TGF-ß1 Induces Changes in the Energy Metabolism of White Adipose Tissue-Derived Human Adult Mesenchymal Stem/Stromal Cells In Vitro
Abstract
1. Introduction
2. Results
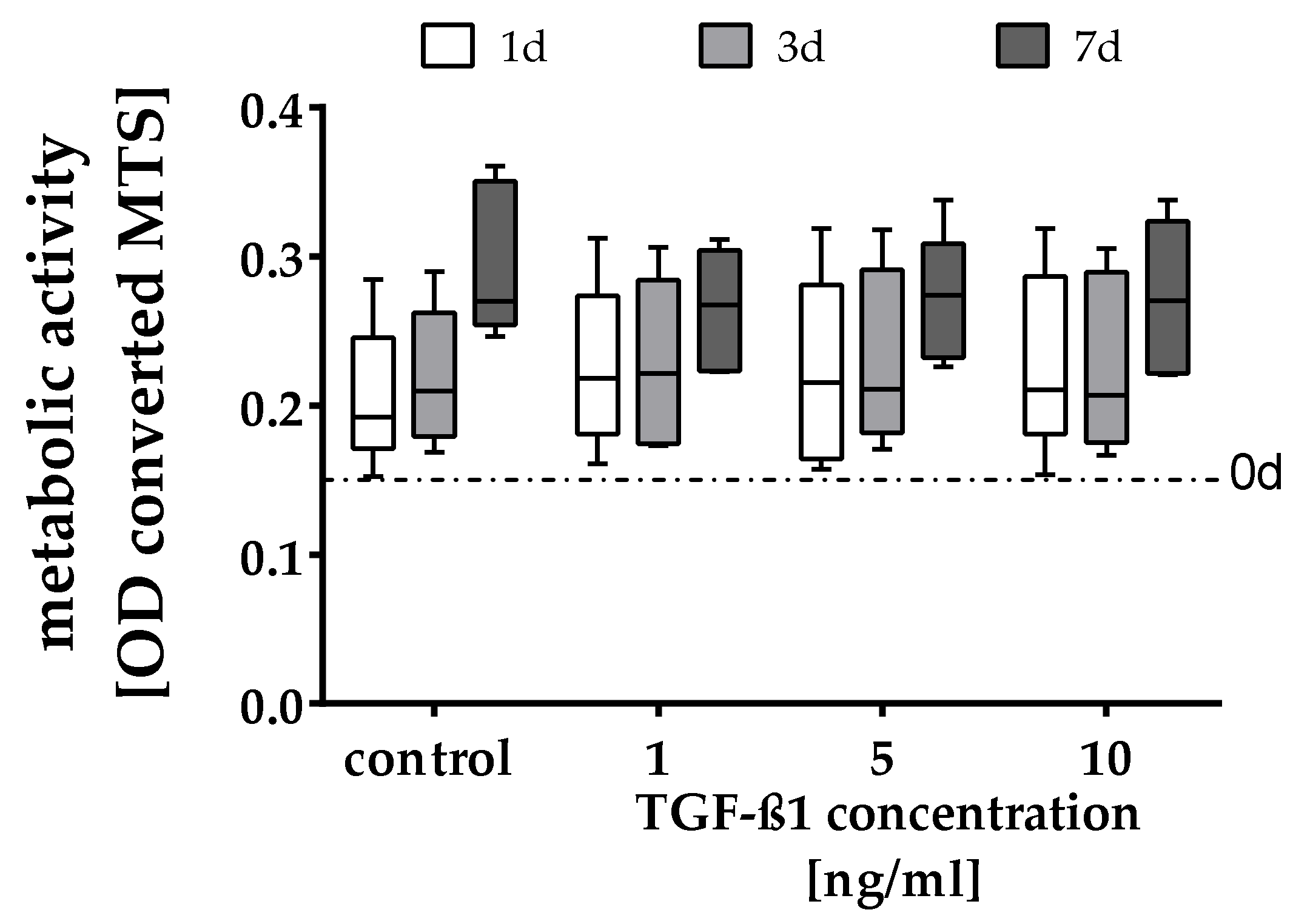
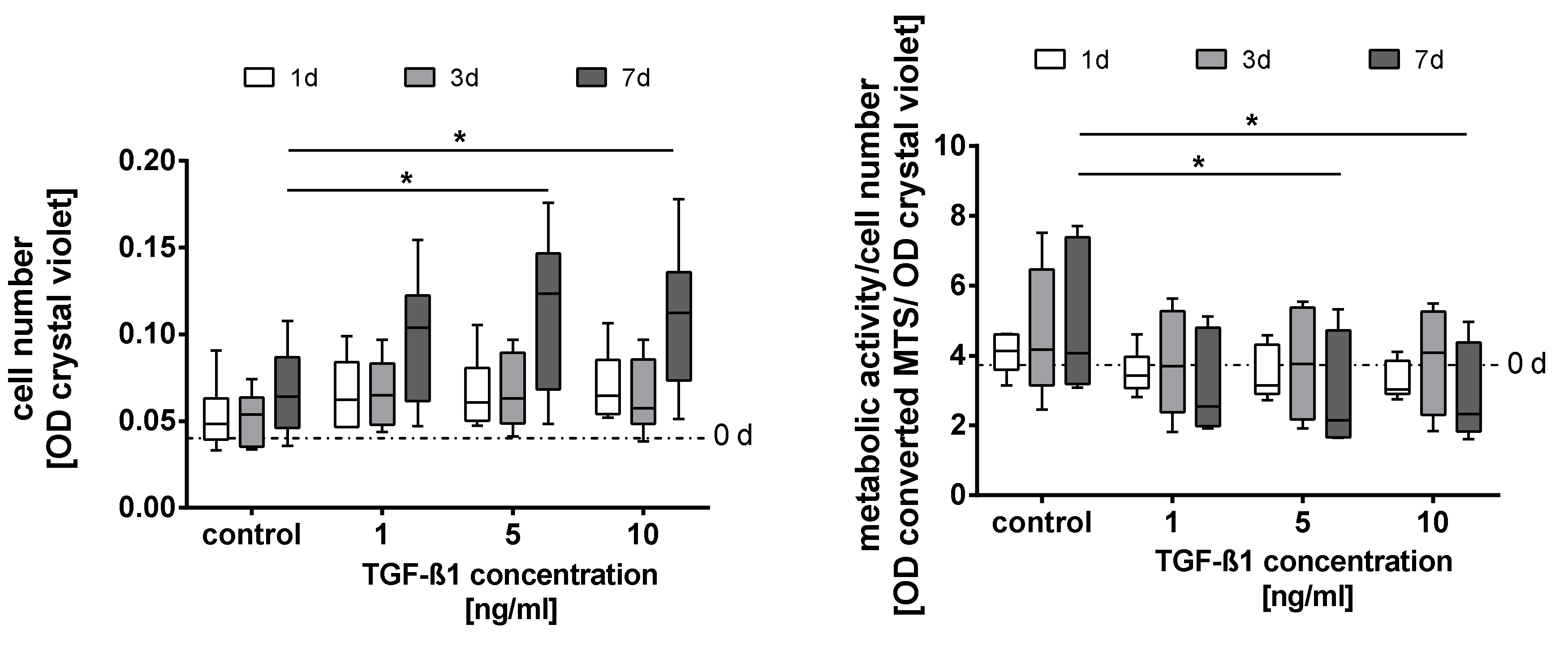
2.1. Quantification of the Cell Number and Metabolic Activity
2.2. Cell Cycle Analyses
2.3. Metabolic Characterization
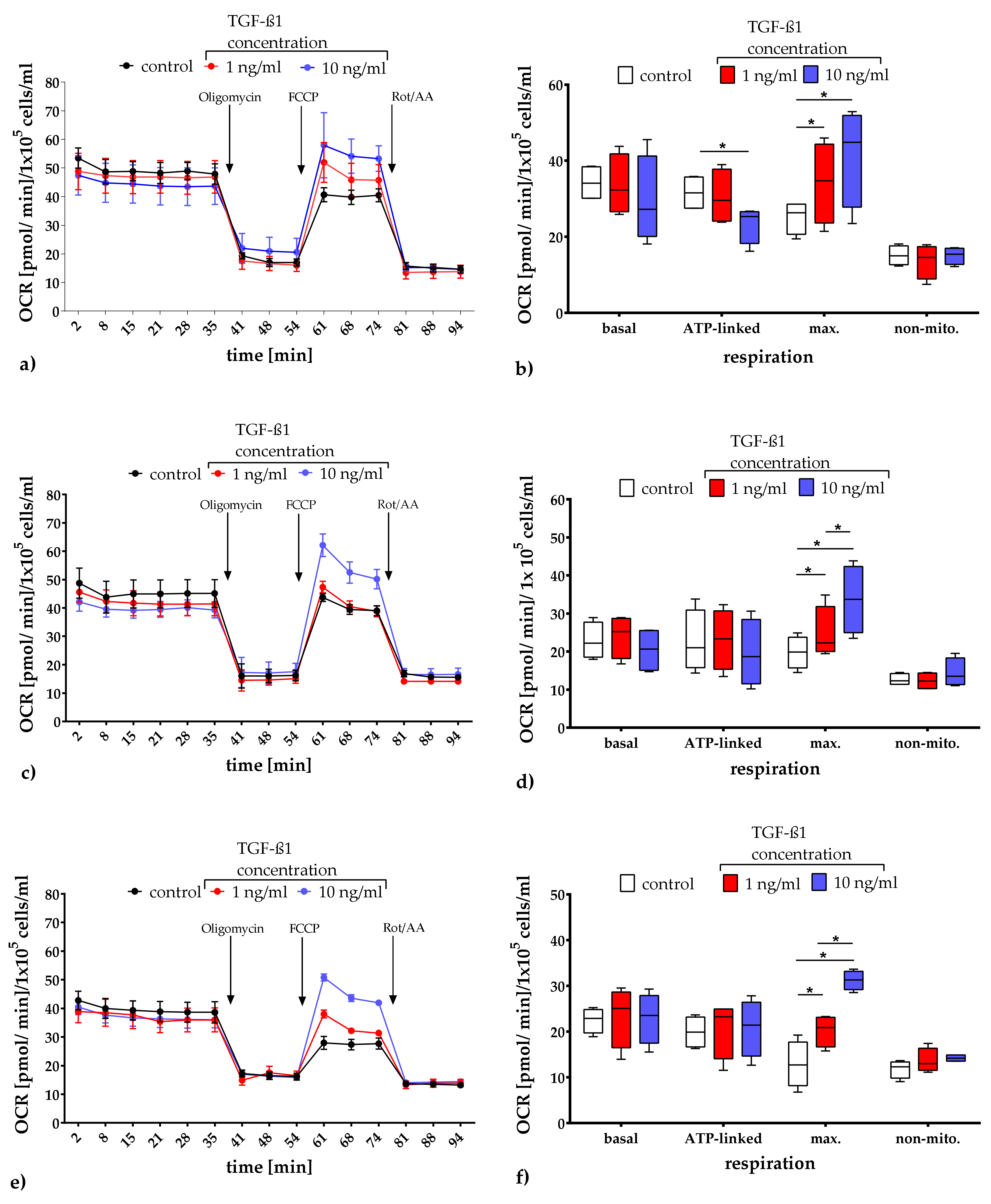
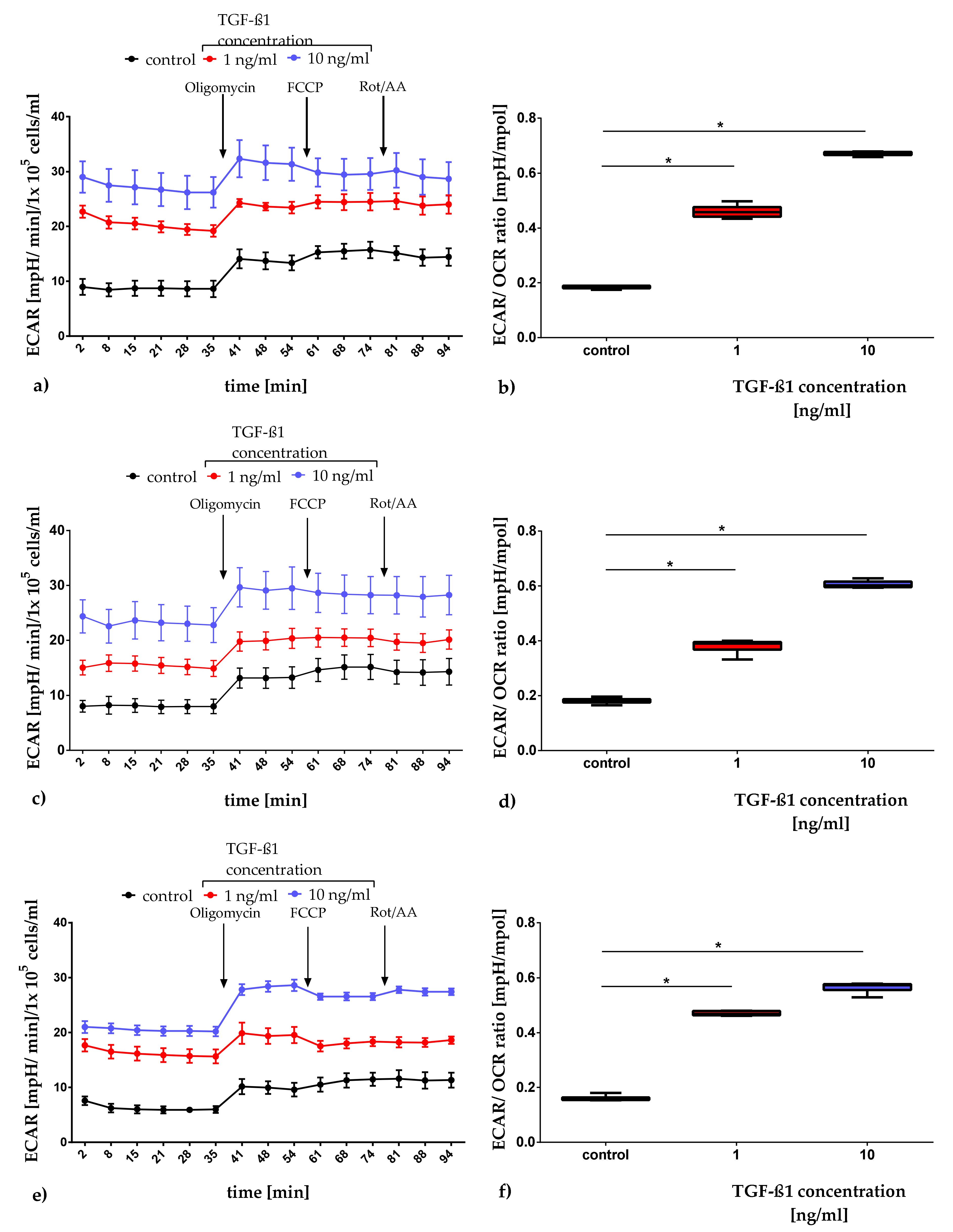
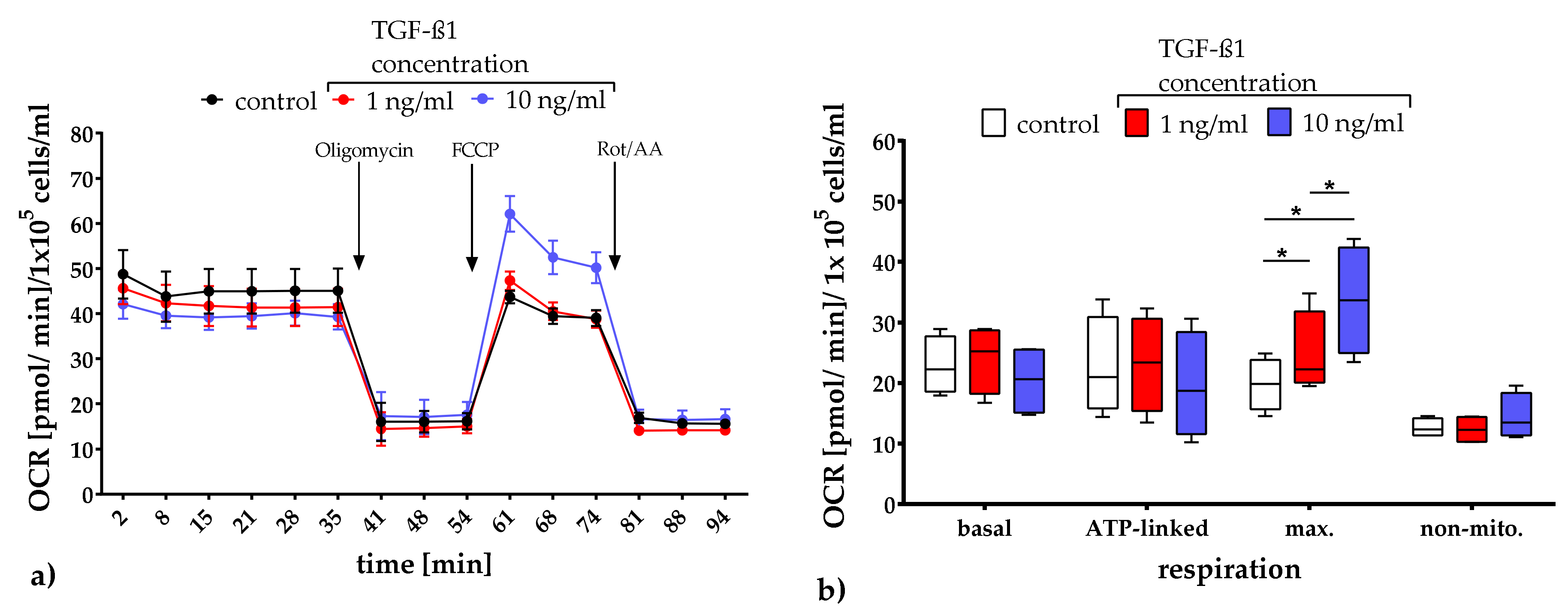
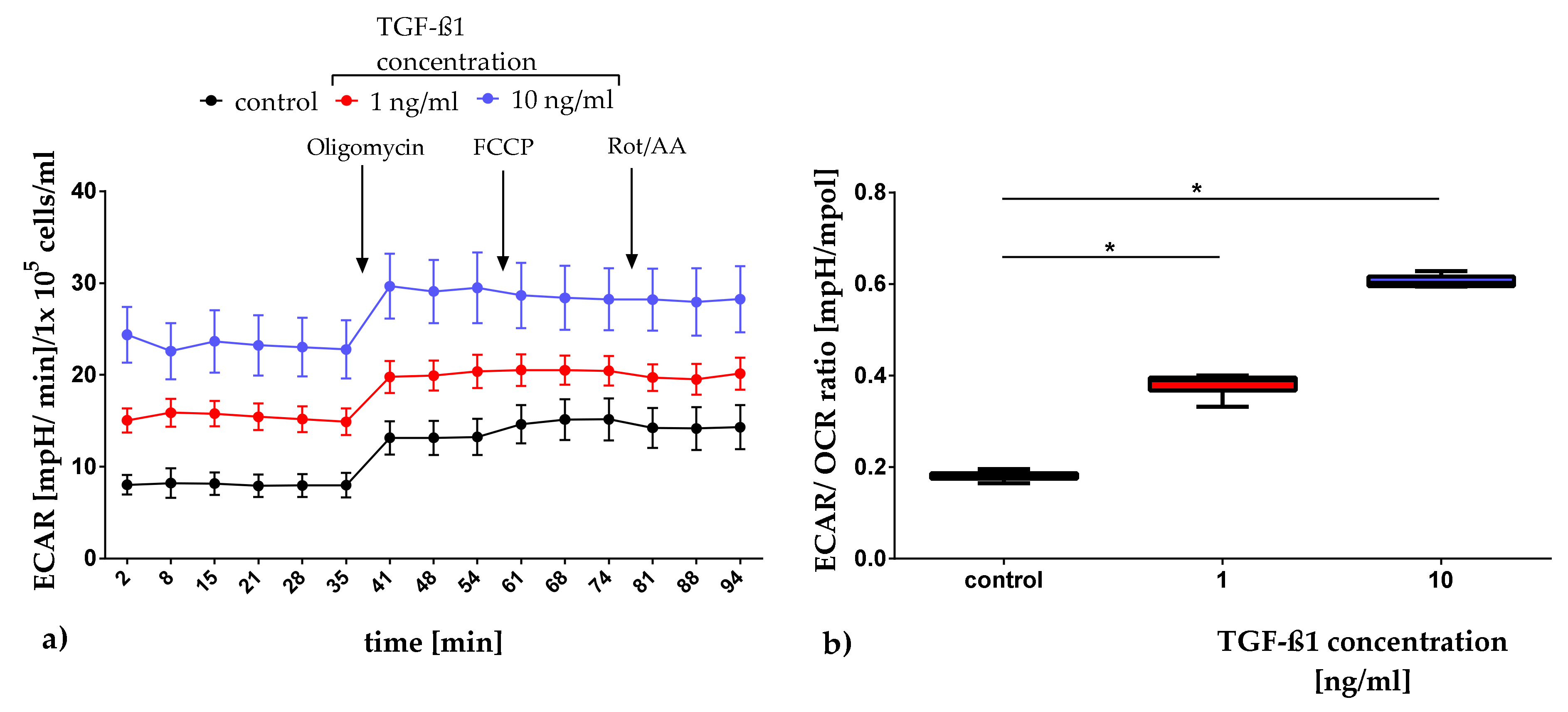
2.3.1. Quantification of Mitochondrial Respiration and Glycolysis
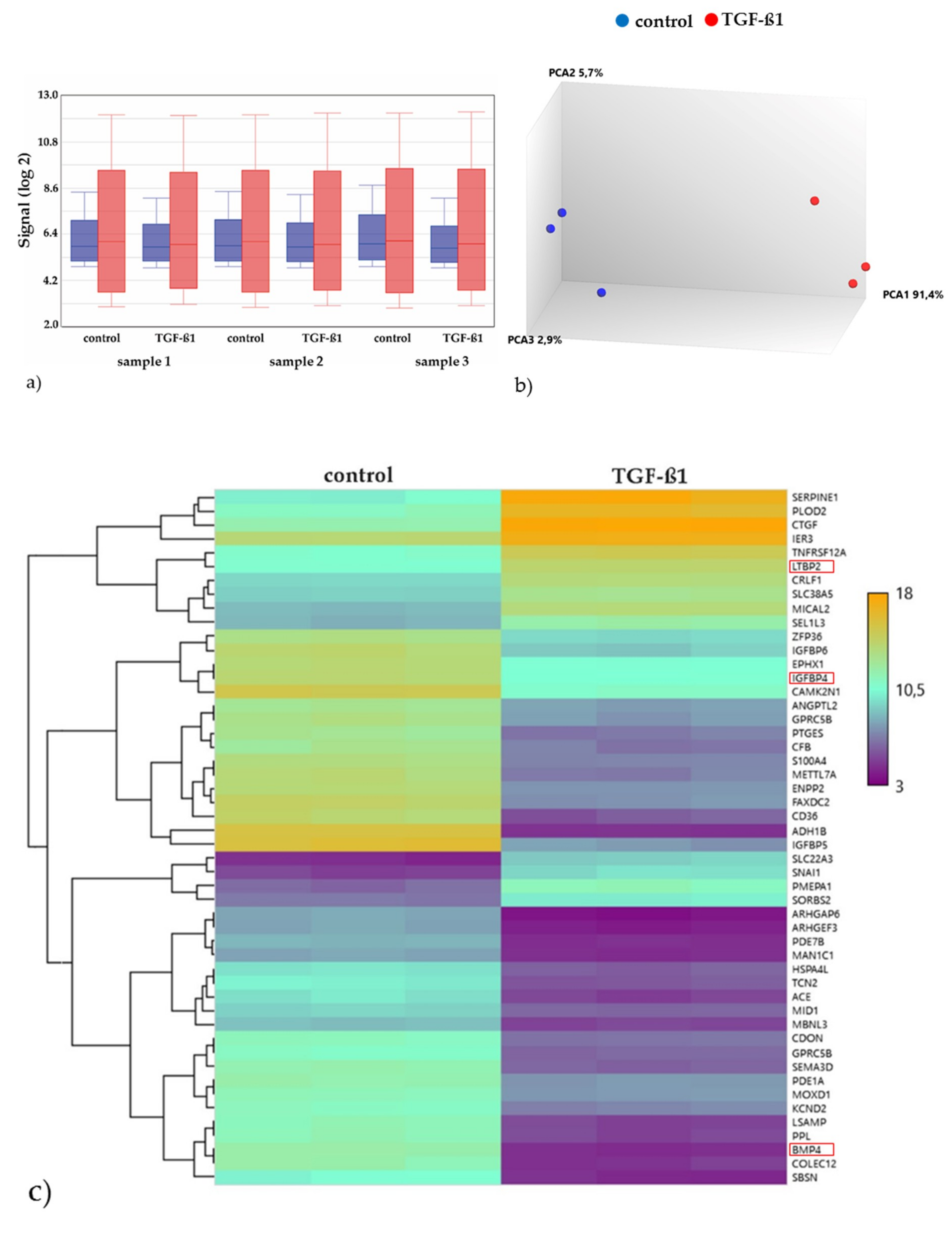
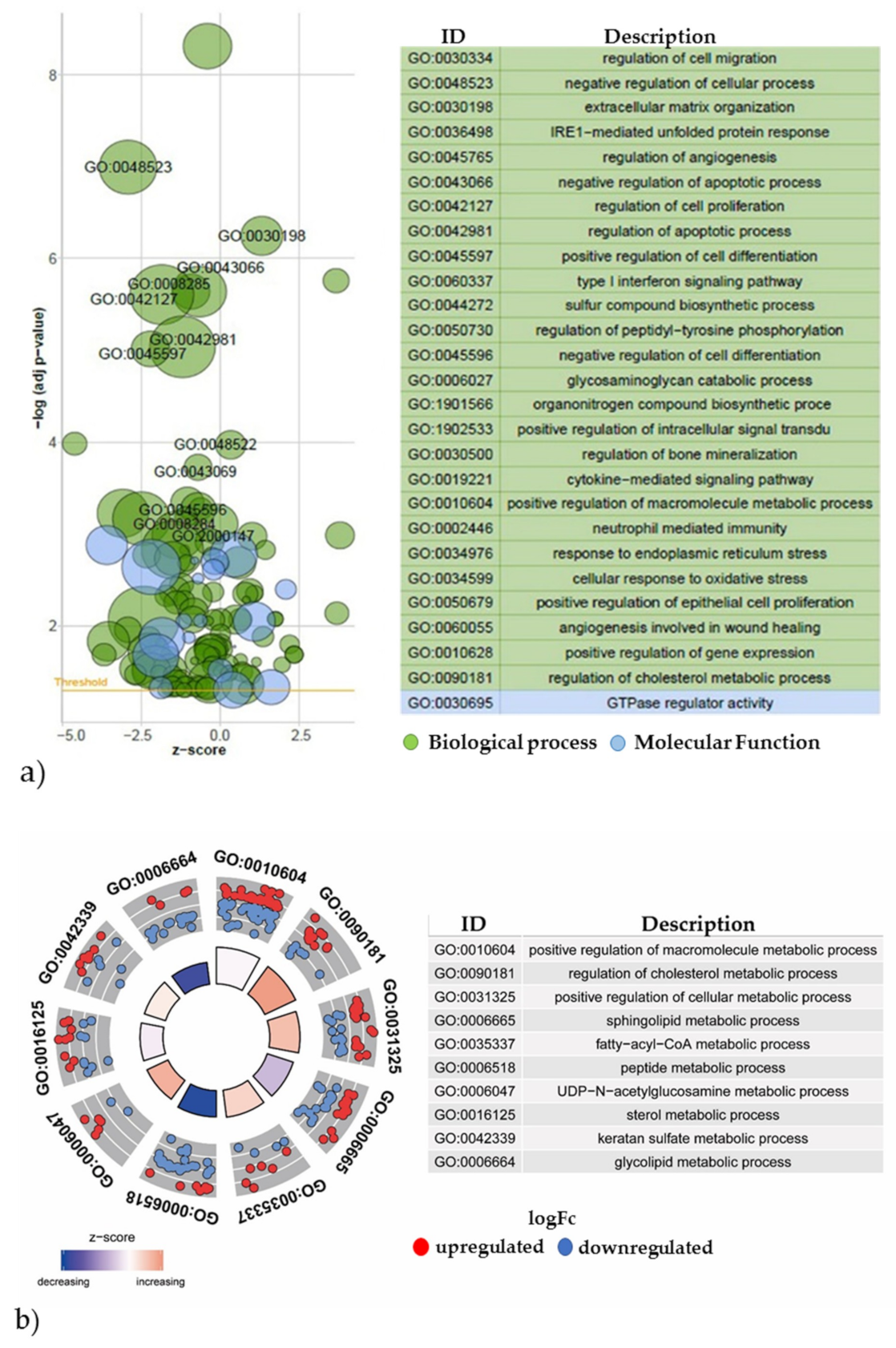
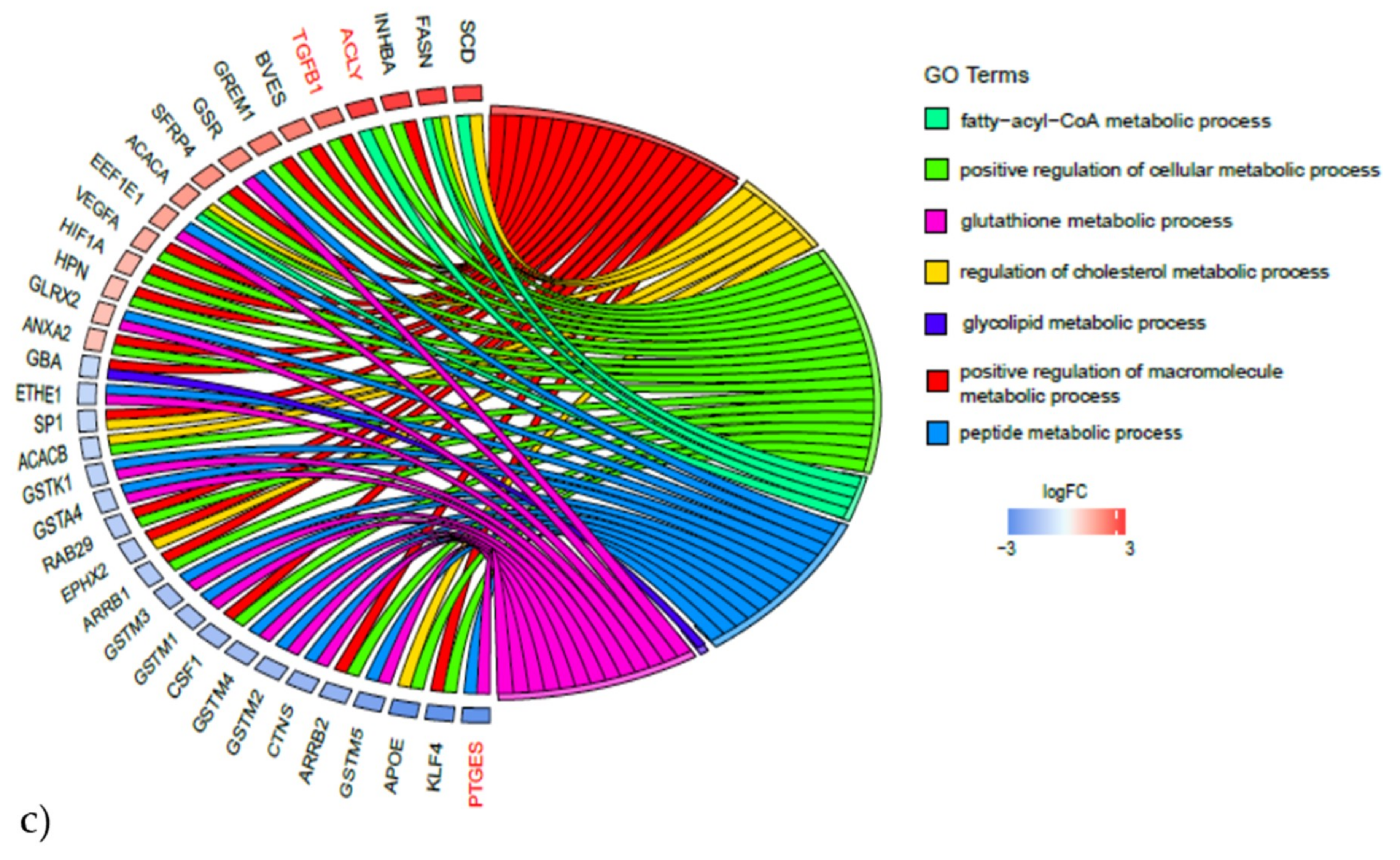
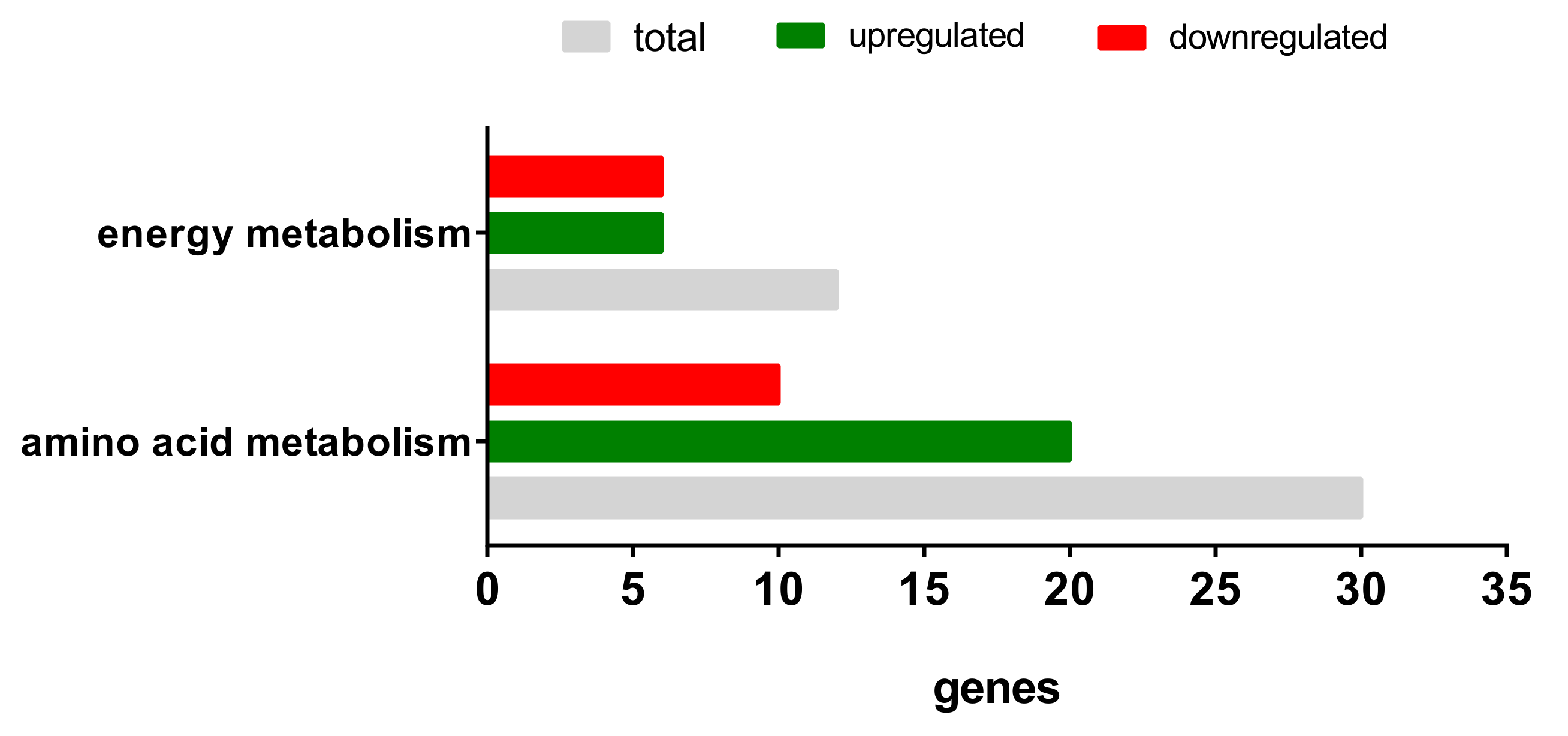
2.3.2. Gene Expression Analyses of the Energy and Amino Acid Metabolism
3. Discussion
4. Materials and Methods
4.1. Cell Isolation and Cultivation
4.2. Quantification of the Cell Number and Metabolic Activity
4.3. Cell Cycle Analyses
4.4. Quantification of Mitochondrial Respiration and Glycolysis
4.5. Gene Expression Analyses
4.5.1. RNA Extraction
4.5.2. Microarray Analysis
4.5.3. Microarray Data Analysis
4.5.4. Gene Expression Network Analysis
4.6. Data Illustration and Statistical Analysis
4.7. Ethic Statement
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
Appendix C
References
- Klaus, S. Adipose tissue as a regulator of energy balance. Curr. Drug Targets 2004. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Spiegelman, B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Barchetta, I.; Cimini, F.A.; Ciccarelli, G.; Baroni, M.G.; Cavallo, M.G. Sick fat: The good and the bad of old and new circulating markers of adipose tissue inflammation. J. Endocrinol. Investig. 2019, 1257–1272. [Google Scholar] [CrossRef]
- Bourin, P.; Bunnell, B.A.; Casteilla, L.; Dominici, M.; Katz, A.J.; March, K.L.; Redl, H.; Rubin, J.P.; Yoshimura, K.; Gimble, J.M. Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: A joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International Society for Cellular Therapy (ISCT). Cytotherapy 2013, 641–648. [Google Scholar] [CrossRef]
- Tallone, T.; Realini, C.; Böhmler, A.; Kornfeld, C.; Vassalli, G.; Moccetti, T.; Bardelli, S.; Soldati, G. Adult human adipose tissue contains several types of multipotent cells. J. Cardiovasc. Transl. Res. 2011, 200–210. [Google Scholar] [CrossRef]
- Zimmerlin, L.; Donnenberg, V.S.; Pfeifer, M.E.; Meyer, E.M.; Péault, B.; Rubin, J.P.; Donnenberg, A.D. Stromal vascular progenitors in adult human adipose tissue. Cytom. Part A J. Int. Soc. Anal. Cytol. 2010, 77, 22–30. [Google Scholar] [CrossRef]
- Zuk, P.A.; Zhu, M.; Mizuno, H.; Huang, J.; Futrell, W.; Katz, A.J.; Benhaim, P.; Lorenz, P.; Hedrick, M.H. Multilineage cells from human adipose tissue: Implications for cell-based therapies. Tissue Eng. 2001, 211–228. [Google Scholar] [CrossRef]
- Grafe, I.; Alexander, S.; Peterson, J.R.; Snider, T.N.; Levi, B.; Lee, B.; Mishina, Y. TGF-β family signaling in mesenchymal differentiation. Cold Spring Harb. Perspect. Biol. 2019, 1–66. [Google Scholar] [CrossRef]
- Lin, G.; Garcia, M.; Ning, H.; Banie, L.; Guo, Y.-L.; Lue, T.F.; Lin, C.-S. Defining stem and progenitor cells within adipose tissue. Stem Cells Dev. 2008, 1053–1063. [Google Scholar] [CrossRef]
- Montserrat, E.R. Adipose tissue: Cell heterogeneity and functional diversity. Endocrinol. Nutr. (Engl. Ed.) 2014, 61, 100–112. [Google Scholar] [CrossRef]
- Löffler, G.; Hauner, H. Adipose tissue development: The role of precursor cells and adipogenic factors. Klin. Wochenschr. 1987, 812–817. [Google Scholar] [CrossRef]
- Assian, R.K.; Komoriya, A.; Meyers, C.A.; Miller, D.M.; Sporn, M.B. Transforming growth factor-ß in human platelets: Identifcation of a major storage site, purification and characterization. J. Biol. Chem. 1983, 7155–7160. [Google Scholar]
- Ibelgaufts, H. (Ed.) Lexikon Zytokine: Mit Tabellen; Medikon: München, Germany, 1992; ISBN 3-923866-46-1. [Google Scholar]
- De Farias, A.V.; Carrillo-Gálvez, A.B.; Martín, F.; Anderson, P. TGF-β and mesenchymal stromal cells in regenerative medicine, autoimmunity and cancer. Cytokine Growth Factor Rev. 2018, 25–37. [Google Scholar] [CrossRef]
- Ng, M.; Fareha, A.; Ooi, S.I.; Aminuddin, B.S.; Ruszymah, B.H.I. Effects of TGF-ß1 on bone marrow stem cells proliferation, osteogenic differentiation and maturation. Regen. Res. 2012, 20–26. [Google Scholar]
- Walenda, G.; Abnaof, K.; Joussen, S.; Meurer, S.; Smeets, H.; Rath, B.; Hoffmann, K.; Fröhlich, H.; Zenke, M.; Weiskirchen, R.; et al. TGF-beta1 does not induce senescence of multipotent mesenchymal stromal cells and has similar effects in early and late passages. PLoS ONE 2013. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, W.; Park, I.; Pins, M.; Kozlowski, J.M.; Jovanovic, B.; Zhang, J.; Lee, C.; Ilio, K. Dual regulation of proliferation and growth arrest in prostatic stromal cells by transforming growth factor-ß1. Endocrinology 2003, 4280–4284. [Google Scholar] [CrossRef] [PubMed]
- Hinz, B.; Phan, S.H.; Thannickal, V.J.; Galli, A.; Bochaton-Piallat, M.; Gabbiani, G. The myofibroblast: One function, multiple origins. Am. J. Pathol. 2007, 1807–1816. [Google Scholar] [CrossRef]
- Hinz, B.; Celetta, G.; Tomasek, J.J.; Gabbiani, G.; Chaponnier, C. Alpha-smooth muscle actin expression upregulates fibroblast contractile activity. Mol. Biol. Cell 2001, 2730–2741. [Google Scholar] [CrossRef]
- Hinz, B.; Gabbiani, G.; Chaponnier, C. The NH2-terminal peptide of alpha-smooth muscle actin inhibits force generation by the myofibroblast in vitro and in vivo. J. Cell Biol. 2002, 657–663. [Google Scholar] [CrossRef]
- Eyden, B. The myofibroblast: Phenotypic characterization as a prerequisite to understanding its functions in translational medicine. J. Cell. Mol. Med. 2008, 22–37. [Google Scholar] [CrossRef]
- Tomasek, J.J.; Gabbiani, G.; Hinz, B.; Chaponnier, C.; Brown, R.A. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 2002, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Ochocki, J.D.; Simon, M.C. Nutrient-sensing pathways and metabolic regulation in stem cells. J. Cell Biol. 2013, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Shyh-Chang, N.; Daley, G.Q. Metabolic switches linked to pluripotency and embryonic stem cell differentiation. Cell Metab. 2015, 349–350. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.; Salamon, A.; Mispagel, S.; Kamp, G.; Peters, K. Energy metabolic capacities of human adipose-derived mesenchymal stromal cells in vitro and their adaptations in osteogenic and adipogenic differentiation. Exp. Cell Res. 2018, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Shares, B.H.; Busch, M.; White, N.; Shum, L.; Eliseev, R.A. Active mitochondria support osteogenic differentiation by stimulating β-catenin acetylation. J. Biol. Chem. 2018, 16019–16027. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013. [Google Scholar] [CrossRef]
- Jian, H.; Shen, X.; Liu, I.; Semenov, M.; He, X.; Wang, X. Smad3-dependent nuclear translocation of ß-catenin is required for TGF-ß1- induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 2006, 666–674. [Google Scholar] [CrossRef]
- Koli, K.; Ryynänen, M.J.; Keski-Oja, J. Latent TGF-beta binding proteins (LTBPs)-1 and -3 coordinate proliferation and osteogenic differentiation of human mesenchymal stem cells. Bone 2008, 679–688. [Google Scholar] [CrossRef]
- Ng, F.; Boucher, S.; Koh, S.; Sastry, K.S.R.; Chase, L.; Lakshmipathy, U.; Choong, C.; Yang, Z.; Vemuri, M.C.; Rao, M.S.; et al. PDGF, TGF-beta, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): Transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood 2008, 295–307. [Google Scholar] [CrossRef]
- Debacq-Chainiaux, F.; Borlon, C.; Pascal, T.; Royer, V.; Eliaers, F.; Ninane, N.; Carrard, G.; Friguet, B.; de Longueville, F.; Boffe, S.; et al. Repeated exposure of human skin fibroblasts to UVB at subcytotoxic level triggers premature senescence through the TGF-beta1 signaling pathway. J. Cell Sci. 2005, 743–758. [Google Scholar] [CrossRef]
- Ito, T.; Sawada, R.; Fuijiwara, Y.; Seyama, Y.; Tsuchiya, T. FGF-2 suppresses cellular senescence of human mesenchymal stem cells by down-regulation of TGF-β2. Biochem. Biophys. Res. Commun. 2007. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J. How cells read TGF-ß signals. Nat. Rev. Mol. Cell Biol. 2000, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Kassem, M.; Keivborg, M.; Eriksen, E.F. Production and action of transforming growth factor-ß in human osteoblast cultures: Dependence on cell differentiation and modulation by calcitriol. Eur. J. Clin. Investig. 2000, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Roostaeian, J.; Carlsen, B.; Simhaee, D.; Jarrahy, R.; Huang, W.; Ishida, K.; Rudkin, G.H.; Yamaguchi, D.T.; Miller, T.A. Characterization of growth and osteogenic differentiation of rabbit bone marrow stromal cells. J. Surg. Res. 2006, 76–83. [Google Scholar] [CrossRef]
- Morton, J.C.; Armstrong, J.A.; Sud, A.; Tepikin, A.V.; Sutton, R.; Criddle, D.N. Altered bioenergetics of blood cell sub-populations in acute pancreatitis patients. J. Clin. Med. 2019. [Google Scholar] [CrossRef]
- Valente, A.J.; Fonseca, J.; Moradi, F.; Foran, G.; Necakov, A.; Stuart, J.A. Mitochondria in health and in sickness: Quantification of mitochondrial network characteristics in health and disease. In Mitochondria in Health and in Sickness; Springer: Singapore, 2019. [Google Scholar]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science 2009, 1029–1033. [Google Scholar] [CrossRef]
- Peters, K.; Kamp, G.; Berz, A.; Unger, R.E.; Barth, S.; Salamon, A.; Rychyly, J.; Kirckpatrick, C.J. Changes in human endothelial cell energy metabolic capacities during in vitro cultivation. The role of “aerobic glycolysis” and proliferation. Cell. Physiol. Biochem. 2009, 483–492. [Google Scholar] [CrossRef]
- Meyer, J.; Engelmann, R.; Kamp, G.; Peters, K. Human adipocytes and CD34+ cells from the stromal vascular fraction of the same adipose tissue differ in their energy metabolic enzyme configuration. Exp. Cell Res. 2019, 47–54. [Google Scholar] [CrossRef]
- Tian, W.N.; Braunstein, L.D.; Pang, J.; Stuhlmeier, K.M.; Xi, Q.C.; Tian, X.; Stanton, R.C. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J. Biol. Chem. 1998, 10609–10617. [Google Scholar] [CrossRef]
- Warbug, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Mischen, B.T.; Follmar, K.E.; Moyer, K.E.; Buehrer, B.; Olbrich, K.C.; Levin, L.S.; Klitzman, B.; Erdmann, D. Metabolic and functional characterization of human adipose-derived stem cells in tissue engineering. Plast. Reconstr. Surg. 2008, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2001, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Landrier, J.-F.; Kasiri, E.; Karkeni, E.; Mihály, J.; Béke, G.; Weiss, K.; Lucas, R.; Aydemir, G.; Salles, J.; Walrand, S.; et al. Reduced adiponectin expression after high-fat diet is associated with selective up-regulation of ALDH1A1 and further retinoic acid receptor signaling in adipose tissue. Fed. Am. Soc. Exp. Biol. 2017, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Haenisch, M.; Treuting, P.M.; Brabb, T.; Goldstein, A.S.; Berkseth, K.; Amory, J.K.; Paik, J. Pharmacological inhibition of ALDH1A enzymes suppresses weight gain in a mouse model of diet-induced obesity. Obes. Res. Clin. Pract. 2018, 93–101. [Google Scholar] [CrossRef]
- Lu, G.; Ota, A.; Ren, S.; Franklin, S.; Rau, C.D.; Ping, P.; Lane, T.F.; Zhou, Z.H.; Reue, K.; Lusis, A.J.; et al. PPM1l encodes an inositol requiring-protein 1 (IRE1) specific phosphatase that regulates the functional outcome of the ER stress response. Mol. Metab. 2008, 405–416. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, J.; Lum, P.Y.; Yang, X.; Pinto, S.; MacNeil, D.J.; Zhang, C.; Lamb, J.; Edwards, S.; Sieberts, S.K.; et al. Variations in DNA elucidate molecular networks that cause disease. Nature 2008, 429–435. [Google Scholar] [CrossRef]
- Hao, Q.; Yadav, R.; Basse, A.L.; Petersen, S.; Sonne, S.B.; Rasmussen, S.; Zhu, Q.; Lu, Z.; Wang, J.; Audouze, K.; et al. Transcriptome profiling of brown adipose tissue during cold exposure reveals extensive regulation of glucose metabolism. Am. J. Physiol. Endocrinol. Metab. 2014, E380–E392. [Google Scholar] [CrossRef]
- Buresova, J.; Janovska, P.; Kuda, O.; Krizova, J.; der Stelt, I.R.; Keijer, J.; Hansikova, H.; Rossmeisl, M.; Kopecky, J. Postnatal induction of muscle fatty acid oxidation in mice differing in propensity to obesity: A role of pyruvate dehydrogenase. Int. J. Obes. 2018, 235–244. [Google Scholar] [CrossRef]
- Kumar, D.; Bansal, G.; Narang, A.; Basak, T.; Abbas, T.; Dash, D. Integrating transcriptome and proteome profiling: Strategies and applications. Proteomics 2016, 2533–2544. [Google Scholar] [CrossRef]
- Peters, K.; Salamon, A.; van Vlierberghe, S.; Rychly, J.; Kreutzer, M.; Neumann, H.G.; Schacht, E.; Dubruel, P. A new approach for adipose tissue regeneration based on human mesenchymal stem cells in contact to hydrogels-an in vitro study. Adv. Eng. Mater. 2009, B155–B161. [Google Scholar] [CrossRef]
- Salamon, A.; Jonitz-Heincke, A.; Adam, S.; Rychly, J.; Müller-Hilke, B.; Bader, R.; Lochner, K.; Peters, K. Articular cartilage-derived cells hold a strong osteogenic differentiation potential in comparison to mesenchymal stem cells in vitro. Exp. Cell Res. 2013, 2856–2865. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.; Didier, N.; Denten, M. Determination of cell number in monolayer cultures. Anal. Biochem. 1986, 109–113. [Google Scholar] [CrossRef]
- Noeske, K. Die Bindung von Kristallviolett an Desoxyribonukleinsäure. Histochemie 1966, 7, 273–287. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific. Transcriptome Analysis Console (TAC) Software; Thermo Fisher Scientific: Waltham, MA, USA, 2019. [Google Scholar]
- Ritchie, M.E.; Phipson, B.; Di, W.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, e47. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 29–34. [Google Scholar] [CrossRef]
- Pico, A.R.; Kelder, T.; van Iersel, M.P.; Hanspers, K.; Conklin, B.R.; Evelo, C. WikiPathways: Pathway editing for the people. PLoS Biol. 2008, e184. [Google Scholar] [CrossRef]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 2129–2141. [Google Scholar] [CrossRef]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 2912–2914. [Google Scholar] [CrossRef]
| Cell Cycles Phases | |||||
|---|---|---|---|---|---|
| sub G1 | G0/G1 | S | G2 | ||
| 0 d | [%] | 1.4 (± 0.1) | 89.5 (± 1.3) | 2.3 (± 0.6) | 6.1 (± 0.5) |
| 1 d | control [%] | 1.9 (± 0.4) | 88.7 (± 1.3) | 2.9 (± 0.3) | 6.0 (± 0.9) |
| TGF-ß1 [%] | 0.9 (± 0.5) | 86.1 (± 3.9) | 5.7 (± 3.1) | 4.1 (± 1.6) | |
| 3 d | control [%] | 2.3 (± 0.2) | 89.3 ( ±1.0) | 2.5 (± 0.5) | 5.8 (± 0.7) |
| TGF-ß1 [%] | 0.9 (± 0.3) | 89.4 (± 1.2) | 2.0 (± 0.4) | 6.3 (± 0.9) | |
| 7 d | control [%] | 2.5 (± 0.2) | 89.9 (± 0.8) | 2.3 (± 0.5) | 5.5 (± 0.5) |
| TGF-ß1 [%] | 1.5 (± 0.2) | 87.9 (± 1.7) | 3.4 (± 1.0) | 6.2 (± 0.5) | |
| Energy Metabolism | Amino Acid Metabolism | ||
|---|---|---|---|
| Upregulated | Downregulated | Upregulated | Downregulated |
| PPP3CA | PPARG | ACLY | ADH1C |
| PPP3R1 | MEF2C | ALDH18A1 | ALDH1A1 |
| PPP3CC | NRF1 | AOC3 | ALDH7A1 |
| PPARD | PPARGC1A | ARG2 | AUH |
| PRKAB2 | SIRT3 | ASNS | FAH |
| PRKAG2 | TFAM | BCAT1 | GLUL |
| CBS | HIBADH | ||
| CTH | MCCC1 | ||
| EPRS | PDK4 | ||
| GCLM | PPM1L | ||
| GLS | |||
| GOT1 | |||
| GPT2 | |||
| GSR | |||
| IARS | |||
| ODC1 | |||
| P4HA2 | |||
| PYCR1 | |||
| SRM | |||
| WARS | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hahn, O.; Ingwersen, L.-C.; Soliman, A.; Hamed, M.; Fuellen, G.; Wolfien, M.; Scheel, J.; Wolkenhauer, O.; Koczan, D.; Kamp, G.; et al. TGF-ß1 Induces Changes in the Energy Metabolism of White Adipose Tissue-Derived Human Adult Mesenchymal Stem/Stromal Cells In Vitro. Metabolites 2020, 10, 59. https://doi.org/10.3390/metabo10020059
Hahn O, Ingwersen L-C, Soliman A, Hamed M, Fuellen G, Wolfien M, Scheel J, Wolkenhauer O, Koczan D, Kamp G, et al. TGF-ß1 Induces Changes in the Energy Metabolism of White Adipose Tissue-Derived Human Adult Mesenchymal Stem/Stromal Cells In Vitro. Metabolites. 2020; 10(2):59. https://doi.org/10.3390/metabo10020059
Chicago/Turabian StyleHahn, Olga, Lena-Christin Ingwersen, Abdelrahman Soliman, Mohamed Hamed, Georg Fuellen, Markus Wolfien, Julia Scheel, Olaf Wolkenhauer, Dirk Koczan, Günter Kamp, and et al. 2020. "TGF-ß1 Induces Changes in the Energy Metabolism of White Adipose Tissue-Derived Human Adult Mesenchymal Stem/Stromal Cells In Vitro" Metabolites 10, no. 2: 59. https://doi.org/10.3390/metabo10020059
APA StyleHahn, O., Ingwersen, L.-C., Soliman, A., Hamed, M., Fuellen, G., Wolfien, M., Scheel, J., Wolkenhauer, O., Koczan, D., Kamp, G., & Peters, K. (2020). TGF-ß1 Induces Changes in the Energy Metabolism of White Adipose Tissue-Derived Human Adult Mesenchymal Stem/Stromal Cells In Vitro. Metabolites, 10(2), 59. https://doi.org/10.3390/metabo10020059






