Extensive Profiling of Polyphenols from Two Trollius Species Using a Combination of Untargeted and Targeted Approaches
Abstract
1. Introduction
2. Results
2.1. Column Selection for Polyphenol Separation
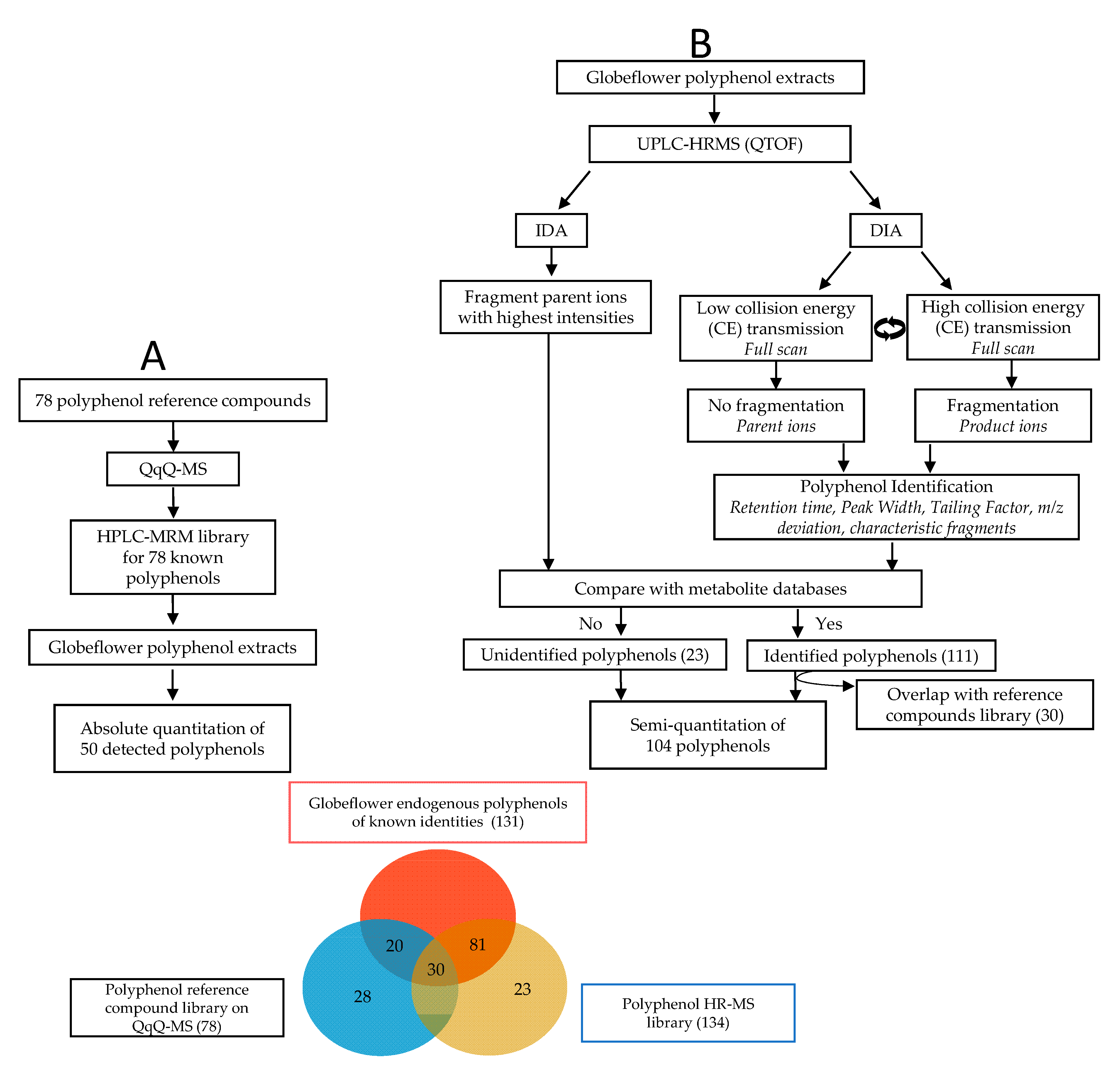
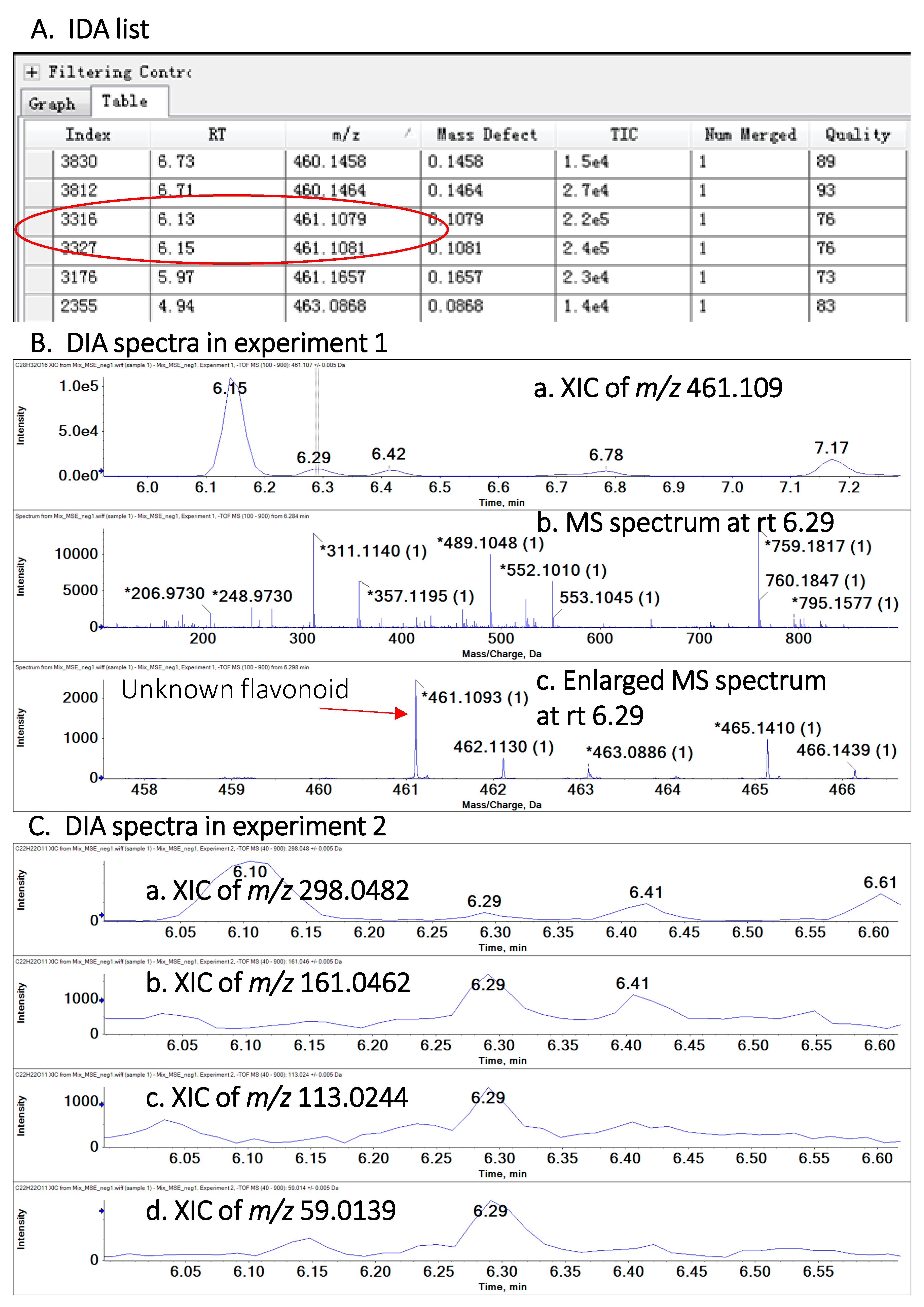
2.2. Identification and Quantification of Polyphenols
2.3. Method Validation
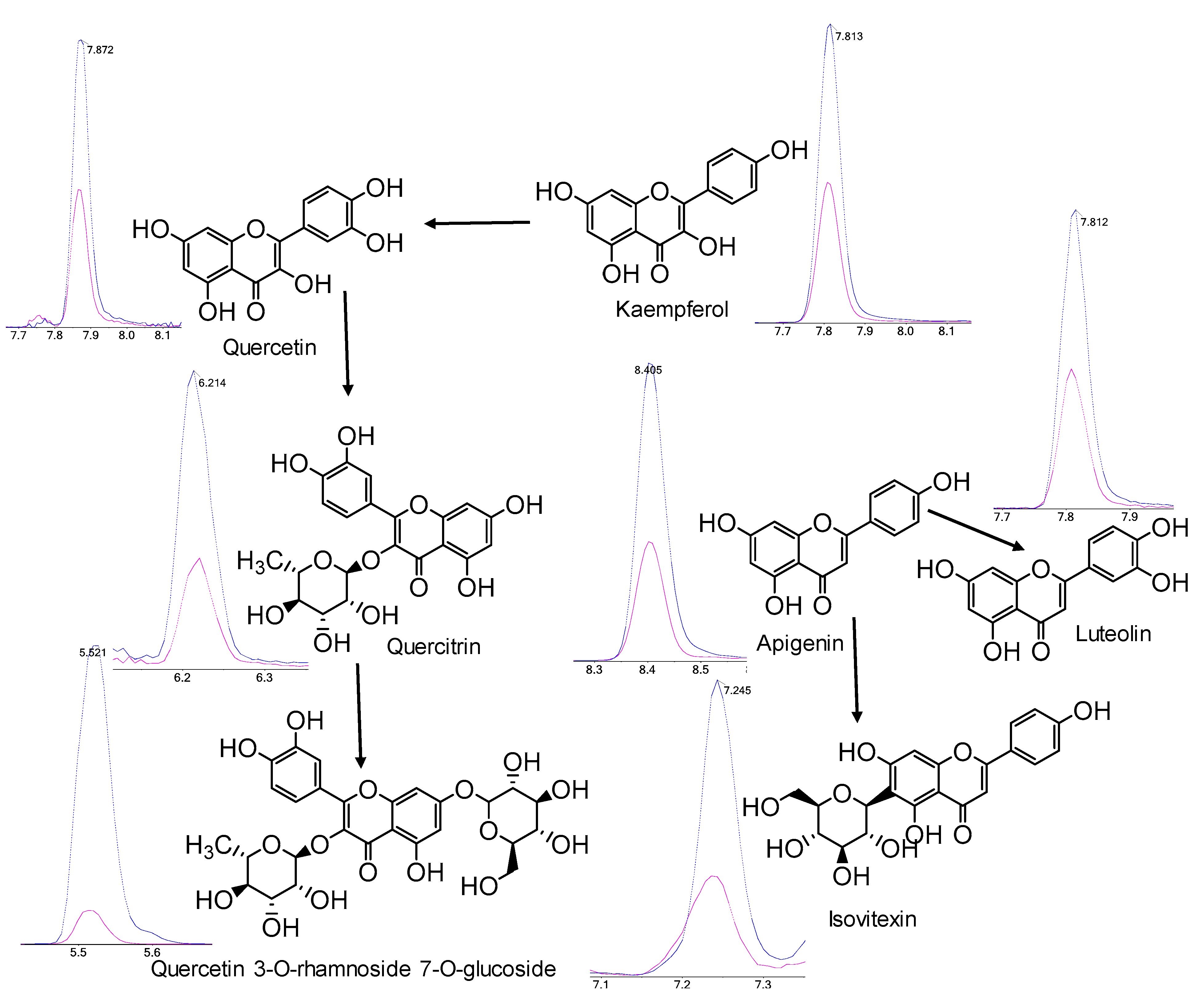
2.4. Polyphenol Profiles of T. chinensis and T. ledebouri
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Polyphenols Extraction and Preparation
4.3. LC/MSMS
4.4. Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Witkowska, B.E. Flavonoids from Trollius europaeus flowers and evaluation of their biological activity. J. Pharm. Pharmacol. 2018, 70, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.A.; Zhao, Y.M.; Yu, N.J. Flavone c-glycosides from Trollius ledebouri reichb. J. Asian Nat. Prod. Res. 2006, 8, 541–544. [Google Scholar]
- Yuan, M.; Wang, R.F.; Liu, L.J.; Yang, X.; Peng, Y.S.; Sun, Z.X. Contribution evaluation of the floral parts to orientin and vitexin concentrations in the flowers of Trollius chinensis. Chin. J. Nat. Med. 2013, 11, 699–704. [Google Scholar]
- Shi, D.; Chen, M.; Liu, L.; Wang, Q.; Liu, S.; Wang, L.; Wang, R. Anti-influenza A virus mechanism of three representative compounds from Flos Trollii via TLRs signaling pathways. J. Ethnopharmacol. 2020, 28, 112634. [Google Scholar]
- Liu, Y.; Tong, J.; Tong, Y.; Li, P.; Cui, X.; Cao, H. In vitro anti-influenza virus effect of total flavonoid from Trollius ledebouri Reichb. J. Int. Med. Res. 2018, 46, 1380–1390. [Google Scholar]
- Davatgaran-Taghipour, Y.; Masoomzadeh, S.; Farzaei, M.H.; Bahramsoltani, R.; Karimi-Soureh, Z.; Rahimi, R.; Abdollahi, M. Polyphenol nanoformulations for cancer therapy: Experimental evidence and clinical perspective. Int. J. Nanomedicine. 2017, 12, 2689–2702. [Google Scholar] [CrossRef]
- An, F.; Yang, G.; Tian, J.; Wang, S. Antioxidant effects of the orientin and vitexin in Trollius chinensis Bunge in D-galactose-aged mice. Neural. Regen. Res. 2012, 7, 2565–2675. [Google Scholar]
- Witkowska, B.E. The genus Trollius-review of pharmacological and chemical research. Phytother. Res. 2015, 29, 475–500. [Google Scholar] [CrossRef]
- Sioud, F.; Amor, S.; Toumia, I.B.; Lahmar, A.; Aires, V.; Chekir-Ghedira, L.; Delmas, D. A new highlight of ephedra alata decne properties as potential adjuvant in combination with cisplatin to induce cell death of 4T1 breast cancer cells in vitro and in vivo. Cells 2020, 9, 362. [Google Scholar] [CrossRef]
- Li, D.Y.; Wei, J.X.; Hua, H.M.; Li, Z.L. Antimicrobial constituents from the flowers of Trollius chinensis. J. Asian Nat. Prod. Res. 2014, 16, 1018–1023. [Google Scholar]
- Li, D.; Wang, Q.; Xu, L.; Li, M.; Jing, X.; Zhang, L. Pharmacokinetic study of three active flavonoid glycosides in rat after intravenous administration of Trollius ledebourii extract by liquid chromatography. Biomed. Chromatogr. 2008, 22, 1130–1136. [Google Scholar]
- Guo, L.; Qiao, S.; Hu, J.; Li, D.; Zheng, S.; Shi, D.; Liu, J.; Wang, R. Investigation of the effective components of the flowers of Trollius chinensis from the perspectives of intestinal bacterial transformation and intestinal absorption. Pharm. Biol. 2017, 55, 1747–1758. [Google Scholar]
- Giampieri, F.; Afrin, S.; Stewart, D.; McDougall, G.J.; Brennan, R.; Blyth, L.; Gasparrini, M.; Mazzoni, L.; Capocasa, F.; Alvarez-Suarez, J.M.; et al. Phytochemical Composition and Cytotoxic Effects on Liver Hepatocellular Carcinoma Cells of Different Berries Following a Simulated In Vitro Gastrointestinal Digestion. Molecules 2018, 23, 1918. [Google Scholar] [CrossRef] [PubMed]
- Shui, G.; Leong, L.P. Screening and identification of antioxidants in biological samples using high-performance liquid chromatography-mass spectrometry and its application on Salacca edulis Reinw. J. Agric. Food Chem. 2005, 53, 880–886. [Google Scholar]
- Peters, K.; Treutler, H.; Döll, S.; Kindt, A.S.D.; Hankemeier, T.; Neumann, S. Chemical Diversity and Classification of Secondary Metabolites in Nine Bryophyte Species. Metabolites 2019, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- La Barbera, G.; Capriotti, A.L.; Cavaliere, C.; Montone, C.M.; Piovesana, S.; Samperi, R.; Zenezini Chiozzi, R.; Laganà, A. Liquid chromatography-high resolution mass spectrometry for the analysis of phytochemicals in vegetal-derived food and beverages. Food Res. Int. 2017, 100, 28–52. [Google Scholar] [CrossRef]
- Gangopadhyay, N.; Rai, D.K.; Brunton, N.P.; Gallagher, E.; Hossain, M.B. Antioxidant-guided isolation and mass spectrometric identification of the major polyphenols in barley (Hordeum vulgare) grain. Food Chem. 2016, 210, 212–220. [Google Scholar] [CrossRef]
- Wu, T.; Lv, H.; Wang, F.; Wang, Y. Characterization of Polyphenols from Lycium ruthenicum Fruit by UPLC-Q-TOF/MS(E) and Their Antioxidant Activity in Caco-2 Cells. J. Agric. Food Chem. 2016, 64, 2280–2288. [Google Scholar]
- Haag, A.M. Mass Analyzers and Mass Spectrometers. Adv. Exp. Med. Biol. 2016, 919, 157–169. [Google Scholar]
- Tsugawa, H.; Satoh, A.; Uchino, H.; Cajka, T.; Arita, M.; Arita, M. Mass Spectrometry Data Repository Enhances Novel Metabolite Discoveries with Advances in Computational Metabolomics. Metabolites 2019, 9, 119. [Google Scholar] [CrossRef]
- Alcoriza-Balaguer, M.I.; García-Cañaveras, J.C.; López, A.; Conde, I.; Juan, O.; Carretero, J.; Lahoz, A. LipidMS: An R Package for Lipid Annotation in Untargeted Liquid Chromatography-Data Independent Acquisition-Mass Spectrometry Lipidomics. Anal. Chem. 2019, 91, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Shan, J.; Tan, X.; Zou, J.; Yin, A.; Cai, B.; Di, L. Effect of chito-oligosaccharide on the oral absorptions of phenolic acids of Flos Lonicerae extract. Phytomedicine 2014, 21, 184–194. [Google Scholar]
- Wu, Y.H.; Zhang, B.Y.; Qiu, L.P.; Guan, R.F.; Ye, Z.H.; Yu, X.P. Structure Properties and Mechanisms of Action of Naturally Originated Phenolic Acids and Their Derivatives against Human Viral Infections. Curr. Med. Chem. 2017, 24, 4279–4302. [Google Scholar]
- Zhou, X.; Chen, X.; Wu, X.; Cao, G.; Zhang, J. Characterization of the chemical composition of white chrysanthemum flowers of Hangzhou by using high-performance ion trap mass spectrometry. J. Sep. Sci. 2016, 39, 1218–1222. [Google Scholar]
- Heyman, H.M.; Senejoux, F.; Seibert, I.; Klimkait, T.; Maharaj, V.J.; Meyer, J.J. Identification of anti-HIV active dicaffeoylquinic and tricaffeoylquinic acids in helichrysum populifolium by NMR-based metabolomic guided fractionation. Fitoterapia 2015, 103, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Cheng, X.; Zhang, X.; Diao, X.; Liang, C.; Zhang, L. Qualitative and Quantitative Analyses of Active Constituents in Trollius ledebourii. J. Chromatogr. Sci. 2018, 56, 619–635. [Google Scholar]
- Forouzesh, A.; Samadi Foroushani, S.; Forouzesh, F.; Zand, E. Reliable Target Prediction of Bioactive Molecules Based on Chemical Similarity Without Employing Statistical Methods. Front. Pharmacol. 2019, 10, 835–850. [Google Scholar] [CrossRef]
- Lam, S.M.; Shui, G. Lipidomics as a principal tool for advancing biomedical research. J. Genet. Genomics. 2013, 40, 375–390. [Google Scholar] [CrossRef]
- Wang, R.X.; Li, B.W.; Lam, S.M.; Shui, G. Integration of lipidomics and metabolomics for in-depth understanding of cellular mechanism and disease progression Journal of Genetics and Genomics. J. Genet. Genom. 2019, in press. [Google Scholar] [CrossRef]
- Tian, H.; Lam, S.M.; Shui, G. Metabolomics, a Powerful Tool for Agricultural Research. Int. J. Mol. Sci. 2016, 17, E1871. [Google Scholar]
- Wu, L.Z.; Zhang, X.P.; Xu, X.D.; Zheng, Q.X.; Yang, J.S.; Ding, W.L. Characterization of aromatic glycosides in the extracts of Trollius species by ultra high-performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight tandem mass spectrometry. J. Pharm. Biomed. Anal. 2013, 75, 55–63. [Google Scholar]
- The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 12 February 2020).

| No. | Characteristic Fragments | Theoretical m/z [M − H] | No | Characteristic Fragments | Theoretical m/z [M − H] |
|---|---|---|---|---|---|
| 1 | C15H10O7 | 301.0354 | 18 | C7H4O3 | 135.0088 |
| 2 | C15H9O7 | 300.0276 | 19 | C9H10O | 133.0659 |
| 3 | C15H12O6 | 287.0561 | 20 | C5H8O4 | 131.0350 |
| 4 | C15H10O6 | 285.0405 | 21 | C6H6O3 | 125.0244 |
| 5 | C14H8O6 | 271.0248 | 22 | C6H4O3 | 123.0088 |
| 6 | C15H10O5 | 269.0455 | 23 | C7H6O2 | 121.0295 |
| 7 | C14H8O5 | 255.0299 | 24 | C5H6O3 | 113.0244 |
| 8 | C13H12O5 | 247.0612 | 25 | C6H4O2 | 107.0139 |
| 9 | C13H14O | 185.0972 | 26 | C5H6O2 | 97.0295 |
| 10 | C8H6O5 | 178.9986 | 27 | C5H4O2 | 95.0139 |
| 11 | C12H12O | 171.0815 | 28 | C6H4O | 91.0190 |
| 12 | C8H4O4 | 163.0037 | 29 | C7H6 | 89.0397 |
| 13 | C6H9O10 | 161.0456 | 30 | C4H4O2 | 83.0139 |
| 14 | C7H4O4 | 151.0037 | 31 | C2H4O2 | 59.0139 |
| 15 | C8H6O3 | 149.0244 | 32 | C3H4O | 55.0190 |
| 16 | C6H9O4 | 144.0428 | 33 | C3H2O | 53.0033 |
| 17 | C7H6O3 | 137.0244 |
| No. | Formula | Molecular Weight | Molecular Assignment |
|---|---|---|---|
| 1 | C7H12O7 | 208.0589 | Glucose + CO |
| 2 | C7H12O6 | 192.0640 | Glucose + C |
| 3 | C6H12O6 | 180.0628 | Glucose |
| 4 | C6H10O7 | 194.0432 | Glucuronic acid |
| 5 | C6H8O6 | 176.0326 | Glucuronic acid − H2O |
| 6 | C4H8O4 | 120.04281 | Glucose − C2H4O2 |
| 7 | C6H10O5 | 162.0523 | Glucose − H2O |
| 8 | C6H11O5. | 163.0612 | Glucose − HO. |
| 9 | C6H12O5 | 164.0679 | Rhamnose |
| 10 | C6H10O4 | 146.0574 | Rhamnose − H2O |
| 11 | C5H10O5 | 150.0523 | Arabinose |
| 12 | C5H8O4 | 132.0417 | Arabinose − H2O or Xylose |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, H.; Zhou, Z.; Shui, G.; Lam, S.M. Extensive Profiling of Polyphenols from Two Trollius Species Using a Combination of Untargeted and Targeted Approaches. Metabolites 2020, 10, 119. https://doi.org/10.3390/metabo10030119
Tian H, Zhou Z, Shui G, Lam SM. Extensive Profiling of Polyphenols from Two Trollius Species Using a Combination of Untargeted and Targeted Approaches. Metabolites. 2020; 10(3):119. https://doi.org/10.3390/metabo10030119
Chicago/Turabian StyleTian, He, Zhiyang Zhou, Guanghou Shui, and Sin Man Lam. 2020. "Extensive Profiling of Polyphenols from Two Trollius Species Using a Combination of Untargeted and Targeted Approaches" Metabolites 10, no. 3: 119. https://doi.org/10.3390/metabo10030119
APA StyleTian, H., Zhou, Z., Shui, G., & Lam, S. M. (2020). Extensive Profiling of Polyphenols from Two Trollius Species Using a Combination of Untargeted and Targeted Approaches. Metabolites, 10(3), 119. https://doi.org/10.3390/metabo10030119





