Integrative Metabolomics to Identify Molecular Signatures of Responses to Vaccines and Infections
Abstract
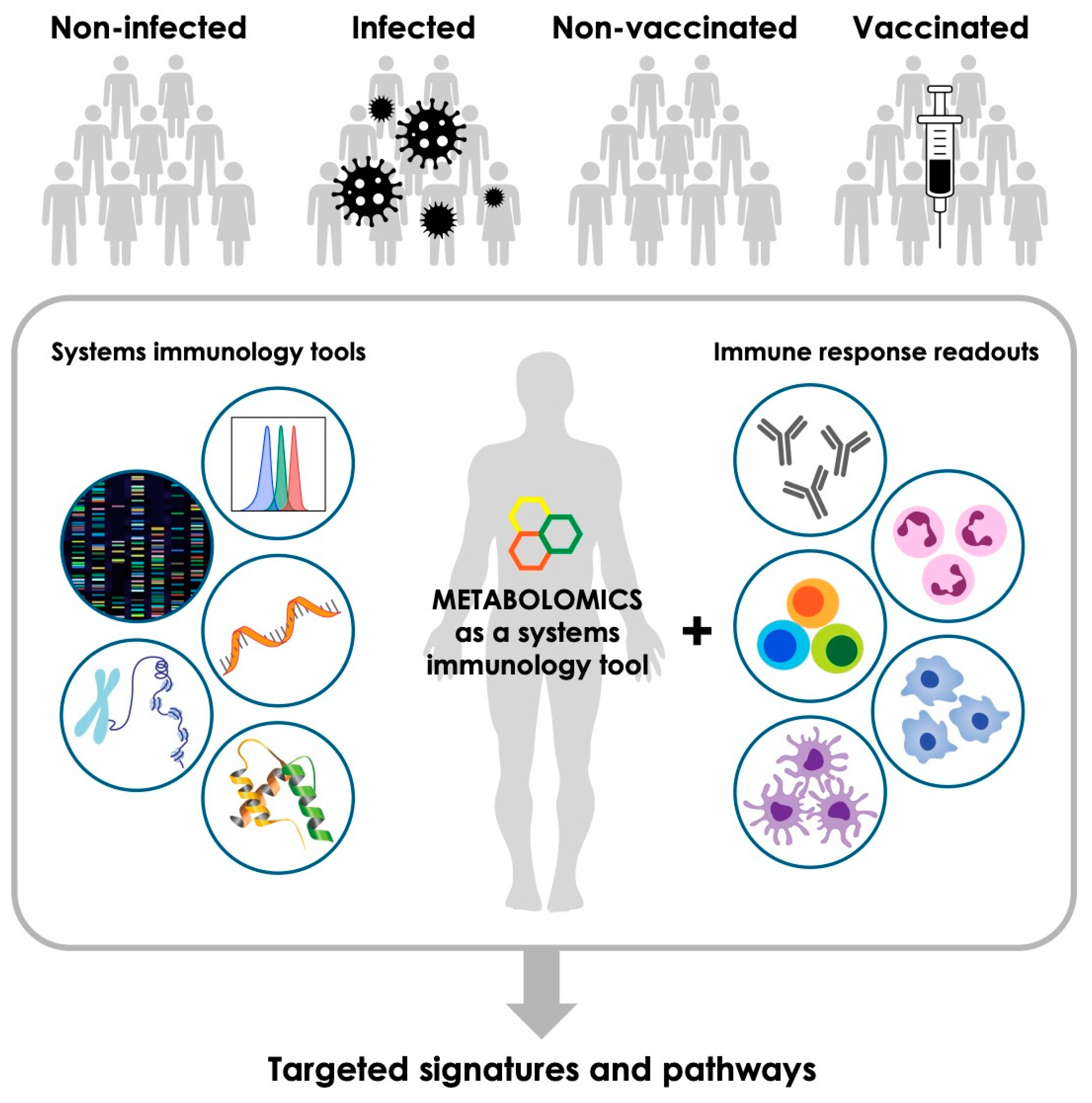
1. Introduction
2. Metabolomics—An Emerging Tool to Complement Other Systems Immunology Platforms
3. Immunometabolism
4. Impact of Infection on Host Metabolic Signatures
5. Metabolic Signatures of Vaccine-Induced Responses
6. Shared Metabolic Pathways Across Infection and Vaccination Studies
7. Integrative Metabolomics—Challenges and Emerging Horizons
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Viant, M.R.; Kurland, I.J.; Jones, M.R.; Dunn, W.B. How close are we to complete annotation of metabolomes? Curr. Opin. Chem. Biol. 2017, 36, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.V.; Hu, Y.J. Integrative Analysis of Multi-omics Data for Discovery and Functional Studies of Complex Human Diseases. Adv. Genet. 2016, 93, 147–190. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, D.E. An Introduction to Metabolic Pathways. S. Dagley, Donald E. Nicholson. Q. Rev. Biol. 1971, 46, 288–290. [Google Scholar] [CrossRef]
- Veenstra, T.D. Metabolomics: The final frontier? Genome Med. 2012, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Xu, Y.; Chen, Y.; Zhan, Y.; Wei, X.; Li, L.; Wang, D.; He, P.; Li, S.; Chen, S. Metabolomics analysis reveals global acetoin stress response of Bacillus licheniformis. Metabolomics 2019, 15, 25. [Google Scholar] [CrossRef] [PubMed]
- Blighe, K.; Chawes, B.L.; Kelly, R.S.; Mirzakhani, H.; McGeachie, M.; Litonjua, A.A.; Weiss, S.T.; Lasky-Su, J.A. Vitamin D prenatal programming of childhood metabolomics profiles at age 3 y. Am. J. Clin. Nutr. 2017, 106, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Li, T.; Song, X.; Huang, L.; Zang, Q.; Xu, J.; Bi, N.; Jiao, G.; Hao, Y.; Chen, Y.; et al. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc. Natl. Acad. Sci. USA 2019, 116, 52–57. [Google Scholar] [CrossRef]
- Liu, N.N.; Acosta-Zaldivar, M.; Qi, W.; Diray-Arce, J.; Walker, L.A.; Kottom, T.J.; Kelly, R.; Yuan, M.; Asara, J.M.; Lasky-Su, J.A.; et al. Phosphoric Metabolites Link Phosphate Import and Polysaccharide Biosynthesis for Candida albicans Cell Wall Maintenance. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Beale, D.J.; Oh, D.Y.; Karpe, A.V.; Tai, C.; Dunn, M.S.; Tilmanis, D.; Palombo, E.A.; Hurt, A.C. Untargeted metabolomics analysis of the upper respiratory tract of ferrets following influenza A virus infection and oseltamivir treatment. Metabolomics 2019, 15, 33. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sullivan, N.L.; Rouphael, N.; Yu, T.; Banton, S.; Maddur, M.S.; McCausland, M.; Chiu, C.; Canniff, J.; Dubey, S.; et al. Metabolic Phenotypes of Response to Vaccination in Humans. Cell 2017, 169, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Abu-Farha, M.; Thanaraj, T.A.; Qaddoumi, M.G.; Hashem, A.; Abubaker, J.; Al-Mulla, F. The Role of Lipid Metabolism in COVID-19 Virus Infection and as a Drug Target. Int. J. Mol. Sci. 2020, 21, 3544. [Google Scholar] [CrossRef] [PubMed]
- Sharma, L.; Prakash, H. Sphingolipids Are Dual Specific Drug Targets for the Management of Pulmonary Infections: Perspective. Front. Immunol. 2017, 8, 378. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.L.; Dickinson, P.; Forster, T.; Craigon, M.; Ross, A.; Khondoker, M.R.; France, R.; Ivens, A.; Lynn, D.J.; Orme, J.; et al. Identification of a human neonatal immune-metabolic network associated with bacterial infection. Nat. Commun. 2014, 5, 4649. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Zhang, K.; Min, J.; Chen, C.; Cao, Y.; Ding, C.; Liu, W.; Li, J. Metabolomic Analysis of Influenza A Virus A/WSN/1933 (H1N1) Infected A549 Cells during First Cycle of Viral Replication. Viruses 2019, 11, 1007. [Google Scholar] [CrossRef] [PubMed]
- Niki, M.; Yoshiyama, T.; Nagai, H.; Miyamoto, Y.; Niki, M.; Oinuma, K.I.; Tsubouchi, T.; Kaneko, Y.; Matsumoto, S.; Sasaki, Y.; et al. Nutritional status positively impacts humoral immunity against its Mycobacterium tuberculosis, disease progression, and vaccine development. PLoS ONE 2020, 15, e0237062. [Google Scholar] [CrossRef]
- German, J.B.; Gillies, L.A.; Smilowitz, J.T.; Zivkovic, A.M.; Watkins, S.M. Lipidomics and lipid profiling in metabolomics. Curr. Opin. Lipidol. 2007, 18, 66–71. [Google Scholar] [CrossRef]
- Chen, R.; Mias, G.I.; Li-Pook-Than, J.; Jiang, L.; Lam, H.Y.; Chen, R.; Miriami, E.; Karczewski, K.J.; Hariharan, M.; Dewey, F.E.; et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012, 148, 1293–1307. [Google Scholar] [CrossRef]
- Petersen, A.K.; Zeilinger, S.; Kastenmuller, G.; Romisch-Margl, W.; Brugger, M.; Peters, A.; Meisinger, C.; Strauch, K.; Hengstenberg, C.; Pagel, P.; et al. Epigenetics meets metabolomics: An epigenome-wide association study with blood serum metabolic traits. Hum. Mol. Genet. 2014, 23, 534–545. [Google Scholar] [CrossRef]
- Nakaya, H.I.; Li, S.; Pulendran, B. Systems vaccinology: Learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Shao, Y.; Zhao, X.; Hong, C.S.; Wang, F.; Lu, X.; Li, J.; Ye, G.; Yan, M.; Zhuang, Z.; et al. Integration of Metabolomics and Transcriptomics Reveals Major Metabolic Pathways and Potential Biomarker Involved in Prostate Cancer. Mol. Cell. Proteom. 2016, 15, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2018, 24, 1248–1259. [Google Scholar] [CrossRef] [PubMed]
- Hakimi, A.A.; Reznik, E.; Lee, C.H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.W.; Chen, Y.T.; Hsieh, Y.J.; Chang, K.P.; Hsueh, P.C.; Chen, T.W.; Yu, J.S.; Chang, Y.S.; Li, L.; Wu, C.C. Integrated analyses utilizing metabolomics and transcriptomics reveal perturbation of the polyamine pathway in oral cavity squamous cell carcinoma. Anal. Chim. Acta 2019, 1050, 113–122. [Google Scholar] [CrossRef]
- Zhang, G.; He, P.; Tan, H.; Budhu, A.; Gaedcke, J.; Ghadimi, B.M.; Ried, T.; Yfantis, H.G.; Lee, D.H.; Maitra, A.; et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res. 2013, 19, 4983–4993. [Google Scholar] [CrossRef]
- Yang, K.; Xia, B.; Wang, W.; Cheng, J.; Yin, M.; Xie, H.; Li, J.; Ma, L.; Yang, C.; Li, A.; et al. A Comprehensive Analysis of Metabolomics and Transcriptomics in Cervical Cancer. Sci. Rep. 2017, 7, 43353. [Google Scholar] [CrossRef]
- Ghosh, N.; Dutta, M.; Singh, B.; Banerjee, R.; Bhattacharyya, P.; Chaudhury, K. Transcriptomics, proteomics and metabolomics driven biomarker discovery in COPD: An update. Expert Rev. Mol. Diagn. 2016, 16, 897–913. [Google Scholar] [CrossRef]
- Kuruvilla, M.E.; Lee, F.E.; Lee, G.B. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin. Rev. Allergy Immunol. 2019, 56, 219–233. [Google Scholar] [CrossRef]
- Kelly, R.S.; Chawes, B.L.; Blighe, K.; Virkud, Y.V.; Croteau-Chonka, D.C.; McGeachie, M.J.; Clish, C.B.; Bullock, K.; Celedon, J.C.; Weiss, S.T.; et al. An Integrative Transcriptomic and Metabolomic Study of Lung Function in Children With Asthma. Chest 2018, 154, 335–348. [Google Scholar] [CrossRef]
- Lee, J.; Banerjee, D. Metabolomics and the Microbiome as Biomarkers in Sepsis. Crit. Care Clin. 2020, 36, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Liao, S.; Li, P.; Zhang, Q.; Lv, Y.; Fu, X.; Yang, M.; Wang, J.; Kong, L. Metabolomics Coupled with Transcriptomics Approach Deciphering Age Relevance in Sepsis. Aging Dis. 2019, 10, 854–870. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.Z.; Chen, G.; Hong, Q.; Huang, S.; Smith, H.M.; Shah, R.D.; Scholz, M.; Ferguson, J.F. Multi-Omic Analysis of the Microbiome and Metabolome in Healthy Subjects Reveals Microbiome-Dependent Relationships Between Diet and Metabolites. Front. Genet. 2019, 10, 454. [Google Scholar] [CrossRef] [PubMed]
- Zierer, J.; Jackson, M.A.; Kastenmuller, G.; Mangino, M.; Long, T.; Telenti, A.; Mohney, R.P.; Small, K.S.; Bell, J.T.; Steves, C.J.; et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018, 50, 790–795. [Google Scholar] [CrossRef]
- Wilmanski, T.; Rappaport, N.; Earls, J.C.; Magis, A.T.; Manor, O.; Lovejoy, J.; Omenn, G.S.; Hood, L.; Gibbons, S.M.; Price, N.D. Blood metabolome predicts gut microbiome alpha-diversity in humans. Nat. Biotechnol. 2019, 37, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Dao, M.C.; Sokolovska, N.; Brazeilles, R.; Affeldt, S.; Pelloux, V.; Prifti, E.; Chilloux, J.; Verger, E.O.; Kayser, B.D.; Aron-Wisnewsky, J.; et al. A Data Integration Multi-Omics Approach to Study Calorie Restriction-Induced Changes in Insulin Sensitivity. Front. Physiol. 2018, 9, 1958. [Google Scholar] [CrossRef] [PubMed]
- Kiebish, M.A.; Cullen, J.; Mishra, P.; Ali, A.; Milliman, E.; Rodrigues, L.O.; Chen, E.Y.; Tolstikov, V.; Zhang, L.; Panagopoulos, K.; et al. Multi-omic serum biomarkers for prognosis of disease progression in prostate cancer. J. Transl. Med. 2020, 18, 10. [Google Scholar] [CrossRef]
- Khan, S.R.; Manialawy, Y.; Wheeler, M.B.; Cox, B.J. Unbiased data analytic strategies to improve biomarker discovery in precision medicine. Drug Discov. Today 2019, 24, 1735–1748. [Google Scholar] [CrossRef]
- Lone, A.M.; Tasken, K. Proinflammatory and immunoregulatory roles of eicosanoids in T cells. Front. Immunol. 2013, 4, 130. [Google Scholar] [CrossRef]
- Yui, K.; Imataka, G.; Nakamura, H.; Ohara, N.; Naito, Y. Eicosanoids Derived From Arachidonic Acid and Their Family Prostaglandins and Cyclooxygenase in Psychiatric Disorders. Curr. Neuropharmacol. 2015, 13, 776–785. [Google Scholar] [CrossRef]
- Wijnands, K.A.; Castermans, T.M.; Hommen, M.P.; Meesters, D.M.; Poeze, M. Arginine and citrulline and the immune response in sepsis. Nutrients 2015, 7, 1426–1463. [Google Scholar] [CrossRef] [PubMed]
- Nikolaus, S.; Schulte, B.; Al-Massad, N.; Thieme, F.; Schulte, D.M.; Bethge, J.; Rehman, A.; Tran, F.; Aden, K.; Hasler, R.; et al. Increased Tryptophan Metabolism Is Associated With Activity of Inflammatory Bowel Diseases. Gastroenterology 2017, 153, 1504–1516. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Y.; Peng, J.H.; Ma, X.J.; Zhang, Y.N.; Zhu, W.; He, X.X.; Ying, L.W.; Bao, Y.Q.; Zhou, J.; Jia, W.P. Metabolic perturbations of post-load hyperglycemia vs. fasting hyperglycemia. Acta Pharmacol. Sin. 2019, 40, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Long, N.P.; Nghi, T.D.; Kang, Y.P.; Anh, N.H.; Kim, H.M.; Park, S.K.; Kwon, S.W. Toward a Standardized Strategy of Clinical Metabolomics for the Advancement of Precision Medicine. Metabolites 2020, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Bergman, M.; Abdul-Ghani, M.; DeFronzo, R.A.; Manco, M.; Sesti, G.; Fiorentino, T.V.; Ceriello, A.; Rhee, M.; Phillips, L.S.; Chung, S.; et al. Review of methods for detecting glycemic disorders. Diabetes Res. Clin. Pract. 2020, 165, 108233. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Shetty, V.; Maresso, A.W. Global metabolomic analysis of a mammalian host infected with Bacillus anthracis. Infect. Immun. 2015, 83, 4811–4825. [Google Scholar] [CrossRef]
- Lee, A.H.; Shannon, C.P.; Amenyogbe, N.; Bennike, T.B.; Diray-Arce, J.; Idoko, O.T.; Gill, E.E.; Ben-Othman, R.; Pomat, W.S.; van Haren, S.D.; et al. Dynamic molecular changes during the first week of human life follow a robust developmental trajectory. Nat. Commun. 2019, 10, 1092. [Google Scholar] [CrossRef]
- Whittaker, E.; Goldblatt, D.; McIntyre, P.; Levy, O. Neonatal Immunization: Rationale, Current State, and Future Prospects. Front. Immunol. 2018, 9, 532. [Google Scholar] [CrossRef]
- Idoko, O.T.; Smolen, K.K.; Wariri, O.; Imam, A.; Shannon, C.P.; Dibassey, T.; Diray-Arce, J.; Darboe, A.; Strandmark, J.; Ben-Othman, R.; et al. Clinical Protocol for a Longitudinal Cohort Study Employing Systems Biol. to Identify Markers of Vaccine Immunogenicity in Newborn Infants in The Gambia and Papua New Guinea. Front. Pediatr. 2020, 8, 197. [Google Scholar] [CrossRef]
- Kumar, V. Immunometabolism: Another Road to Sepsis and Its Therapeutic Targeting. Inflammation 2019, 42, 765–788. [Google Scholar] [CrossRef]
- Conti, M.G.; Angelidou, A.; Diray-Arce, J.; Smolen, K.K.; Lasky-Su, J.; De Curtis, M.; Levy, O. Immunometabolic approaches to prevent, detect, and treat neonatal sepsis. Pediatr. Res. 2020, 87, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Joosten, L.A.; Latz, E.; Mills, K.H.; Natoli, G.; Stunnenberg, H.G.; O’Neill, L.A.; Xavier, R.J. Trained immunity: A program of innate immune memory in health and disease. Science 2016, 352, aaf1098. [Google Scholar] [CrossRef] [PubMed]
- Dang, E.V.; Barbi, J.; Yang, H.Y.; Jinasena, D.; Yu, H.; Zheng, Y.; Bordman, Z.; Fu, J.; Kim, Y.; Yen, H.R.; et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 2011, 146, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Ahmed, R. AMPK: A metabolic switch for CD8+ T-cell memory. Eur. J. Immunol. 2013, 43, 878–881. [Google Scholar] [CrossRef]
- Lochner, M.; Berod, L.; Sparwasser, T. Fatty acid metabolism in the regulation of T cell function. Trends Immunol. 2015, 36, 81–91. [Google Scholar] [CrossRef]
- O’Neill, L.A.J.; Kishton, R.J.; Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016, 16, 553–565. [Google Scholar] [CrossRef]
- Mayer, K.A.; Stöckl, J.; Zlabinger, G.J.; Gualdoni, G.A. Hijacking the Supplies: Metabolism as a Novel Facet of Virus-Host Interaction. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Eisenreich, W.; Rudel, T.; Heesemann, J.; Goebel, W. How Viral and Intracellular Bacterial Pathogens Reprogram the Metabolism of Host Cells to Allow Their Intracellular Replication. Front. Cell. Infect. Microbiol. 2019, 9, 42. [Google Scholar] [CrossRef]
- Robinson, J.I.; Weir, W.H.; Crowley, J.R.; Hink, T.; Reske, K.A.; Kwon, J.H.; Burnham, C.D.; Dubberke, E.R.; Mucha, P.J.; Henderson, J.P. Metabolomic networks connect host-microbiome processes to human Clostridioides difficile infections. J. Clin. Investig. 2019, 129, 3792–3806. [Google Scholar] [CrossRef]
- Zhou, P.; Zhou, N.; Shao, L.; Li, J.; Liu, S.; Meng, X.; Duan, J.; Xiong, X.; Huang, X.; Chen, Y.; et al. Diagnosis of Clostridium difficile infection using an UPLC-MS based metabolomics method. Metabolomics 2018, 14, 102. [Google Scholar] [CrossRef]
- Lussu, M.; Camboni, T.; Piras, C.; Serra, C.; Del Carratore, F.; Griffin, J.; Atzori, L.; Manzin, A. (1)H NMR spectroscopy-based metabolomics analysis for the diagnosis of symptomatic E. coli-associated urinary tract infection (UTI). BMC Microbiol. 2017, 17, 201. [Google Scholar] [CrossRef] [PubMed]
- Puebla-Barragan, S.; Renaud, J.; Sumarah, M.; Reid, G. Malodorous biogenic amines in Escherichia coli-caused urinary tract infections in women-a metabolomics approach. Sci. Rep. 2020, 10, 9703. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.M.; Siddiqa, A.; Jones, D.P.; Liu, K.; Kempker, R.R.; Nizam, A.; Shah, N.S.; Ismail, N.; Ouma, S.G.; Tukvadze, N.; et al. Tryptophan catabolism reflects disease activity in human tuberculosis. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, L.P.; Fischer, S.M.; Marrero, J.; Nathan, C.; Ehrt, S.; Rhee, K.Y. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 2010, 17, 1122–1131. [Google Scholar] [CrossRef]
- Cho, Y.; Park, Y.; Sim, B.; Kim, J.; Lee, H.; Cho, S.-N.; Kang, Y.A.; Lee, S.-G. Identification of serum biomarkers for active pulmonary tuberculosis using a targeted metabolomics approach. Sci. Rep. 2020, 10, 3825. [Google Scholar] [CrossRef]
- Frediani, J.K.; Jones, D.P.; Tukvadze, N.; Uppal, K.; Sanikidze, E.; Kipiani, M.; Tran, V.T.; Hebbar, G.; Walker, D.I.; Kempker, R.R.; et al. Plasma metabolomics in human pulmonary tuberculosis disease: A pilot study. PLoS ONE 2014, 9, e108854. [Google Scholar] [CrossRef]
- Zhou, A.; Ni, J.; Xu, Z.; Wang, Y.; Lu, S.; Sha, W.; Karakousis, P.C.; Yao, Y.-F. Application of (1)h NMR spectroscopy-based metabolomics to sera of tuberculosis patients. J. Proteome Res. 2013, 12, 4642–4649. [Google Scholar] [CrossRef]
- Weiner, J., 3rd; Parida, S.K.; Maertzdorf, J.; Black, G.F.; Repsilber, D.; Telaar, A.; Mohney, R.P.; Arndt-Sullivan, C.; Ganoza, C.A.; Fae, K.C.; et al. Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PLoS ONE 2012, 7, e40221. [Google Scholar] [CrossRef]
- Sun, L.; Li, J.Q.; Ren, N.; Qi, H.; Dong, F.; Xiao, J.; Xu, F.; Jiao, W.W.; Shen, C.; Song, W.Q.; et al. Utility of Novel Plasma Metabolic Markers in the Diagnosis of Pediatric Tuberculosis: A Classification and Regression Tree Analysis Approach. J. Proteome Res. 2016, 15, 3118–3125. [Google Scholar] [CrossRef]
- Weiner, J., 3rd; Maertzdorf, J.; Sutherland, J.S.; Duffy, F.J.; Thompson, E.; Suliman, S.; McEwen, G.; Thiel, B.; Parida, S.K.; Zyla, J.; et al. Metabolite changes in blood predict the onset of tuberculosis. Nat. Commun. 2018, 9, 5208. [Google Scholar] [CrossRef]
- Duffy, F.J.; Weiner, J., 3rd; Hansen, S.; Tabb, D.L.; Suliman, S.; Thompson, E.; Maertzdorf, J.; Shankar, S.; Tromp, G.; Parida, S.; et al. Immunometabolic Signatures Predict Risk of Progression to Active Tuberculosis and Disease Outcome. Front. Immunol. 2019, 10, 527. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; VanderVen, B.C.; Fahey, R.J.; Russell, D.G. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J. Biol. Chem. 2013, 288, 6788–6800. [Google Scholar] [CrossRef] [PubMed]
- Kinsella, R.J.; Fitzpatrick, D.A.; Creevey, C.J.; McInerney, J.O. Fatty acid biosynthesis in Mycobacterium tuberculosis: Lateral gene transfer, adaptive evolution, and gene duplication. Proc. Natl. Acad. Sci. USA 2003, 100, 10320–10325. [Google Scholar] [CrossRef] [PubMed]
- MacGurn, J.A.; Cox, J.S. A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect. Immun. 2007, 75, 2668–2678. [Google Scholar] [CrossRef] [PubMed]
- Garg, S.K.; Volpe, E.; Palmieri, G.; Mattei, M.; Galati, D.; Martino, A.; Piccioni, M.S.; Valente, E.; Bonanno, E.; De Vito, P.; et al. Sphingosine 1–Phosphate Induces Antimicrobial Activity Both In Vitro and In Vivo. J. Infect. Dis. 2004, 189, 2129–2138. [Google Scholar] [CrossRef]
- Shrinet, J.; Shastri, J.S.; Gaind, R.; Bhavesh, N.S.; Sunil, S. Serum metabolomics analysis of patients with chikungunya and dengue mono/co-infections reveals distinct metabolite signatures in the three disease conditions. Sci. Rep. 2016, 6, 36833. [Google Scholar] [CrossRef]
- Cui, L.; Lee, Y.H.; Kumar, Y.; Xu, F.; Lu, K.; Ooi, E.E.; Tannenbaum, S.R.; Ong, C.N. Serum metabolome and lipidome changes in adult patients with primary dengue infection. PLoS Negl. Trop. Dis. 2013, 7, e2373. [Google Scholar] [CrossRef]
- Cui, L.; Lee, Y.H.; Thein, T.L.; Fang, J.; Pang, J.; Ooi, E.E.; Leo, Y.S.; Ong, C.N.; Tannenbaum, S.R. Serum Metabolomics Reveals Serotonin as a Predictor of Severe Dengue in the Early Phase of Dengue Fever. PLoS Negl. Trop. Dis. 2016, 10, e0004607. [Google Scholar] [CrossRef]
- Cui, L.; Pang, J.; Lee, Y.H.; Ooi, E.E.; Ong, C.N.; Leo, Y.S.; Tannenbaum, S.R. Serum metabolome changes in adult patients with severe dengue in the critical and recovery phases of dengue infection. PLoS Negl. Trop. Dis. 2018, 12, e0006217. [Google Scholar] [CrossRef]
- Dickens, A.M.; Anthony, D.C.; Deutsch, R.; Mielke, M.M.; Claridge, T.D.; Grant, I.; Franklin, D.; Rosario, D.; Marcotte, T.; Letendre, S.; et al. Cerebrospinal fluid metabolomics implicate bioenergetic adaptation as a neural mechanism regulating shifts in cognitive states of HIV-infected patients. AIDS 2015, 29, 559–569. [Google Scholar] [CrossRef]
- Chan, E.Y.; Sutton, J.N.; Jacobs, J.M.; Bondarenko, A.; Smith, R.D.; Katze, M.G. Dynamic host energetics and cytoskeletal proteomes in human immunodeficiency virus type 1-infected human primary CD4 cells: Analysis by multiplexed label-free mass spectrometry. J. Virol. 2009, 83, 9283–9295. [Google Scholar] [CrossRef]
- Hollenbaugh, J.A.; Munger, J.; Kim, B. Metabolite profiles of human immunodeficiency virus infected CD4+ T cells and macrophages using LC-MS/MS analysis. Virology 2011, 415, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Scarpellini, B.; Zanoni, M.; Sucupira, M.C.; Truong, H.M.; Janini, L.M.; Segurado, I.D.; Diaz, R.S. Plasma Metabolomics Biosignature According to HIV Stage of Infection, Pace of Disease Progression, Viremia Level and Immunological Response to Treatment. PLoS ONE 2016, 11, e0161920. [Google Scholar] [CrossRef] [PubMed]
- Serezani, C.H.; Ballinger, M.N.; Aronoff, D.M.; Peters-Golden, M. Cyclic AMP: Master regulator of innate immune cell function. Am. J. Respir. Cell Mol. Biol. 2008, 39, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Banoei, M.M.; Vogel, H.J.; Weljie, A.M.; Kumar, A.; Yende, S.; Angus, D.C.; Winston, B.W.; Canadian Critical Care Translational Biology Group. Plasma metabolomics for the diagnosis and prognosis of H1N1 influenza pneumonia. Crit. Care 2017, 21, 97. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Fang, P.; He, R.; Li, M.; Yu, H.; Zhou, L.; Yi, Y.; Wang, F.; Rong, Y.; Zhang, Y.; et al. O-GlcNAc transferase promotes influenza A virus-induced cytokine storm by targeting interferon regulatory factor-5. Sci. Adv. 2020, 6, eaaz7086. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zheng, D.; Lee, Y.H.; Chan, T.K.; Kumar, Y.; Ho, W.E.; Chen, J.Z.; Tannenbaum, S.R.; Ong, C.N. Metabolomics Investigation Reveals Metabolite Mediators Associated with Acute Lung Injury and Repair in a Murine Model of Influenza Pneumonia. Sci. Rep. 2016, 6, 26076. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; Hlh Across Speciality Collaboration, U.K. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Song, J.W.; Lam, S.M.; Fan, X.; Cao, W.J.; Wang, S.Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.P.; Xu, Z.; et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 2020, 32, 188–202.e5. [Google Scholar] [CrossRef]
- Hong, W. Combating COVID-19 with Chloroquine. J. Mol. Cell Biol. 2020, 12, 249–250. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Shu, T.; Yang, X.; Song, J.-X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef]
- Bogdan, C.; Rollinghoff, M.; Diefenbach, A. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr. Opin. Immunol. 2000, 12, 64–76. [Google Scholar] [CrossRef]
- Appelberg, R. Macrophage nutriprive antimicrobial mechanisms. J. Leukoc. Biol. 2006, 79, 1117–1128. [Google Scholar] [CrossRef]
- Spooner, R.; Yilmaz, O. The role of reactive-oxygen-species in microbial persistence and inflammation. Int. J. Mol. Sci. 2011, 12, 334–352. [Google Scholar] [CrossRef]
- Nathan, C.; Shiloh, M.U. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad Sci. USA 2000, 97, 8841–8848. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Escoll, P.; Buchrieser, C. Metabolic reprogramming of host cells upon bacterial infection: Why shift to a Warburg-like metabolism? FEBS J. 2018, 285, 2146–2160. [Google Scholar] [CrossRef]
- Hu, J.; Jin, K.; He, Z.G.; Zhang, H. Citrate lyase CitE in Mycobacterium tuberculosis contributes to mycobacterial survival under hypoxic conditions. PLoS ONE 2020, 15, e0230786. [Google Scholar] [CrossRef]
- Dai, Z.L.; Li, X.L.; Xi, P.B.; Zhang, J.; Wu, G.; Zhu, W.Y. L-Glutamine regulates amino acid utilization by intestinal bacteria. Amino Acids 2013, 45, 501–512. [Google Scholar] [CrossRef]
- Eisenreich, W.; Heesemann, J.; Rudel, T.; Goebel, W. Metabolic host responses to infection by intracellular bacterial pathogens. Front. Cell. Infect. Microbiol. 2013, 3, 24. [Google Scholar] [CrossRef] [PubMed]
- Marrero, J.; Rhee, K.Y.; Schnappinger, D.; Pethe, K.; Ehrt, S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. USA 2010, 107, 9819–9824. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479–480, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Useros, N.; Naranjo-Gomez, M.; Erkizia, I.; Puertas, M.C.; Borras, F.E.; Blanco, J.; Martinez-Picado, J. HIV and mature dendritic cells: Trojan exosomes riding the Trojan horse? PLoS Pathog. 2010, 6, e1000740. [Google Scholar] [CrossRef] [PubMed]
- Sitole, L.J.; Williams, A.A.; Meyer, D. Metabonomic analysis of HIV-infected biofluids. Mol. Biosyst. 2013, 9, 18–28. [Google Scholar] [CrossRef]
- Nakaya, H.I.; Pulendran, B. Vaccinology in the era of high-throughput biology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370. [Google Scholar] [CrossRef]
- Nakaya, H.I.; Pulendran, B. Systems vaccinology: Its promise and challenge for HIV vaccine development. Curr. Opin. HIV AIDS 2012, 7, 24–31. [Google Scholar] [CrossRef]
- Pulendran, B.; Li, S.; Nakaya, H.I. Systems Vaccinology. Immunity 2010, 33, 516–529. [Google Scholar] [CrossRef]
- Borriello, F.; van Haren, S.D.; Levy, O. First International Precision Vaccines Conference: Multidisciplinary Approaches to Next-Generation Vaccines. Msphere 2018, 3. [Google Scholar] [CrossRef]
- Hagan, T.; Cortese, M.; Rouphael, N.; Boudreau, C.; Linde, C.; Maddur, M.S.; Das, J.; Wang, H.; Guthmiller, J.; Zheng, N.Y.; et al. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell 2019, 178, 1313–1328 e1313. [Google Scholar] [CrossRef]
- Khan, A.; Shin, O.S.; Na, J.; Kim, J.K.; Seong, R.K.; Park, M.S.; Noh, J.Y.; Song, J.Y.; Cheong, H.J.; Park, Y.H.; et al. A Systems Vaccinology Approach Reveals the Mechanisms of Immunogenic Responses to Hantavax Vaccination in Humans. Sci. Rep. 2019, 9, 4760. [Google Scholar] [CrossRef] [PubMed]
- Dhur, A.; Galan, P.; Hercberg, S. Folate status and the immune system. Prog. Food Nutr. Sci. 1991, 15, 43–60. [Google Scholar] [PubMed]
- Oh, K.Y.; Kang, M.J.; Choi, W.A.; Kwon, J.W.; Kim, B.J.; Yu, J.; Hong, S.J. Association Between Serum IgE Levels and the CTLA4 + 49A/G and FCER1B-654C/T Polymorphisms in Korean Children With Asthma. Allergy Asthma Immunol. Res. 2010, 2, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Manzetti, S.; Zhang, J.; van der Spoel, D. Thiamin function, metabolism, uptake, and transport. Biochemistry 2014, 53, 821–835. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Chitranshi, N.; Agarwal, A.K. Significance and biological importance of pyrimidine in the microbial world. Int. J. Med. Chem. 2014, 2014, 202784. [Google Scholar] [CrossRef]
- Ferreira, A.M.; Ferrari, M.I.; Trostchansky, A.; Batthyany, C.; Souza, J.M.; Alvarez, M.N.; Lopez, G.V.; Baker, P.R.; Schopfer, F.J.; O’Donnell, V.; et al. Macrophage activation induces formation of the anti-inflammatory lipid cholesteryl-nitrolinoleate. Biochem. J. 2009, 417, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Naito, T.; Irie, H.; Tsujimoto, K.; Ikeda, K.; Arakawa, T.; Koyama, A.H. Antiviral effect of arginine against herpes simplex virus type 1. Int. J. Mol. Med. 2009, 23, 495–499. [Google Scholar] [CrossRef]
- Goll, J.B.; Li, S.; Edwards, J.L.; Bosinger, S.E.; Jensen, T.L.; Wang, Y.; Hooper, W.F.; Gelber, C.E.; Sanders, K.L.; Anderson, E.J.; et al. Transcriptomic and Metabolic Responses to a Live-Attenuated Francisella tularensis Vaccine. Vaccines 2020, 8, 412. [Google Scholar] [CrossRef]
- Kuhtreiber, W.M.; Tran, L.; Kim, T.; Dybala, M.; Nguyen, B.; Plager, S.; Huang, D.; Janes, S.; Defusco, A.; Baum, D.; et al. Long-term reduction in hyperglycemia in advanced type 1 diabetes: The value of induced aerobic glycolysis with BCG vaccinations. NPJ Vaccines 2018, 3, 23. [Google Scholar] [CrossRef]
- McClenathan, B.M.; Stewart, D.A.; Spooner, C.E.; Pathmasiri, W.W.; Burgess, J.P.; McRitchie, S.L.; Choi, Y.S.; Sumner, S.C. Metabolites as biomarkers of adverse reactions following vaccination: A pilot study using nuclear magnetic resonance metabolomics. Vaccine 2017, 35, 1238–1245. [Google Scholar] [CrossRef]
- McClenathan, B.M.; Edwards, K.M. Vaccine safety: An evolving evidence-based science. Br. J. Clin. Pharmacol. 2019, 85, 2649–2651. [Google Scholar] [CrossRef] [PubMed]
- Arts, R.J.; Joosten, L.A.; Netea, M.G. Immunometabolic circuits in trained immunity. Semin. Immunol. 2016, 28, 425–430. [Google Scholar] [CrossRef]
- Li, P.; Yin, Y.L.; Li, D.; Kim, S.W.; Wu, G. Amino acids and immune function. Br. J. Nutr. 2007, 98, 237–252. [Google Scholar] [CrossRef] [PubMed]
- Wu, G. Amino acids: Metabolism, functions, and nutrition. Amino Acids 2009, 37, 1–17. [Google Scholar] [CrossRef]
- Ren, W.; Rajendran, R.; Zhao, Y.; Tan, B.; Wu, G.; Bazer, F.W.; Zhu, G.; Peng, Y.; Huang, X.; Deng, J.; et al. Amino Acids As Mediators of Metabolic Cross Talk between Host and Pathogen. Front. Immunol. 2018, 9, 319. [Google Scholar] [CrossRef]
- Ren, W.; Duan, J.; Yin, J.; Liu, G.; Cao, Z.; Xiong, X.; Chen, S.; Li, T.; Yin, Y.; Hou, Y.; et al. Dietary L-glutamine supplementation modulates microbial community and activates innate immunity in the mouse intestine. Amino Acids 2014, 46, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Koeken, V.; Lachmandas, E.; Riza, A.; Matzaraki, V.; Li, Y.; Kumar, V.; Oosting, M.; Joosten, L.A.B.; Netea, M.G.; van Crevel, R. Role of Glutamine Metabolism in Host Defense Against Mycobacterium tuberculosis Infection. J. Infect. Dis. 2019, 219, 1662–1670. [Google Scholar] [CrossRef] [PubMed]
- Karinch, A.M.; Pan, M.; Lin, C.M.; Strange, R.; Souba, W.W. Glutamine metabolism in sepsis and infection. J. Nutr. 2001, 131, 2535S–2538S; discussion 2550S–2551S. [Google Scholar] [CrossRef]
- Ma, E.H.; Bantug, G.; Griss, T.; Condotta, S.; Johnson, R.M.; Samborska, B.; Mainolfi, N.; Suri, V.; Guak, H.; Balmer, M.L.; et al. Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metab. 2017, 25, 345–357. [Google Scholar] [CrossRef]
- Passalacqua, K.D.; Lu, J.; Goodfellow, I.; Kolawole, A.O.; Arche, J.R.; Maddox, R.J.; Carnahan, K.E.; O’Riordan, M.X.D.; Wobus, C.E. Glycolysis Is an Intrinsic Factor for Optimal Replication of a Norovirus. mBio 2019, 10. [Google Scholar] [CrossRef]
- Hayek, I.; Fischer, F.; Schulze-Luehrmann, J.; Dettmer, K.; Sobotta, K.; Schatz, V.; Kohl, L.; Boden, K.; Lang, R.; Oefner, P.J.; et al. Limitation of TCA Cycle Intermediates Represents an Oxygen-Independent Nutritional Antibacterial Effector Mechanism of Macrophages. Cell Rep. 2019, 26, 3502–3510. [Google Scholar] [CrossRef] [PubMed]
- Ulas, T.; Pirr, S.; Fehlhaber, B.; Bickes, M.S.; Loof, T.G.; Vogl, T.; Mellinger, L.; Heinemann, A.S.; Burgmann, J.; Schoning, J.; et al. S100-alarmin-induced innate immune programming protects newborn infants from sepsis. Nat. Immunol. 2017, 18, 622–632. [Google Scholar] [CrossRef]
- Dreschers, S.; Ohl, K.; Lehrke, M.; Mollmann, J.; Denecke, B.; Costa, I.; Vogl, T.; Viemann, D.; Roth, J.; Orlikowsky, T.; et al. Impaired cellular energy metabolism in cord blood macrophages contributes to abortive response toward inflammatory threats. Nat. Commun. 2019, 10, 1685. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Ren, W.; Huang, X.; Deng, J.; Li, T.; Yin, Y. Potential Mechanisms Connecting Purine Metabolism and Cancer Therapy. Front. Immunol. 2018, 9, 1697. [Google Scholar] [CrossRef] [PubMed]
- Yegutkin, G.G. Nucleotide-and nucleoside-converting ectoenzymes: Important modulators of purinergic signalling cascade. Biochim. Biophys. Acta 2008, 1783, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Rai, B.; Kaur, J.; Jacobs, R.; Anand, S.C. Adenosine deaminase in saliva as a diagnostic marker of squamous cell carcinoma of tongue. Clin. Oral Investig. 2011, 15, 347–349. [Google Scholar] [CrossRef]
- Passos, D.F.; Bernardes, V.M.; da Silva, J.L.G.; Schetinger, M.R.C.; Leal, D.B.R. Adenosine signaling and adenosine deaminase regulation of immune responses: Impact on the immunopathogenesis of HIV infection. Purinergic Signal. 2018, 14, 309–320. [Google Scholar] [CrossRef]
- Staley, C.; Weingarden, A.R.; Khoruts, A.; Sadowsky, M.J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl. Microbiol. Biotechnol. 2017, 101, 47–64. [Google Scholar] [CrossRef]
- Oh, J.Z.; Ravindran, R.; Chassaing, B.; Carvalho, F.A.; Maddur, M.S.; Bower, M.; Hakimpour, P.; Gill, K.P.; Nakaya, H.I.; Yarovinsky, F.; et al. TLR5-Mediated Sensing of Gut Microbiota Is Necessary for Antibody Responses to Seasonal Influenza Vaccination. Immunity 2014, 41, 478–492. [Google Scholar] [CrossRef]
- Kloverpris, H.N.; Kazer, S.W.; Mjosberg, J.; Mabuka, J.M.; Wellmann, A.; Ndhlovu, Z.; Yadon, M.C.; Nhamoyebonde, S.; Muenchhoff, M.; Simoni, Y.; et al. Innate Lymphoid Cells Are Depleted Irreversibly during Acute HIV-1 Infection in the Absence of Viral Suppression. Immunity 2016, 44, 391–405. [Google Scholar] [CrossRef]
- Clish, C.B. Metabolomics: An emerging but powerful tool for precision medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Satoh, A.; Uchino, H.; Cajka, T.; Arita, M.; Arita, M. Mass Spectrometry Data Repository Enhances Novel Metabolite Discoveries with Advances in Computational Metabolomics. Metabolites 2019, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Deelen, J.; Kettunen, J.; Fischer, K.; van der Spek, A.; Trompet, S.; Kastenmuller, G.; Boyd, A.; Zierer, J.; van den Akker, E.B.; Ala-Korpela, M.; et al. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat. Commun. 2019, 10, 3346. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration, HHS. Medical devices; clinical chemistry and clinical toxicology devices; classification of newborn screening test systems for amino acids, free carnitine, and acylcarnitines using tandem mass spectrometry. Final rule. Fed. Regist. 2004, 69, 68254–68255. [Google Scholar]
- Patel, R. Maldi-tof ms for the diagnosis of infectious diseases. Clin. Chem. 2015, 61, 100–111. [Google Scholar] [CrossRef]
- Herman, W.A.; Devey, G.B. Future Trends in Medical Device Technologies: A Ten-Year Forecast. Food Drug Adm. Cent. Devices Radiol. Health 2011. [Google Scholar]
- Wilson, I.D. Metabolic phenotyping by liquid chromatography-mass spectrometry to study human health and disease. Anal. Chem. 2015, 87, 2519. [Google Scholar] [CrossRef]
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic phenotyping in health and disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef]
- Fernandez-Garcia, M.; Rojo, D.; Rey-Stolle, F.; Garcia, A.; Barbas, C. Metabolomic-Based Methods in Diagnosis and Monitoring Infection Progression. Exp. Suppl. 2018, 109, 283–315. [Google Scholar] [CrossRef]
- Zurfluh, S.; Baumgartner, T.; Meier, M.A.; Ottiger, M.; Voegeli, A.; Bernasconi, L.; Neyer, P.; Mueller, B.; Schuetz, P. The role of metabolomic markers for patients with infectious diseases: Implications for risk stratification and therapeutic modulation. Expert Rev. Anti Infect. Ther. 2018, 16, 133–142. [Google Scholar] [CrossRef]
- Beger, R.D.; Schmidt, M.A.; Kaddurah-Daouk, R. Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine. Metabolites 2020, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Andorf, S.; Gomes, L.; Dunn, P.; Schaefer, H.; Pontius, J.; Berger, P.; Desborough, V.; Smith, T.; Campbell, J.; et al. ImmPort: Disseminating data to the public for the future of immunology. Immunol. Res. 2014, 58, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Dunn, P.; Thomas, C.G.; Smith, B.; Schaefer, H.; Chen, J.; Hu, Z.; Zalocusky, K.A.; Shankar, R.D.; Shen-Orr, S.S.; et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data 2018, 5, 180015. [Google Scholar] [CrossRef] [PubMed]
| Pathogen Type Target | Pathogen (Infection) | Biosample Type | Technique Used | Examples of Metabolites or Metabolic Pathways Perturbed | References |
|---|---|---|---|---|---|
| Bacteria | Clostridioides difficile (infection) | stool | LC–MS, GC–MS | 2-hydroxy-4-methypentanoic acid, 2TMS derivative, 4-methylpentanoic acid, allo-isoleucine, bile acids, chenodeoxycholic acid, cholenic acids, choleonoic acid, eicosatrienoic acid, fatty acids, fructose, glyceryl glycoside, isoleucine, lysophosphatidylcholine (16:0), phenylalanine, propylene glycol, ribitol, sphingolipid, sphingomyelin, tyrosine, tyrosol | [59,60] |
| Bacteria | Escherichia coli (associated urinary tract infection) | urine | H-NMR, LC–MS | acetate, amines, aspartic acid, cadaverine, citrate, glutamic acid, glycine, hippurate, trimethylamine, trimethylamine n-oxide | [61,62] |
| Bacteria | Mycobacterium tuberculosis (tuberculosis) | plasma, serum | LC–MS, FIA–MS, GC–MS | amino-acyl tRNA, asparagine, aspartate, citrulline, cysteine, gamma-glutamylglutamine, fatty acid metabolism, glutamate, glutamine, histidine, inosine, kynurenine, lysophosphatidylcholines, medium chain fatty acid, lysosome pathway, mannose methionine, protein digestion pathway, sphingolipid, sphingosine-1-phosphate, sulfoxymethionine, tryptophan, urea | [13,63,64,65,66,67,68,69,70,71,72,73,74,75] |
| Virus | Alphavirus (Chikungunya) | serum | H-NMR | 2-hydroxycaproic acid, azelaic acid, carnitine, d-maltose, ethanol, galactitol, galactose metabolism and citrate cycle, gluconolactone, glycine, mandelic acid, methylguanidine, serine, threonine metabolism | [76] |
| Virus | Flavivirus (Dengue) | serum | GC–MS, LC–MS | acylcarnitines, amino acids, bile acids, chenodeoxyglycocholic acid, galactose and pyrimidine, glycine, glyoxylate and dicarboxylate, kynurenine, pentose phosphate pathway, phospholipids, propanoate, purines, serine, serotonin, starch and sucrose, threonine, uric acid | [76,77,78,79] |
| Virus | Lentivirus (human immunodeficiency virus/HIV) | CSF, CD4+ T cells, plasma | H-NMR, LC–MS, targeted LC–MS | acetate, citrate, creatine, dicarboxylicacylcarnitines, dopamine, glucose, glycerophospholipids, glycolysis, L-aspartate plasmalogen/plasminogen, lysophospholipids, methylglutarylcarnitine, phosphatidylcholines, sphingomyelin, sphingosine-1-phosphate, TCA cycle | [80,81,82,83] |
| Virus | Alphainfluenza virus (Influenza) | plasma | H-NMR, GC–MS | amino acids and ketone bodies, cAMP, glucose, glutathione, lipid, N-acetylglucosamine(O-GlcNAc), purine | [84,85,86,87] |
| Virus | SARS-CoV-2 (COVID19) | plasma, serum | LC–MS | Bile acids, bile acids, bilirubin, diacylglycerols, free fatty acid, glucose, glucuronate, glycerol 3-phosphate, kynurenine, lysophosphotidylcholines, malic acid, monosialodihexosylganglioside, phosphatidylcholines, sphingomyelin, triglycerides, tryptophan | [14,88,89,90,91,92] |
| Pathogen Type Target | Microbial Target | Vaccine Formulation Studied | Biosample Type | Technique Used | Examples of Metabolites or Metabolic Pathways Perturbed | References |
|---|---|---|---|---|---|---|
| Bacterium | Francisella tularensis | F. tularensis (LVS-DynPort Vaccine) | plasma | LC–MS | 2-oxocarboxylic acid, asparagine, glycolysis, purine, pyruvate, TCA cycle | [118] |
| Bacterium | M. tuberculosis | BCG (Connaught strain) | serum | LC–MS | 1,5-anhydroglucitol, alpha-ketobutyrate, de novo purine synthesis, glucose processing metabolites, methylguanine, N6-carbomoyltheronyladenosine | [119] |
| Virus | Hantavirus | Hantavax (GreenCross) | serum | LC–MS | 16-hydroxyplamitate, arachidonic acid, arginine, benzoate, chenodeoxycholic acid, cholesteryl nitrolinoate, cystathionine, glutamine and citrulline, glycine, histidine, homocysteine, indole 3-acetaldehyde, isoleucine, leucine, methionine, methyl palmitate, N-stearoyl, octanoylcarnitine, phenylalanine, threonine, tryptophan, tyrosine, ubiquinone-9, valine | [111] |
| Virus | Influenza | Fluzone (2014–2015, 2015–2016) *co-administered with antibiotics | plasma | LC–MS | primary and secondary bile acids, tryptophan metabolism | [110] |
| Virus | Varicella zoster (Shingles) | Zostavax | plasma | LC–MS | aldarate, ascorbate, gluconeogenesis, glycolysis, inositol phosphate, propanoate, sterol, TCA cycle, tryptophan | [11] |
| Virus | Variola virus (Small Pox) | DryVax or ACAM 2000 | serum | H-NMR | 2-aminobutyrate, alanine, choline, creatinine, fructose, glutamate, glutamine, histidine, lactate, threonine, lysine, methionine, propylene glycol, serine | [120] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diray-Arce, J.; Conti, M.G.; Petrova, B.; Kanarek, N.; Angelidou, A.; Levy, O. Integrative Metabolomics to Identify Molecular Signatures of Responses to Vaccines and Infections. Metabolites 2020, 10, 492. https://doi.org/10.3390/metabo10120492
Diray-Arce J, Conti MG, Petrova B, Kanarek N, Angelidou A, Levy O. Integrative Metabolomics to Identify Molecular Signatures of Responses to Vaccines and Infections. Metabolites. 2020; 10(12):492. https://doi.org/10.3390/metabo10120492
Chicago/Turabian StyleDiray-Arce, Joann, Maria Giulia Conti, Boryana Petrova, Naama Kanarek, Asimenia Angelidou, and Ofer Levy. 2020. "Integrative Metabolomics to Identify Molecular Signatures of Responses to Vaccines and Infections" Metabolites 10, no. 12: 492. https://doi.org/10.3390/metabo10120492
APA StyleDiray-Arce, J., Conti, M. G., Petrova, B., Kanarek, N., Angelidou, A., & Levy, O. (2020). Integrative Metabolomics to Identify Molecular Signatures of Responses to Vaccines and Infections. Metabolites, 10(12), 492. https://doi.org/10.3390/metabo10120492







