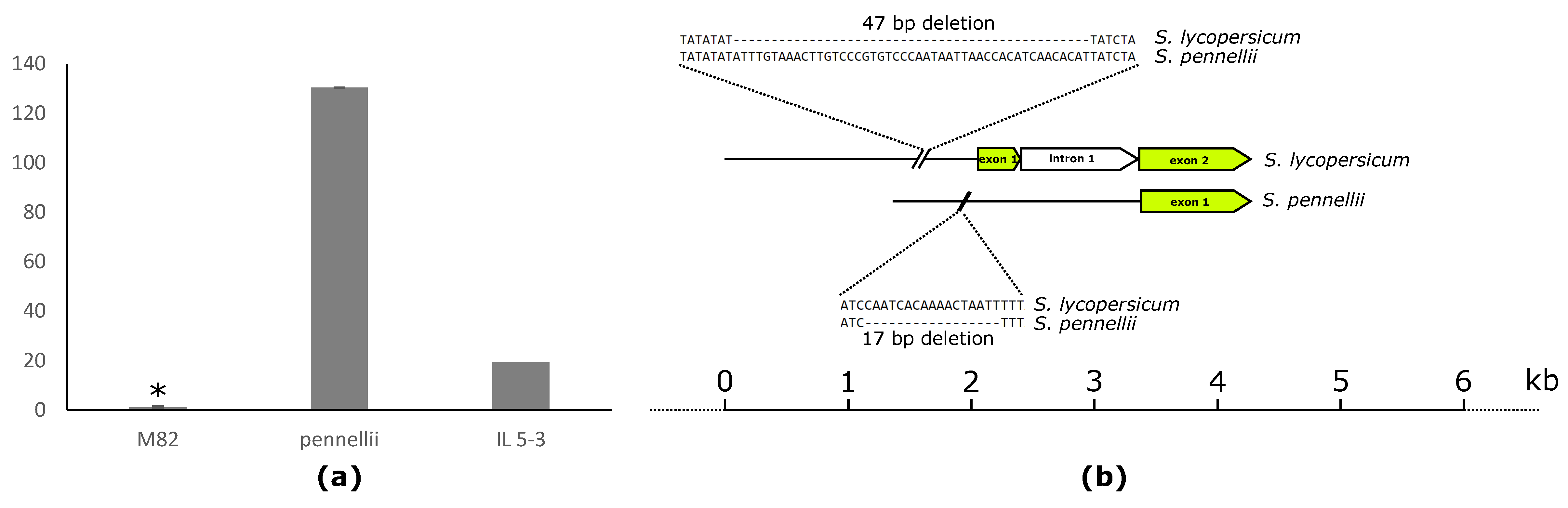
2.5. Leaf-Specific Lipid-Related Genes Show Many Negative Correlations with Lipophilic Compounds
3. Discussion
3.1. Network Analysis: Correlation between Metabolic and Phenotypic Traits
3.2. Network Analysis: Combining Metabolite Profiling and Expression for Gene Discovery
4. Materials and Methods
4.1. Plant Material
4.2. Correlation Analysis
4.3. Promoter Analysis
4.4. Trait Classes Used for Correlations
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pott, D.M.; Osorio, S.; Vallarino, J.G. From Central to Specialized Metabolism: An Overview of Some Secondary Compounds Derived from the Primary Metabolism for Their Role in Conferring Nutritional and Organoleptic Characteristics to Fruit. Front. Plant Sci. 2019, 10, 835. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Strack, D. Phytochemistry meets genome analysis, and beyond. Phytochemistry 2003, 62, 815–816. [Google Scholar] [CrossRef]
- Rai, A.; Saito, K.; Yamazaki, M. Integrated omics analysis of specialized metabolism in medicinal plants. Plant J. 2017, 90, 764–787. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Fernie, A.R.; Luo, J. Exploring the Diversity of Plant Metabolism. Trends Plant Sci. 2019, 24, 83–98. [Google Scholar] [CrossRef]
- Fernie, A.R.; Trethewey, R.N.; Krotzky, A.J.; Willmitzer, L. Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 763–769. [Google Scholar] [CrossRef]
- Maeda, H.A. Evolutionary Diversification of Primary Metabolism and Its Contribution to Plant Chemical Diversity. Front. Plant Sci. 2019, 10, 881. [Google Scholar] [CrossRef]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef]
- Kessler, A.; Kalske, A. Plant Secondary Metabolite Diversity and Species Interactions. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 115–138. [Google Scholar] [CrossRef]
- Martin, C. The interface between plant metabolic engineering and human health. Curr. Opin. Biotechnol. 2013, 24, 344–353. [Google Scholar] [CrossRef]
- Tieman, D.M.; Zeigler, M.; Schmelz, E.A.; Taylor, M.G.; Bliss, P.; Kirst, M.; Klee, H.J. Identification of loci affecting flavour volatile emissions in tomato fruits. J. Exp. Bot. 2006, 57, 887–896. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Saito, K. Metabolomics for unknown plant metabolites. Anal. Bioanal. Chem. 2013, 405, 5005–5011. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, L.; Zhao, Y.; Zhao, C.; Chen, X.; Liu, P.; Zhou, H.; Zhang, J.; Hu, C.; Chen, A.; et al. Metabolic changes in primary, secondary, and lipid metabolism in tobacco leaf in response to topping. Anal. Bioanal. Chem. 2018, 410, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Ohlrogge, J.; Browse, J. Lipid biosynthesis. Plant Cell 1995, 7, 957–970. [Google Scholar] [PubMed]
- Aid, F. Plant Lipid Metabolism. Lipid Metabolism 2019. [Google Scholar] [CrossRef]
- Zhu, G.; Wang, S.; Huang, Z.; Zhang, S.; Liao, Q.; Zhang, C.; Lin, T.; Qin, M.; Peng, M.; Yang, C.; et al. Rewiring of the Fruit Metabolome in Tomato Breeding. Cell 2018, 172, 249–261. [Google Scholar] [CrossRef]
- Eshed, Y.; Zamir, D. An Introgression Line Population of Lycopersicon Pennellii in the Cultivated Tomato Enables the Identification and Fine Mapping of Yield-Associated Qtl. Genetics 1995, 141, 1147–1162. [Google Scholar]
- Bolger, A.; Scossa, F.; Bolger, M.E.; Lanz, C.; Maumus, F.; Tohge, T.; Quesneville, H.; Alseekh, S.; Sorensen, I.; Lichtenstein, G.; et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 2014, 46, 1034–1038. [Google Scholar] [CrossRef]
- Lippman, Z.B.; Semel, Y.; Zamir, D. An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr. Opin. Genet. Dev. 2007, 17, 545–552. [Google Scholar] [CrossRef]
- Kimura, S.; Sinha, N. Tomato (Solanum lycopersicum): A Model Fruit-Bearing Crop. CSH Protoc. 2008. [Google Scholar] [CrossRef]
- Raiola, A.; Rigano, M.M.; Calafiore, R.; Frusciante, L.; Barone, A. Enhancing the Health-Promoting Effects of Tomato Fruit for Biofortified Food. Mediators Inflamm. 2014, 2014. [Google Scholar] [CrossRef]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.-P.; Matros, A.; Peterek, S.; Schijlen, E.G.W.M.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Alseekh, S.; Tohge, T.; Wendenberg, R.; Scossa, F.; Omranian, N.; Li, J.; Kleessen, S.; Giavalisco, P.; Pleban, T.; Mueller-Roeber, B.; et al. Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 2015, 27, 485–512. [Google Scholar] [CrossRef] [PubMed]
- Mutschler, M.A.; Doerge, R.W.; Liu, S.C.; Kuai, J.P.; Liedl, B.E.; Shapiro, J.A. QTL analysis of pest resistance in the wild tomato Lycopersicon pennellii: QTLs controlling acylsugar level and composition. Theor. Appl. Genet. 1996, 92, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, S.; Cin, V.D.; Fei, Z.; Li, H.; Bliss, P.; Taylor, M.G.; Klee, H.J.; Tieman, D.M. Flavour compounds in tomato fruits: Identification of loci and potential pathways affecting volatile composition. J. Exp. Bot. 2009, 60, 325–337. [Google Scholar] [CrossRef]
- Tieman, D.; Zhu, G.; Resende, M.F.R.; Lin, T.; Nguyen, C.; Bies, D.; Rambla, J.L.; Beltran, K.S.O.; Taylor, M.; Zhang, B.; et al. A chemical genetic roadmap to improved tomato flavor. Science 2017, 355, 391–394. [Google Scholar] [CrossRef]
- Adato, A.; Mandel, T.; Mintz-Oron, S.; Venger, I.; Levy, D.; Yativ, M.; Domínguez, E.; Wang, Z.; Vos, R.C.H.D.; Jetter, R.; et al. Fruit-Surface Flavonoid Accumulation in Tomato Is Controlled by a SlMYB12-Regulated Transcriptional Network. PLoS Genet. 2009, 5, e1000777. [Google Scholar] [CrossRef]
- Wu, S.; Alseekh, S.; Cuadros-Inostroza, A.; Fusari, C.M.; Mutwil, M.; Kooke, R.; Keurentjes, J.B.; Fernie, A.R.; Willmitzer, L.; Brotman, Y. Combined Use of Genome-Wide Association Data and Correlation Networks Unravels Key Regulators of Primary Metabolism in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006363. [Google Scholar] [CrossRef]
- Kruger, N.J.; Ratcliffe, R.G. Pathways and fluxes: Exploring the plant metabolic network. J. Exp. Bot. 2012, 63, 2243–2246. [Google Scholar] [CrossRef]
- Toubiana, D.; Fernie, A.R.; Nikoloski, Z.; Fait, A. Network analysis: Tackling complex data to study plant metabolism. Trends Biotechnol. 2013, 31, 29–36. [Google Scholar] [CrossRef]
- Schauer, N.; Semel, Y.; Roessner, U.; Gur, A.; Balbo, I.; Carrari, F.; Pleban, T.; Perez-Melis, A.; Bruedigam, C.; Kopka, J.; et al. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 2006, 24, 447–454. [Google Scholar] [CrossRef]
- Toubiana, D.; Semel, Y.; Tohge, T.; Beleggia, R.; Cattivelli, L.; Rosental, L.; Nikoloski, Z.; Zamir, D.; Fernie, A.R.; Fait, A. Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations. PLoS Genet. 2012, 8, e1002612. [Google Scholar] [CrossRef] [PubMed]
- Sales-Campos, H.; de Souza, P.R.; Peghini, B.C.; da Silva, J.S.; Cardoso, C.R. An overview of the modulatory effects of oleic acid in health and disease. Mini Rev. Med. Chem. 2013, 13, 201–210. [Google Scholar] [PubMed]
- Farvid, M.S.; Ding, M.; Pan, A.; Sun, Q.; Chiuve, S.E.; Steffen, L.M.; Willett, W.C.; Hu, F.B. Dietary linoleic acid and risk of coronary heart disease: A systematic review and meta-analysis of prospective cohort studies. Circulation 2014, 130, 1568–1578. [Google Scholar] [CrossRef]
- Weselake, R.J.; Taylor, D.C.; Rahman, M.H.; Shah, S.; Laroche, A.; McVetty, P.B.E.; Harwood, J.L. Increasing the flow of carbon into seed oil. Biotechnol. Adv. 2009, 27, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Lepiniec, L. Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 2010, 49, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Aluyor, E.O.; Ori-Jesu, M. The use of antioxidants in vegetable oils–A review. Afr. J. Biotechnol. 2008, 7, 4836–4842. [Google Scholar]
- Botineștean, C.; Hădărugă, N.G.; Hădărugă, D.I.; Jianu, I. Fatty Acids Composition by Gas Chromatography – Mass Spectrometry (GC-MS) and most important physical- chemicals parameters of Tomato Seed Oil. J. Agroaliment. Process. Technol. 2012, 18, 89–94. [Google Scholar]
- Nunes-Nesi, A.; Alseekh, S.; de Oliveira Silva, F.M.; Omranian, N.; Lichtenstein, G.; Mirnezhad, M.; González, R.R.R.; Sabio y Garcia, J.; Conte, M.; Leiss, K.A.; et al. Identification and characterization of metabolite quantitative trait loci in tomato leaves and comparison with those reported for fruits and seeds. Metabolomics 2019, 15, 46. [Google Scholar] [CrossRef]
- Tohge, T.; Fernie, A.R. Metabolomics-Inspired Insight into Developmental, Environmental and Genetic Aspects of Tomato Fruit Chemical Composition and Quality. Plant Cell Physiol. 2015, 56, 1681–1696. [Google Scholar] [CrossRef]
- Vallarino, J.G.; Yeats, T.H.; Maximova, E.; Rose, J.K.; Fernie, A.R.; Osorio, S. Postharvest changes in LIN5-down-regulated plants suggest a role for sugar deficiency in cuticle metabolism during ripening. Phytochemistry 2017, 142, 11–20. [Google Scholar] [CrossRef]
- Mintz-Oron, S.; Mandel, T.; Rogachev, I.; Feldberg, L.; Lotan, O.; Yativ, M.; Wang, Z.; Jetter, R.; Venger, I.; Adato, A.; et al. Gene Expression and Metabolism in Tomato Fruit Surface Tissues. Plant Physiol. 2008, 147, 823–851. [Google Scholar] [CrossRef] [PubMed]
- España, L.; Heredia-Guerrero, J.A.; Reina-Pinto, J.J.; Fernández-Muñoz, R.; Heredia, A.; Domínguez, E. Transient Silencing of CHALCONE SYNTHASE during Fruit Ripening Modifies Tomato Epidermal Cells and Cuticle Properties1[C][W]. Plant Physiol. 2014, 166, 1371–1386. [Google Scholar] [CrossRef] [PubMed]
- Heredia, A.; Heredia-Guerrero, J.A.; Domínguez, E. CHS silencing suggests a negative cross-talk between wax and flavonoid pathways in tomato fruit cuticle. Plant Signal. Behav. 2015, 10, e1019979. [Google Scholar] [CrossRef] [PubMed]
- Garbowicz, K.; Liu, Z.; Alseekh, S.; Tieman, D.; Taylor, M.; Kuhalskaya, A.; Ofner, I.; Zamir, D.; Klee, H.J.; Fernie, A.R.; et al. Quantitative Trait Loci Analysis Identifies a Prominent Gene Involved in the Production of Fatty Acid-Derived Flavor Volatiles in Tomato. Mol. Plant 2018, 11, 1147–1165. [Google Scholar] [CrossRef]
- Schauer, N.; Semel, Y.; Balbo, I.; Steinfath, M.; Repsilber, D.; Selbig, J.; Pleban, T.; Zamir, D.; Fernie, A.R. Mode of Inheritance of Primary Metabolic Traits in Tomato. Plant Cell 2008, 20, 509–523. [Google Scholar] [CrossRef]
- Tomato Functional Genomics Database (TFGD). Available online: http://ted.bti.cornell.edu/ (accessed on 13 February 2020).
- Chitwood, D.H.; Kumar, R.; Headland, L.R.; Ranjan, A.; Covington, M.F.; Ichihashi, Y.; Fulop, D.; Jiménez-Gómez, J.M.; Peng, J.; Maloof, J.N.; et al. A Quantitative Genetic Basis for Leaf Morphology in a Set of Precisely Defined Tomato Introgression Lines. Plant Cell 2013, 25, 2465–2481. [Google Scholar] [CrossRef]
- Do, P.T.; Prudent, M.; Sulpice, R.; Causse, M.; Fernie, A.R. The Influence of Fruit Load on the Tomato Pericarp Metabolome in a Solanum chmielewskii Introgression Line Population. Plant Physiol. 2010, 154, 1128–1142. [Google Scholar] [CrossRef]
- Ballester, A.-R.; Tikunov, Y.; Molthoff, J.; Grandillo, S.; Viquez-Zamora, M.; de Vos, R.; de Maagd, R.A.; van Heusden, S.; Bovy, A.G. Identification of Loci Affecting Accumulation of Secondary Metabolites in Tomato Fruit of a Solanum lycopersicum × Solanum chmielewskii Introgression Line Population. Front. Plant Sci. 2016, 7, 1428. [Google Scholar] [CrossRef]
- Tieman, D.; Taylor, M.; Schauer, N.; Fernie, A.R.; Hanson, A.D.; Klee, H.J. Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. USA 2006, 103, 8287–8292. [Google Scholar] [CrossRef]
- Mageroy, M.H.; Tieman, D.M.; Floystad, A.; Taylor, M.G.; Klee, H.J. A Solanum lycopersicum catechol-O-methyltransferase involved in synthesis of the flavor molecule guaiacol. Plant J. 2012, 69, 1043–1051. [Google Scholar] [CrossRef]
- Rambla, J.L.; Tikunov, Y.M.; Monforte, A.J.; Bovy, A.G.; Granell, A. The expanded tomato fruit volatile landscape. J. Exp. Bot. 2014, 65, 4613–4623. [Google Scholar] [CrossRef] [PubMed]
- Schilmiller, A.; Shi, F.; Kim, J.; Charbonneau, A.L.; Holmes, D.; Daniel Jones, A.; Last, R.L. Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J. 2010, 62, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Schilmiller, A.L.; Charbonneau, A.L.; Last, R.L. Identification of a BAHD acetyltransferase that produces protective acyl sugars in tomato trichomes. Proc. Natl. Acad. Sci. USA 2012, 109, 16377–16382. [Google Scholar] [CrossRef] [PubMed]
- Sallaud, C.; Rontein, D.; Onillon, S.; Jabès, F.; Duffé, P.; Giacalone, C.; Thoraval, S.; Escoffier, C.; Herbette, G.; Leonhardt, N.; et al. A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 2009, 21, 301–317. [Google Scholar] [CrossRef]
- Gonzales-Vigil, E.; Hufnagel, D.E.; Kim, J.; Last, R.L.; Barry, C.S. Evolution of TPS20-related terpene synthases influences chemical diversity in the glandular trichomes of the wild tomato relative Solanum habrochaites. Plant J. 2012, 71, 921–935. [Google Scholar] [CrossRef]
- Matsuba, Y.; Nguyen, T.T.H.; Wiegert, K.; Falara, V.; Gonzales-Vigil, E.; Leong, B.; Schäfer, P.; Kudrna, D.; Wing, R.A.; Bolger, A.M.; et al. Evolution of a complex locus for terpene biosynthesis in solanum. Plant Cell 2013, 25, 2022–2036. [Google Scholar] [CrossRef]
- Tikunov, Y.M.; Molthoff, J.; de Vos, R.C.H.; Beekwilder, J.; van Houwelingen, A.; van der Hooft, J.J.J.; Nijenhuis-de Vries, M.; Labrie, C.W.; Verkerke, W.; van de Geest, H.; et al. Non-smoky glycosyltransferase1 prevents the release of smoky aroma from tomato fruit. Plant Cell 2013, 25, 3067–3078. [Google Scholar] [CrossRef]
- Yeats, T.H.; Buda, G.J.; Wang, Z.; Chehanovsky, N.; Moyle, L.C.; Jetter, R.; Schaffer, A.A.; Rose, J.K.C. The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. Plant J. 2012, 69, 655–666. [Google Scholar] [CrossRef]
- Sauvage, C.; Segura, V.; Bauchet, G.; Stevens, R.; Do, P.T.; Nikoloski, Z.; Fernie, A.R.; Causse, M. Genome-Wide Association in Tomato Reveals 44 Candidate Loci for Fruit Metabolic Traits1. Plant Physiol. 2014, 165, 1120–1132. [Google Scholar] [CrossRef]
- Ye, J.; Li, W.; Ai, G.; Li, C.; Liu, G.; Chen, W.; Wang, B.; Wang, W.; Lu, Y.; Zhang, J.; et al. Genome-wide association analysis identifies a natural variation in basic helix-loop-helix transcription factor regulating ascorbate biosynthesis via D-mannose/L-galactose pathway in tomato. PLoS Genet. 2019, 15, e1008149. [Google Scholar] [CrossRef]
- Ye, J.; Wang, X.; Hu, T.; Zhang, F.; Wang, B.; Li, C.; Yang, T.; Li, H.; Lu, Y.; Giovannoni, J.J.; et al. An InDel in the Promoter of Al-ACTIVATED MALATE TRANSPORTER9 Selected during Tomato Domestication Determines Fruit Malate Contents and Aluminum Tolerance. Plant Cell 2017, 29, 2249–2268. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Moreno, J.-P.; Levy-Samoha, D.; Malitsky, S.; Monforte, A.J.; Orzaez, D.; Aharoni, A.; Granell, A. Uncovering tomato quantitative trait loci and candidate genes for fruit cuticular lipid composition using the Solanum pennellii introgression line population. J. Exp. Bot. 2017, 68, 2703–2716. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ofner, I.; Lashbrooke, J.; Pleban, T.; Aharoni, A.; Zamir, D. Solanum pennellii backcross inbred lines (BILs) link small genomic bins with tomato traits. Plant J. 2016, 87, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Fridman, E.; Pleban, T.; Zamir, D. A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. USA 2000, 97, 4718–4723. [Google Scholar] [CrossRef]
- Lin, W.; Oliver, D.J. Role of triacylglycerols in leaves. Plant Sci. 2008, 175, 233–237. [Google Scholar] [CrossRef]
- Chapman, K.D.; Dyer, J.M.; Mullen, R.T. Biogenesis and functions of lipid droplets in plants. J. Lipid Res. 2012, 53, 215–226. [Google Scholar] [CrossRef]
- Chapman, K.D.; Dyer, J.M.; Mullen, R.T. Commentary: Why don’t plant leaves get fat? Plant Sci. 2013, 207, 128–134. [Google Scholar] [CrossRef]
- Dowhan, W. Understanding phospholipid function: Why are there so many lipids? J. Biol. Chem. 2017, 292, 10755–10766. [Google Scholar] [CrossRef]
- Luo, J.; Butelli, E.; Hill, L.; Parr, A.; Niggeweg, R.; Bailey, P.; Weisshaar, B.; Martin, C. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: Expression in fruit results in very high levels of both types of polyphenol. Plant J. 2008, 56, 316–326. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Alseekh, S.; Tohge, T.; Rallapalli, G.; Luo, J.; Kawar, P.G.; Hill, L.; Santino, A.; Fernie, A.R.; et al. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015, 6, 8635. [Google Scholar] [CrossRef]
- Cook, D.; Grierson, D.; Jones, C.; Wallace, A.; West, G.; Tucker, G. Modification of fatty acid composition in tomato (Lycopersicon esculentum) by expression of a borage delta6-desaturase. Mol. Biotechnol. 2002, 21, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Lara, I.; Heredia, A.; Domínguez, E. Shelf Life Potential and the Fruit Cuticle: The Unexpected Player. Front. Plant Sci. 2019, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.P.; et al. Acyl-Lipid Metabolism. Arabidopsis Book 2013, 11, e0161. [Google Scholar] [CrossRef] [PubMed]
- Acyl-[acyl-carrier-protein] hydrolase - Solanum lycopersicum (Tomato). Available online: https://www.uniprot.org/uniprot/A0A3Q7J279 (accessed on 20 January 2020).
- Angkawijaya, A.E.; Nguyen, V.C.; Nakamura, Y. Enhanced root growth in phosphate-starved Arabidopsis by stimulating de novo phospholipid biosynthesis through the overexpression of LYSOPHOSPHATIDIC ACID ACYLTRANSFERASE 2 (LPAT2). Plant Cell Environ. 2017, 40, 1807–1818. [Google Scholar] [CrossRef] [PubMed]
- Misra, N.; Panda, P.K.; Parida, B.K. Genome-wide identification and evolutionary analysis of algal LPAT genes involved in TAG biosynthesis using bioinformatic approaches. Mol. Biol. Rep. 2014, 41, 8319–8332. [Google Scholar] [CrossRef]
- Chen, X.; Snyder, C.L.; Truksa, M.; Shah, S.; Weselake, R.J. sn-Glycerol-3-phosphate acyltransferases in plants. Plant Signal. Behav. 2011, 6, 1695–1699. [Google Scholar] [CrossRef]
- Lytovchenko, A.; Eickmeier, I.; Pons, C.; Osorio, S.; Szecowka, M.; Lehmberg, K.; Arrivault, S.; Tohge, T.; Pineda, B.; Anton, M.T.; et al. Tomato Fruit Photosynthesis Is Seemingly Unimportant in Primary Metabolism and Ripening but Plays a Considerable Role in Seed Development. Plant Physiol. 2011, 157, 1650–1663. [Google Scholar] [CrossRef]
- Osorio, S.; Ruan, Y.-L.; Fernie, A.R. An update on source-to-sink carbon partitioning in tomato. Front. Plant Sci. 2014, 5, 516. [Google Scholar] [CrossRef]
- Hölzl, G.; Witt, S.; Kelly, A.A.; Zähringer, U.; Warnecke, D.; Dörmann, P.; Heinz, E. Functional differences between galactolipids and glucolipids revealed in photosynthesis of higher plants. Proc. Natl. Acad. Sci. USA 2006, 103, 7512–7517. [Google Scholar] [CrossRef]
- Uncharacterized protein–Solanum lycopersicum (Tomato). Available online: https://www.uniprot.org/uniprot/A0A3Q7I392 (accessed on 20 January 2020).
- AT2G33150(PKT3). Available online: https://www.arabidopsis.org/servlets/TairObject?type=locus&name=At2g33150 (accessed on 18 March 2020).
- Sol Genomics Network. Available online: https://www.solgenomics.net/locus/22428/view (accessed on 16 March 2020).
- Sol Genomics Network. Available online: https://www.solgenomics.net/locus/21871/view (accessed on 16 March 2020).
- Sol Genomics Network. Available online: https://www.solgenomics.net/locus/32411/view (accessed on 16 March 2020).
- Sol Genomics Network. Available online: https://www.solgenomics.net/locus/35352/view (accessed on 16 March 2020).
- Rowe, H.C.; Hansen, B.G.; Halkier, B.A.; Kliebenstein, D.J. Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 2008, 20, 1199–1216. [Google Scholar] [CrossRef]
- Lisec, J.; Römisch-Margl, L.; Nikoloski, Z.; Piepho, H.-P.; Giavalisco, P.; Selbig, J.; Gierl, A.; Willmitzer, L. Corn hybrids display lower metabolite variability and complex metabolite inheritance patterns. Plant J. 2011, 68, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Carreno-Quintero, N.; Acharjee, A.; Maliepaard, C.; Bachem, C.W.B.; Mumm, R.; Bouwmeester, H.; Visser, R.G.F.; Keurentjes, J.J.B. Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiol. 2012, 158, 1306–1318. [Google Scholar] [CrossRef] [PubMed]
- Semel, Y.; Nissenbaum, J.; Menda, N.; Zinder, M.; Krieger, U.; Issman, N.; Pleban, T.; Lippman, Z.; Gur, A.; Zamir, D. Overdominant quantitative trait loci for yield and fitness in tomato. Proc. Natl. Acad. Sci. USA 2006, 103, 12981–12986. [Google Scholar] [CrossRef] [PubMed]
| MGDGs | DGDGs | Phospholipids | TAGs | DAGs | |
|---|---|---|---|---|---|
| Acyl sugars | 100 | 96 | 50 | 78 | 100 |
| Flavonoids | 99 | 93 | 51 | 88 | 97 |
| Hydroxycinnamate derivatives | 93 | 94 | 48 | 82 | 95 |
| N-containing compounds | 93 | 92 | 49 | 78 | 90 |
| Unspecified | 100 | 99 | 50 | 85 | 100 |
| Amino acid | 100 | 100 | 45 | 71 | 100 |
| Glycoalkaloids | 100 | 100 | 50 | 78 | 100 |
| Others (phenolics) | 98 | 98 | 53 | 79 | 100 |
| MGDGs | DGDGs | Phospholipids | TAGs | DAGs | |
|---|---|---|---|---|---|
| MGDGs | 99 | 99 | 75 | 96 | 90 |
| DGDGs | 99 | 99 | 70 | 87 | 100 |
| Phospholipids | 75 | 70 | 78 | 60 | 65 |
| TAGs | 96 | 87 | 60 | 90 | 85 |
| DAGs | 90 | 100 | 65 | 85 | 100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuhalskaya, A.; Wijesingha Ahchige, M.; Perez de Souza, L.; Vallarino, J.; Brotman, Y.; Alseekh, S. Network Analysis Provides Insight into Tomato Lipid Metabolism. Metabolites 2020, 10, 152. https://doi.org/10.3390/metabo10040152
Kuhalskaya A, Wijesingha Ahchige M, Perez de Souza L, Vallarino J, Brotman Y, Alseekh S. Network Analysis Provides Insight into Tomato Lipid Metabolism. Metabolites. 2020; 10(4):152. https://doi.org/10.3390/metabo10040152
Chicago/Turabian StyleKuhalskaya, Anastasiya, Micha Wijesingha Ahchige, Leonardo Perez de Souza, José Vallarino, Yariv Brotman, and Saleh Alseekh. 2020. "Network Analysis Provides Insight into Tomato Lipid Metabolism" Metabolites 10, no. 4: 152. https://doi.org/10.3390/metabo10040152
APA StyleKuhalskaya, A., Wijesingha Ahchige, M., Perez de Souza, L., Vallarino, J., Brotman, Y., & Alseekh, S. (2020). Network Analysis Provides Insight into Tomato Lipid Metabolism. Metabolites, 10(4), 152. https://doi.org/10.3390/metabo10040152





