Genetic Variability in Antioxidative and Inflammatory Pathways Modifies the Risk for PCOS and Influences Metabolic Profile of the Syndrome
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics of PCOS Patients
2.2. Polymorphism Frequencies in PCOS Patients and Controls and PCOS Risk
2.3. Interactions between CARD8, NLRP3, IL1B, and IL6 Polymorphisms and PCOS Risk
2.4. Polymorphisms in Genes Related to OS and Clinical Manifestations of PCOS
2.5. Polymorphisms in Genes Related to Inflammation and Clinical Manifestations of PCOS
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sheehan, M.T. Polycystic Ovarian Syndrome: Diagnosis and Management. Clin. Med. Res. 2004, 2, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Branavan, U.; Muneeswaran, K.; Wijesundera, S.; Jayakody, S.; Chandrasekharan, V.; Wijeyaratne, C. Identification of Selected Genetic Polymorphisms in Polycystic Ovary Syndrome in Sri Lankan Women Using Low Cost Genotyping Techniques. PLoS ONE 2018, 13, e0209830. [Google Scholar] [CrossRef] [PubMed]
- Brännström, M. Potential Role of Cytokines in Ovarian Physiology: The Case for Interleukin-1. In The Ovary, 2nd ed.; Elsevier: London, UK, 2004; pp. 261–271. [Google Scholar] [CrossRef]
- Ebejer, K.; Calleja-Agius, J. The Role of Cytokines in Polycystic Ovarian Syndrome. Gynecol. Endocrinology 2013, 29, 536–540. [Google Scholar] [CrossRef]
- Artimani, T.; Karimi, J.; Mehdizadeh, M.; Yavangi, M.; Khanlarzadeh, E.; Ghorbani, M.; Asadi, S.; Kheiripour, N. Evaluation of Pro-Oxidant-Antioxidant Balance (PAB) and Its Association with Inflammatory Cytokines in Polycystic Ovary Syndrome (PCOS). Gynecol. Endocrinol. 2018, 34, 148–152. [Google Scholar] [CrossRef]
- Gao, L.; Gu, Y.; Yin, X. High Serum Tumor Necrosis Factor-Alpha Levels in Women with Polycystic Ovary Syndrome: A Meta-Analysis. PLoS ONE 2016, 11, e0164021. [Google Scholar] [CrossRef]
- Peng, Z.; Sun, Y.; Lv, X.; Zhang, H.; Liu, C.; Dai, S. Interleukin-6 Levels in Women with Polycystic Ovary Syndrome: A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0148531. [Google Scholar] [CrossRef]
- Mu, Y.; Liu, J.; Wang, B.; Wen, Q.; Wang, J.; Yan, J.; Zhou, S.; Ma, X.; Cao, Y. Interleukin 1 Beta (IL-1β) Promoter C [-511] T Polymorphism but Not C [+3953] T Polymorphism Is Associated with Polycystic Ovary Syndrome. Endocrine 2010, 37, 71–75. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, H.; Kouadir, M.; Song, H.; Shi, F. Recent Advances in the Mechanisms of NLRP3 Inflammasome Activation and Its Inhibitors. Cell Death Dis. 2019, 10, 128. [Google Scholar] [CrossRef]
- Ito, S.; Hara, Y.; Kubota, T. CARD8 Is a Negative Regulator for NLRP3 Inflammasome, but Mutant NLRP3 in Cryopyrin-Associated Periodic Syndromes Escapes the Restriction. Arthritis Res. Ther. 2014, 16, R52. [Google Scholar] [CrossRef]
- Klen, J.; Goričar, K.; Janež, A.; Dolžan, V. NLRP3 Inflammasome Polymorphism and Macrovascular Complications in Type 2 Diabetes Patients. J. Diabetes Res. 2015, 2015, 616747. [Google Scholar] [CrossRef][Green Version]
- Desai, V.; Prasad, N.R.; Manohar, S.M.; Sachan, A.; Lakshmi Narasimha, S.R.P.V.; Bitla, A.R.R. Oxidative Stress in Non-Obese Women with Polycystic Ovarian Syndrome. J. Clin. Diagnostic Res. 2014, 8, CC01–CC03. [Google Scholar] [CrossRef]
- Murri, M.; Luque-ramírez, M.; Insenser, M.; Ojeda-ojeda, M.; Escobar-morreale, H.F. Circulating Markers of Oxidative Stress and Polycystic Ovary Syndrome (Pcos): A Systematic Review and Meta-Analysis. Hum. Reprod. Update 2013, 19, 268–288. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, M.A.; Al-Farsi, Y.M.; Al-Khaduri, M.M.; Saleh, J.; Waly, M.I. Polycystic Ovarian Syndrome Is Linked to Increased Oxidative Stress in Omani Women. Int. J. Womens Health 2018, 10, 763–771. [Google Scholar] [CrossRef]
- Gyftaki, R.; Gougoura, S.; Kalogeris, N.; Loi, V.; Koukoulis, G.; Vryonidou, A. Oxidative Stress Markers in Women with Polycystic Ovary Syndrome without Insulin Resistance. Endocr. Abstr. 2018, 56, P950. [Google Scholar] [CrossRef]
- Blair, S.A.; Kyaw-Tun, T.; Young, I.S.; Phelan, N.A.; Gibney, J.; McEneny, J. Oxidative Stress and Inflammation in Lean and Obese Subjects with Polycystic Ovary Syndrome. J. Reprod. Med. 2013, 58, 107–114. [Google Scholar]
- Zhao, Y.; Zhang, C.; Huang, Y.; Yu, Y.; Li, R.; Li, M.; Liu, N.; Liu, P.; Qiao, J. Up Regulated Expression of Wnt5a Increases Inflammation and Oxidative Stress via PI3K/AKT/NFêB Signaling in the Granulosa Cells of PCOS Patients. J. Clin. Endocrinol. Metab. 2015, 100, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Kitani, A.; Similuk, M.; Oler, A.J.; Albenberg, L.; Kelsen, J.; Aktay, A.; Quezado, M.; Yao, M.; Montgomery-Recht, K.; et al. Loss-of-Function CARD8 Mutation Causes NLRP3 Inflammasome Activation and Crohn’s Disease. J. Clin. Investig. 2018, 128, 1793–1806. [Google Scholar] [CrossRef]
- Zhang, Z.T.; Ma, X.J.; Zong, Y. Is the CARD8 rs2043211 polymorphism associated with susceptibility to Chron’s disease? A meta-analysis. Autoimmunity. 2015, 48, 524–531. [Google Scholar] [CrossRef]
- Cullen, S.P.; Kearney, C.J.; Clancy, D.M.; Martin, S.J. Diverse Activators of the NLRP3 Inflammasome Promote IL-1β Secretion by Triggering Necrosis. Cell Rep. 2015, 11, 1535–1548. [Google Scholar] [CrossRef]
- Souza de Lima, D.; Ogusku, M.M.; Sadahiro, A.; Pontillo, A. Inflammasome Genetics Contributes to the Development and Control of Active Pulmonary Tuberculosis. Infect. Genet. Evol. 2016, 41, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Ravimohan, S.; Nfanyana, K.; Tamuhla, N.; Tiemessen, C.T.; Weissman, D.; Bisson, G.P. Common Variation in NLRP3 is Associated with Early Death and Elevated Inflammasome Biomarkers Among Advanced HIV/TB Co-Infected Patients in Botswana. Open Forum Infect Dis. 2018, 5, ofy075. [Google Scholar] [CrossRef] [PubMed]
- Pontillo, A.; Carvalho, M.S.; Kamada, A.J.; Moura, R.; Schindler, H.C.; Duarte, A.J.S.; Crovella, S. Susceptibility to Mycobacterium Tuberculosis Infection in HIV-Positive Patients Is Associated with CARD8 Genetic Variant. J. Acquir. Immune Defic. Syndr. 2013, 63, 147–151. [Google Scholar] [CrossRef]
- Pontillo, A.; Bricher, P.; Leal, V.N.C.; Lima, S.; Souza, P.R.E.; Crovella, S. Role of Inflammasome Genetics in Susceptibility to HPV Infection and Cervical Cancer Development. J. Med. Virol. 2016, 88, 1646–1651. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, W.C.; Oshiro, T.M.; de Sá, D.C.; Franco, D.D.G.S.; Festa Neto, C.; Pontillo, A. Genotyping and Differential Expression Analysis of Inflammasome Genes in Sporadic Malignant Melanoma Reveal Novel Contribution of CARD8, IL1B and IL18 in Melanoma Susceptibility and Progression. Cancer Genet. 2016, 210, 474–480. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, H.; Bai, H.; Huang, W.; Zhang, R.; Tan, J.; Guan, L.; Fan, P. Association of SOD2 A16V and PON2 S311C Polymorphisms with Polycystic Ovary Syndrome in Chinese Women. J. Endocrinol. Investig. 2019. [Google Scholar] [CrossRef]
- Paludo, F.J.O.; Bristot, I.J.; Alho, C.S.; Gelain, D.P.; Moreira, J.C.F. Effects of 47C Allele (Rs4880) of the SOD2 Gene in the Production of Intracellular Reactive Species in Peripheral Blood Mononuclear Cells with and without Lipopolysaccharides Induction. Free Radic. Res. 2014, 48, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Dornelles, E.; Mânica-Cattani, M.F.; Algarve, T.D.; De Souza Filho, O.C.; Sagrillo, M.R.; Garcia, L.F.M.H.; Da Cruz, I.B.M. Influence of Val16Ala SOD2 Polymorphism on the In-Vitro Effect of Clomiphene Citrate in Oxidative Metabolism. Reprod. Biomed. Online 2012. [Google Scholar] [CrossRef]
- Makki, K.; Froguel, P.; Wolowczuk, I. Adipose Tissue in Obesity-Related Inflammation and Insulin Resistance: Cells, Cytokines, and Chemokines. ISRN Inflamm. 2013, 139239. [Google Scholar] [CrossRef]
- Borst, S.E. The Role of TNF-α in Insulin Resistance. Endocrine 2004. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Arner, P.; Caro, J.F.; Atkinson, R.L.; Spiegelman, B.M. Increased Adipose Tissue Expression of Tumor Necrosis Factor-α in Human Obesity and Insulin Resistance. J. Clin. Investig. 1995, 95, 2409–2415. [Google Scholar] [CrossRef]
- Bouhaha, R.; Baroudi, T.; Ennafaa, H.; Vaillant, E.; Abid, H.; Sassi, R.; Vatin, V.; Froguel, P.; Gaaied, A.B.; Meyre, D.; et al. Study of TNFα-308G/A and IL6-174G/C Polymorphisms in Type 2 Diabetes and Obesity Risk in the Tunisian Population. Clin. Biochem. 2010. [Google Scholar] [CrossRef]
- Mahajan, A.; Tabassum, R.; Srechavali, E.; Dwivedi, O.P.; Chauhan, G.; Tandon, N.; Bharadwaj, D. Obesity-Dependent Association of TNF-LTA Locus with Type 2 Diabetes in North Indians. J. Mol. Med. 2010, 88, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Hebert-Schuster, M.; Fabre, E.E.; Nivet-Antoine, V. Catalase Polymorphisms and Metabolic Diseases. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Góth, L.; Nagy, T.; Kósa, Z.; Fejes, Z.; Bhattoa, H.P.; Paragh, G.; Káplár, M. Effects of Rs769217 and Rs1001179 Polymorphisms of Catalase Gene on Blood Catalase, Carbohydrate and Lipid Biomarkers in Diabetes Mellitus. Free Radic. Res. 2012, 46, 1249–1257. [Google Scholar] [CrossRef]
- Pask, R.; Cooper, J.D.; Walker, N.M.; Nutland, S.; Hutchings, J.; Dunger, D.B.; Nejentsev, S.; Todd, J.A. No Evidence for a Major Effect of Two Common Polymorphisms of the Catalase Gene in Type 1 Diabetes Susceptibility. Diabetes. Metab. Res. Rev. 2006, 22, 356–360. [Google Scholar] [CrossRef]
- Batool, A.; Jahan, N.; Sun, Y.; Hanif, A.; Xue, H. Genetic Association of IDE, POU2F1, PON1, IL1α and IL1β with Type 2 Diabetes in Pakistani Population. Mol. Biol. Rep. 2014, 41, 3063–3069. [Google Scholar] [CrossRef]
- Wang, J.; Ming Yang, M.; Rong, S.S.; Ng, T.K.; Li, Y.B.; Liu, X.M. Association of Paraoxonase Gene Polymorphisms with Diabetic Nephropathy and Retinopathy. Mol. Med. Rep. 2013, 8, 1845–1851. [Google Scholar] [CrossRef][Green Version]
- Odawara, M.; Tachi, Y.; Yamashita, K. Paraoxonase Polymorphism (Gln 192 -Arg) Is Associated with Coronary Heart Disease in Japanese Noninsulin-Dependent Diabetes Mellitus. J. Clin. Endocrinol. Metab. 1997, 82, 2257–2260. [Google Scholar] [CrossRef]
- Bhaskar, S.; Ganesan, M.; Chandak, G.R.; Mani, R.; Idris, M.M.; Khaja, N.; Gulla, S.; Kumar, U.; Movva, S.; Vattam, K.K.; et al. Association of PON1 and APOA5 Gene Polymorphisms in a Cohort of Indian Patients Having Coronary Artery Disease with and without Type 2 Diabetes. Genet. Test. Mol. Biomark. 2011, 15, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Rajska, A.; Buszewska-Forajta, M.; Rachoń, D.; Markuszewski, M.J. Metabolomic Insight into Polycystic Ovary Syndrome—An Overview. Int. J. Mol. Sci. 2020, 21, 4853. [Google Scholar] [CrossRef] [PubMed]
- Murri, M.; Insenser, M.; Escobar-Morreale, H.F. Metabolomics in Polycystic Ovary Syndrome. Clin. Chim. Acta 2014, 429, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Buszewska-Forajta, M.; Rachoń, D.; Stefaniak, A.; Wawrzyniak, R.; Konieczna, A.; Kowalewska, A.; Markuszewski, M.J. Identification of the Metabolic Fingerprints in Women with Polycystic Ovary Syndrome Using the Multiplatform Metabolomics Technique. J. Steroid Biochem. Mol. Biol. 2019, 186, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Azziz, R. Introduction: Determinants of Polycystic Ovary Syndrome. Fertil. Steril. 2016, 106, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Teede, H.J.; Misso, M.L.; Costello, M.F.; Dokras, A.; Laven, J.; Moran, L.; Piltonen, T.; Norman, R.J.; Andersen, M.; Azziz, R.; et al. Recommendations from the International Evidence-Based Guideline for the Assessment and Management of Polycystic Ovary Syndrome. Hum. Reprod. 2018. [Google Scholar] [CrossRef]
| Characteristic | Median Value (25–75%) |
|---|---|
| Age (years) | 30 (25–35.5) |
| Anthropometric Characteristics | |
| Body mass (kg) | 100.1 (85.1–111.2) |
| BMI (kg/m2) | 35.8 (31.8–39.9) |
| Waist circumference (cm) | 112 (102–121.8) |
| VAT mass (g) | 760 (580.3–965.0) |
| VAT volume (cm3) | 819.5 (627.8–1038.5) |
| VAT surface (cm2) | 157 (117–199) |
| OGTT | |
| Glucose 0 min OGTT (mmol/L) Reference: 3.6–6.1 | 5.2 (4.8–5.6) |
| Glucose 30 min OGTT (mmol/L) | 8.3 (6.9–9.4) |
| Glucose 60 min OGTT (mmol/L) | 8.3 (6.8–9.8) |
| Glucose 90 min OGTT (mmol/L) | 7.6 (5.9–9.1) |
| Glucose 120 min OGTT (mmol/L) | 6.6 (5.5–7.9) |
| Insulin 0 min OGTT (mU/L) Reference: 2–17.2 | 12.4 (7.5–20.0) |
| Insulin 30 min OGTT (mU/L) | 71.2 (45.2–104.5) |
| Insulin 60 min OGTT (mU/L) | 97.3 (63.3–131.5) |
| Insulin 90 min OGTT (mU/L) | 89.9 (62.1–121) |
| Insulin 120 min OGTT (mU/L) | 78 (53.3–123.5) |
| HOMA IR Reference: <2.0 | 2.8 (1.7–5.0) |
| Endocrine Characteristics | |
| DHEAS * (μmol/L) Reference: 0.95–11.67 | 5.8 (4.0–7.4) |
| Total testosteron (nmol/L) Reference: <2.53 | 1.8 (1.2–2.6) |
| Free testosteron (pmol/L) Reference: follicular phase: 1.56–11.00 | 7 (4.6–8.7) |
| SHBG* (nmol/L) Reference: 18–144 | 24 (18–35) |
| Androstenedion (nmol/L) Reference: 0.7–10.8 | 8.7 (6.5–11.2) |
| LH (IU/L) Reference: follicular phase: 1.1–11.6; | 5.4 (3.0–8.7) |
| FSH (mIU/L) Reference: follicular phase: 2.8–11.3; | 4.8 (3.8–6.6) |
| Gene | SNP | Genotype | Number of Controls (%) | Number of Patients (%) | OR (95% CI) | p | MAF |
|---|---|---|---|---|---|---|---|
| Genes Related to OS | |||||||
| CAT | rs1001179 c.-330C>T | CC | 40 (49.4) | 96 (57.1) | Reference | 0.284 | |
| CT | 36 (44.4) | 65 (38.7) | 0.75 (0.43–1.30) | 0.310 | |||
| TT | 5 (6.2) | 7 (4.2) | 0.58 (0.17–1.95) | 0.381 | |||
| CT+TT | 41 (50.6) | 72 (42.9) | 0.73 (0.43–1.25) | 0.250 | |||
| SOD2 | rs4880 p.Ala16Val | CC | 27 (32.9) | 35 (20.8) | Reference | 0.445 | |
| CT | 37 (45.1) | 91 (54.2) | 1.90 (1.01–3.57) | 0.047 | |||
| TT | 18 (22.0) | 42 (25.0) | 1.80 (0.85–3.80) | 0.123 | |||
| CT+TT | 55 (67.1) | 133 (79.2) | 1.87 (1.03–3.37) | 0.039 | |||
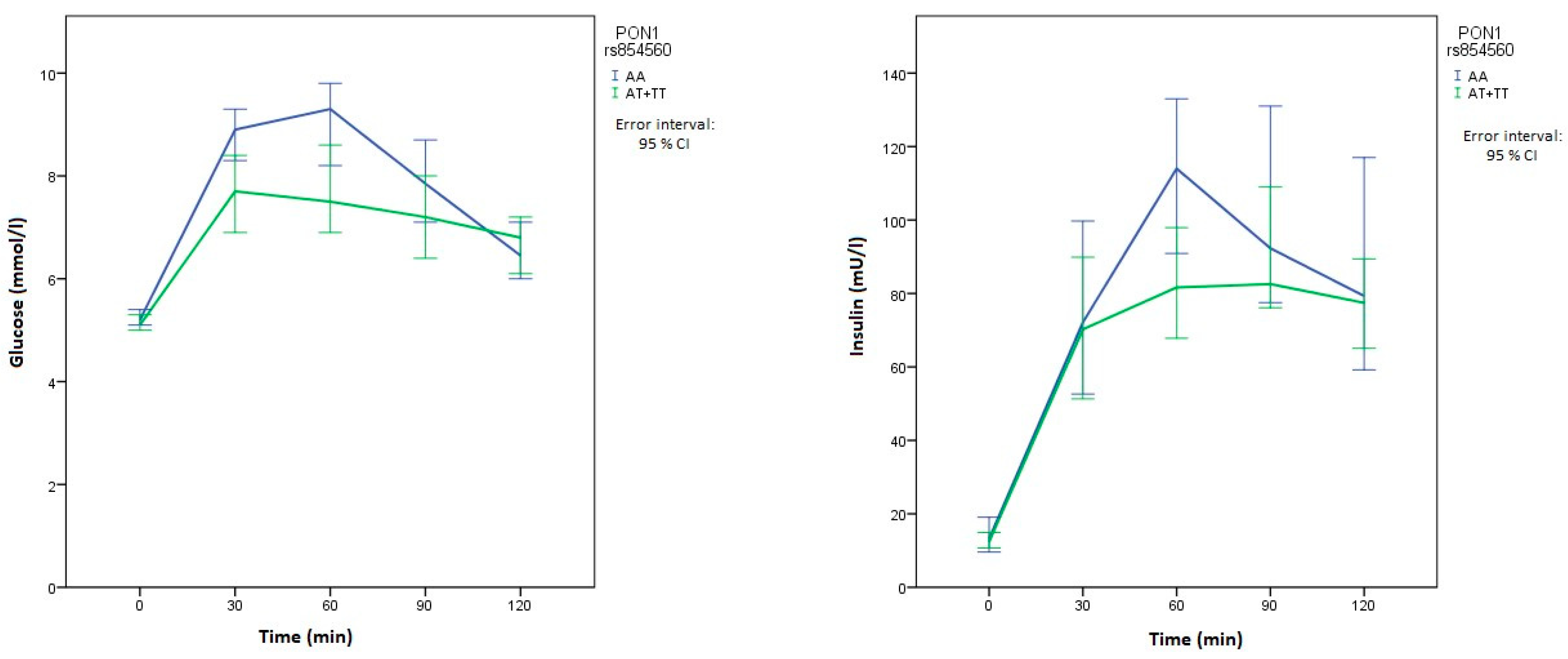
| PON1 | rs854560 p.Leu55Met | AA | 30 (36.1) | 75 (45.2) | Reference | 0.373 | |
| AT | 44 (53.0) | 71 (42.8) | 0.65 (0.37–1.14) | 0.130 | |||
| TT | 9 (10.8) | 20 (12.0) | 0.89 (0.36–2.17) | 0.796 | |||
| AT+TT | 53 (63.8) | 91 (54.8) | 0.69 (0.40–1.18) | 0.174 | |||
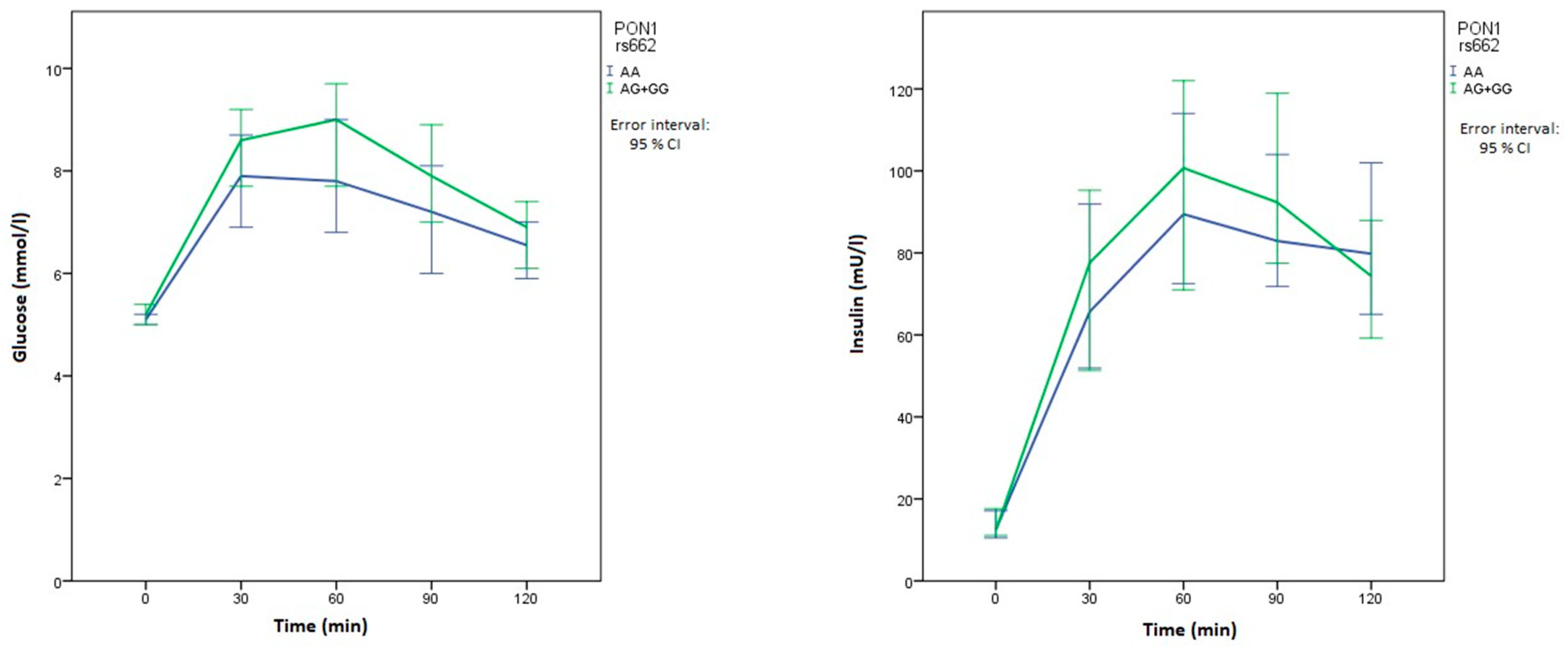
| PON1 | rs662 p.Gln192Arg | AA | 44 (53.0) | 82 (49.1) | Reference | 0.265 | |
| AG | 34 (41.0) | 73 (43.7) | 1.15 (0.67–1.99) | 0.612 | |||
| GG | 5 (6.0) | 12 (7.2) | 1.29 (0.43–3.89) | 0.654 | |||
| AG+GG | 39 (47.0) | 85 (50.9) | 1.17 (0.69–1.98) | 0.560 | |||
| Genes Related to Inflammation | |||||||
| CARD8 | rs2043211 p.Cys10Ter | AA | 30 (36.1) | 78 (46.4) | Reference | 0.428 | |
| AT | 35 (42.2) | 72 (42.9) | 0.79 (0.44–1.42) | 0.431 | |||
| TT | 18 (21.7) | 18 (10.7) | 0.38 (0.18–0.84) | 0.016 | |||
| AT+TT | 53 (63.9) | 90 (53.6) | 0.65 (0.38–1.12) | 0.123 | |||
| NLRP3 | rs35829419 p.Gln705Lys | CC | 76 (91.6) | 145 (86.3) | Reference | 0.042 | |
| CA | 7 (8.4) | 22 (13.1) | / | ||||
| AA | 0 (0.0) | 1 (0.6) | |||||
| CA+AA | 7 (8.4) | 23 (13.7) | 1.72 (0.71–4.20) | 0.231 | |||
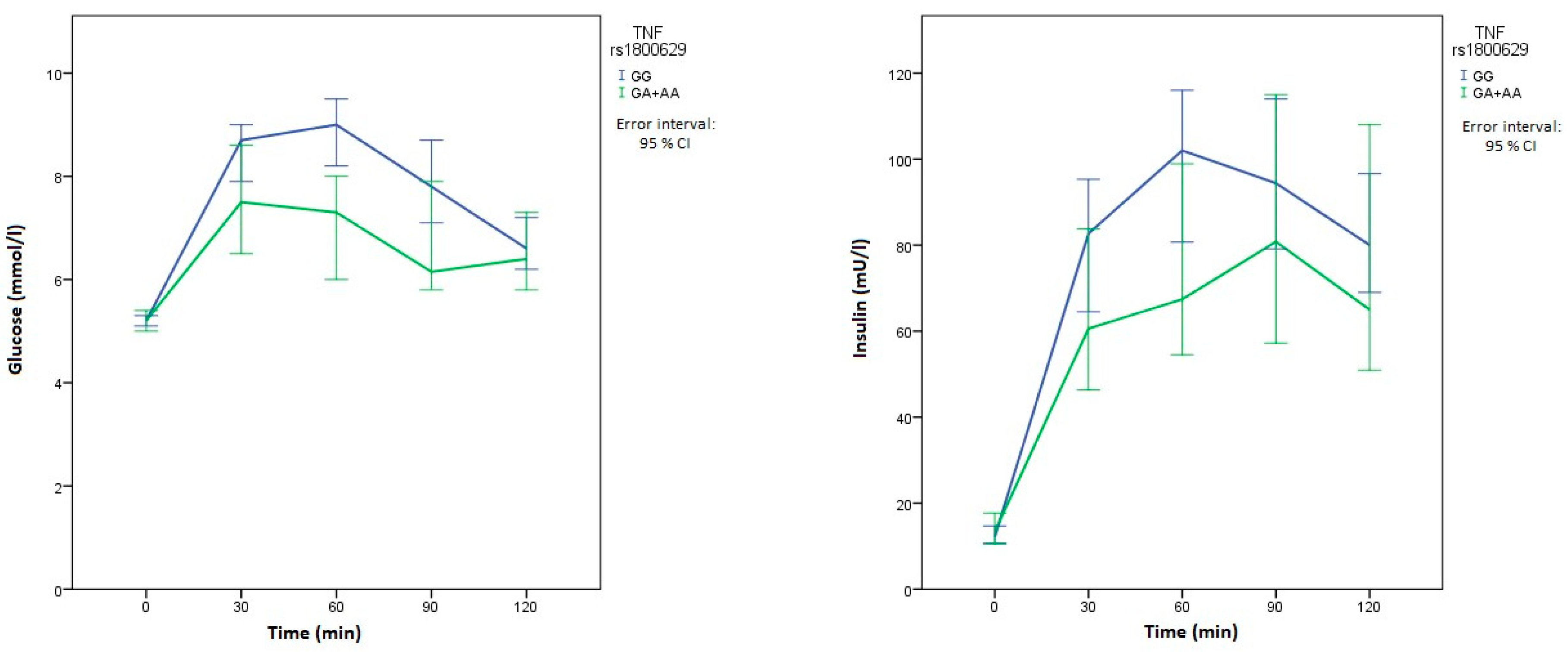
| TNF | rs1800629 c.-308 G>A | GG | 61 (77.2) | 120 (73.6) | Reference | 0.120 | |
| GA | 17 (21.5) | 41 (25.2) | / | ||||
| AA | 1 (1.3) | 2 (1.2) | |||||
| GA+AA | 18 (22.8) | 43 (26.4) | 1.21 (0.65–2.28) | 0.546 | |||
| IL1B | rs1143623 c.-1560G>C | GG | 43 (51.8) | 105 (62.1) | Reference | 0.271 | |
| GC | 35 (42.2) | 51 (30.2) | 0.60 (0.34–1.04) | 0.070 | |||
| CC | 5 (6.0) | 13 (7.7) | 1.06 (0.36–3.17) | 0.910 | |||
| GC+CC | 40 (48.2) | 64 (37.9) | 0.66 (0.39–1.11) | 0.119 | |||
| IL1B | rs16944 c.-598T>C | TT | 9 (11.1) | 20 (11.9) | 0.76 (0.31–1.84) | 0.541 | 0.630 * |
| TC | 42 (51.9) | 60 (35.7) | 0.49 (0.27–0.86) | 0.014 | |||
| CC | 30 (37.0) | 88 (52.4) | Reference | ||||
| TC+CC | 72 (88.9) | 148 (88.1) | 0.54 (0.31–0.92) | 0.024 | |||
| IL6 | rs1800795 c.-174G>C | GG | 34 (41.0) | 60 (35.7) | Reference | 0.373 | |
| GC | 36 (43.4) | 87 (51.8) | 1.37 (0.77–2.43) | 0.282 | |||
| CC | 13 (15.7) | 21 (12.5) | 0.92 (0.41–2.06) | 0.831 | |||
| GC+CC | 49 (59.1) | 108 (64.3) | 1.25 (0.73–2.14) | 0.419 | |||
| Interaction | OR (95% CI) | p |
|---|---|---|
| CARD8 rs2043211 and IL1B rs1143623 | 1.27 (0.43–3.80) | 0.666 |
| CARD8 rs2043211 and IL1B rs16944 | 0.67 (0.22–2.06) | 0.480 |
| CARD8 rs2043211 and IL6 rs1800795 | 0.26 (0.09–0.81) | 0.020 |
| NLRP3 rs35829419 and IL1B rs1143623 | 0.71 (0.31–1.62) | 0.412 |
| NLRP3 rs35829419 and IL1B rs16944 | 1.21 (0.18–8.28) | 0.843 |
| NLRP3 rs35829419 and IL6 rs1800795 | 1.41 (0.23–8.58) | 0.707 |
| IL1B rs1143623 and IL6 rs1800795 | 3.17 (1.04–9.65) | 0.042 |
| IL1B rs16944 and IL6 rs1800795 | 0.44 (0.14–1.37) | 0.158 |
| Characteristic | Genotype * | CAT rs1001179 | p | SOD2 rs4880 | p | PON1 rs854560 | p | PON1 rs662 | p |
|---|---|---|---|---|---|---|---|---|---|
| Median Value (25–75%) | Median Value (25–75%) | Median Value (25–75%) | Median Value (25–75%) | ||||||
| Anthropometric Characteristics | |||||||||
| Body mass (kg) | XX | 101.7 (86.9–111.8) | 0.371 | 100 (85.8–105.7) | 0.365 | 100.2 (82–110.1) | 0.198 | 101 (89.9–112.1) | 0.147 |
| Xx+xx | 97.9 (83–110.4) | 100.6 (84.9–112) | 100 (89.8–112) | 100 (82–110) | |||||
| BMI (kg/m2) | XX | 36.9 (32–41) | 0.106 | 35.5 (31.4–39.9) | 0.598 | 36 (30.8–39.9) | 0.631 | 36.6 (33.2–40.4) | 0.071 |
| Xx+xx | 35.5 (31.2–38.6) | 36 (31.8–39.9) | 35.8 (32.7–39.8) | 34.6 (30.8–39.1) | |||||
| Waist circumference (cm) | XX | 112.5 (103.3–123.8) | 0.220 | 108 (102.5–118.8) | 0.416 | 112 (99–124.5) | 0.728 | 112.5 (102.8–122.6) | 0.448 |
| Xx+xx | 108.5 (100–119.5) | 113 (101.5–123) | 111.5 (103.8–121) | 110 (101–121) | |||||
| VAT mass (g) | XX | 797 (614–990) | 0.227 | 852 (746–990) | 0.162 | 793 (616–968) | 0.490 | 694.5 (561.3–914) | 0.195 |
| Xx+xx | 685 (535–879) | 744 (565–959.5) | 756 (523–970.5) | 810 (651–971) | |||||
| VAT volume (cm3) | XX | 857 (664–1070) | 0.266 | 921 (807–1070) | 0.141 | 857 (666–1046) | 0.421 | 751 (606.5–987.5) | 0.231 |
| Xx+xx | 740 (578–950) | 781.5 (610.8–1018) | 810.5 (565.5–1044.5) | 874 (704–1050) | |||||
| VAT surface (cm2) | XX | 165 (127.3–205) | 0.107 | 177 (155–205) | 0.126 | 162 (127.3–198.3) | 0.534 | 145 (116–201) | 0.487 |
| Xx+xx | 140 (110–177) | 148 (116–194) | 155.5 (108.8–200.5) | 167 (124–199) | |||||
| OGTT | |||||||||
| Glucose 0 min OGTT (mmol/L) | XX | 5.2 (4.9–5.6) | 0.051 | 5.4 (5.1–5.7) | 0.038 | 5.2 (4.9–5.6) | 0.558 | 5.1 (4.8–5.4) | 0.353 |
| Xx+xx | 5.1 (4.8–5.4) | 5.1 (4.8–5.5) | 5.1 (4.8–5.5) | 5.2 (4.9–5.7) | |||||
| Glucose 30 min OGTT (mmol/L) | XX | 8.2 (6.9–9.4) | 0.821 | 8.2 (6.7–9.3) | 0.474 | 8.9 (7.9–9.5) | 0.006 | 7.9 (6.5–9.3) | 0.063 |
| Xx+xx | 8.4 (6.9–9.4) | 8.4 (7–9.4) | 7.7 (6.5–9.3) | 8.6 (7.5–10.2) | |||||
| Glucose 60 min OGTT (mmol/L) | XX | 8.7 (6.8–9.9) | 0.886 | 8.1 (6.7–9.7) | 0.661 | 9.3 (8–10.6) | 0.006 | 7.8 (6.3–9.7) | 0.069 |
| Xx+xx | 8.3 (7.1–9.7) | 8.5 (6.8–10.1) | 7.5 (6–9.7) | 9 (7.2–11.2) | |||||
| Glucose 90 min OGTT (mmol/L) | XX | 7.6 (5.7–9) | 0.583 | 7.4 (5.9–8.9) | 0.681 | 7.9 (7–9.2) | 0.166 | 7.2 (5.7–8.7) | 0.066 |
| Xx+xx | 7.7 (6.3–9.3) | 7.8 (6.2–9.1) | 7.2 (5.7–9.1) | 7.9 (6.5–9.6) | |||||
| Glucose 120 min OGTT (mmol/L) | XX | 6.8 (5.6–8) | 0.158 | 6.6 (5.8–7.8) | 0.677 | 6.5 (5.6–7.5) | 0.660 | 6.6 (5.5–7.6) | 0.359 |
| Xx+xx | 6.3 (5.4–7.6) | 6.6 (5.5–7.9) | 6.8 (5.5–7.9) | 6.9 (5.5–8.1) | |||||
| Insulin 0 min OGTT (mU/L) | XX | 15.2 (11.1–21.6) | 0.001 | 11.8 (9.3–21.1) | 0.822 | 13.2 (6.7–22.3) | 0.628 | 12.5 (7.7–20.1) | 0.873 |
| Xx+xx | 9.3 (5.9–17.1) | 12.5 (7.3–20) | 12.4 (7.9–19.8) | 12.4 (7.5–20) | |||||
| Insulin 30 min OGTT (mU/L) | XX | 74.9 (48.1–102) | 0.413 | 67.6 (43.5–95.9) | 0.869 | 72.2 (45.7–110.3) | 0.598 | 65.7 (46.2–106) | 0.961 |
| Xx+xx | 68.4 (42.8–108.8) | 71.3 (45.1–108.8) | 70.3 (44–104.3) | 77.6 (43.2–106.3) | |||||
| Insulin 60 min OGTT (mU/L) | XX | 87.2 (64.7–125.5) | 0.833 | 78.8 (56.6–138.3) | 0.878 | 114 (76.1–154.8) | 0.024 | 89.5 (63.7–130) | 0.716 |
| Xx+xx | 99.2 (61.5–136.8) | 97.5 (63.2–129.8) | 81.7 (56.3–116) | 100.7 (59.7–139.5) | |||||
| Insulin 90 min OGTT (mU/L) | XX | 91.2 (67.1–119.5) | 0.836 | 82.9 (67.1–118.5) | 0.811 | 92.3 (69.6–154) | 0.327 | 82.9 (62.2–117.3) | 0.426 |
| Xx+xx | 86.6 (57.7–139.5) | 90.6 (59.2–125) | 82.6 (60.5–118.3) | 92.3 (61.5–145) | |||||
| Insulin 120 min OGTT (mU/L) | XX | 87 (56.9–136) | 0.065 | 79.8 (54.8–138.3) | 0.869 | 79.3 (50.8–133.8) | 0.607 | 79.8 (51.3–137) | 0.598 |
| Xx+xx | 66.1 (49.3–94.3) | 77.5 (52–121.8) | 77.5 (53.7–121.3) | 74.4 (54.2–124) | |||||
| HOMA IR | XX | 3.3 (2.5–5.4) | 0.001 | 3 (2.2–5.2) | 0.607 | 2.9 (1.6–5.4) | 0.605 | 2.8 (1.7–4.6) | 0.792 |
| Xx+xx | 2.3 (1.3–3.5) | 2.8 (1.7–4.8) | 2.8 (1.8–4.5) | 2.8 (1.8–5.1) | |||||
| Characteristic | Genotype * | CARD8 rs2043211 | NLRP3 rs35829419 | TNF rs1800629 | IL1B rs1143623 | IL1B rs16944 ** | IL6 rs1800795 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median Value (25–75%) | p | Median Value (25–75%) | p | Median Value (25–75%) | p | Median Value (25–75%) | p | Median Value (25–75%) | p | Median Value (25–75%) | p | ||
| Anthropometric Characteristics | |||||||||||||
| Body mass (kg) | XX | 101 (89.7–113.1) | 0.226 | 100 (85–112) | 0.990 | 101 (84.6–112.2) | 0.801 | 99.9 (86.6–110.6) | 0.776 | 100.5 (83–111.5) | 0.702 | 101(86.6–112) | 0.659 |
| Xx+xx | 98 (84.8–110.6) | 101(85.6–106) | 99.7(88.9–108.5) | 101.7(83.8–112.5) | 100 (87.4–110.7) | 100(84.5–110.4) | |||||||
| BMI (kg/m2) | XX | 36.6 (32.1–40.3) | 0.272 | 35.8 (31.7–40) | 0.551 | 36 (31.8–39.9) | 0.828 | 36 (31.8–40) | 0.995 | 35.5 (31.1–39.5) | 0.346 | 35.7 (31.9–40) | 0.821 |
| Xx+xx | 35.5 (31.6–39.8) | 37.5 (32.2–39.8) | 34.6 (32.1–39.9) | 35.7 (31.7–39.9) | 37 (31.9–40.3) | 36 (31.1–39.7) | |||||||
| Waist circumference (cm) | XX | 112 (102–127) | 0.527 | 110.3(101.3–121) | 0.097 | 112 (102–123) | 0.567 | 110.8(100.8–121) | 0.183 | 112(103–123) | 0.469 | 112.5(102–120.5) | 0.696 |
| Xx+xx | 111.5(101.8–119.5) | 117 (109–125) | 110 (102–119) | 114 (104–128.8) | 111(100–121) | 111 (101–122.8) | |||||||
| VAT mass (g) | XX | 759 (573.5–989) | 0.669 | 758 (580.3–959.5) | 0.634 | 797 (614–988) | 0.111 | 755 (588.5–898) | 0.542 | 710(541.5–979.5) | 0.363 | 698.5(521.5–949) | 0.533 |
| Xx+xx | 761 (581–937) | 804 (562–1015) | 699(514.8–852) | 795(567.8–1003.5) | 805.5(611–952.5) | 761 (612–969.5) | |||||||
| VAT volume (cm3) | XX | 818 (620–1069) | 0.768 | 816 (627.8–1018) | 0.613 | 862 (664–1068) | 0.073 | 810.5(636.8–954.5) | 0.503 | 768(585.5–1059) | 0.403 | 745 (563.5–975.5) | 0.423 |
| Xx+xx | 823 (628.5–1013) | 869 (607–1097) | 747(556.3–909.8) | 859.5(613.5–1085) | 843.5(661–1003.5) | 823 (662–1048) | |||||||
| VAT surface (cm2) | XX | 155 (116–199.5) | 0.864 | 156 (117–194) | 0.568 | 165 (124–201) | 0.202 | 153.5(115.5–179.8) | 0.272 | 152.5(113.8–205) | 0.718 | 144 (110.5–204.5) | 0.739 |
| Xx+xx | 158.5(122.3–199.5) | 167 (116–211) | 144 (108–177) | 165 (118.5–211) | 156.5(124.8–187.5) | 157 (125.5–200) | |||||||
| OGTT | |||||||||||||
| Glucose 0 min OGTT (mmol/L) | XX | 5.1 (4.8–5.6) | 0.847 | 5.2 (4.8–5.6) | 0.568 | 5.2 (4.9–5.6) | 0.762 | 5.1 (4.8–5.6) | 0.547 | 5.1 (4.9–5.5) | 0.616 | 5.2 (4.9–5.6) | 0.572 |
| Xx+xx | 5.2 (4.9–5.6) | 5.2 (4.9–5.7) | 5.2 (4.7–5.7) | 5.2 (4.9–5.5) | 5.2 (4.8–5.7) | 5.1 (4.8–5.6) | |||||||
| Glucose 30 min OGTT (mmol/L) | XX | 8.4 (6.9–9.3) | 0.833 | 8.4 (6.9–9.3) | 0.620 | 8.7 (7.3–10.1) | 0.020 | 8.4 (6.7–9.3) | 0.552 | 7.9 (7–10) | 0.840 | 8.1 (6.9–9.5) | 0.492 |
| Xx+xx | 8.3 (7.1–9.6) | 8.1 (6.9–10.7) | 7.5 (6.3–9) | 8.1 (7.1–10.5) | 8.6 (6.9–9.3) | 8.5 (7–9.4) | |||||||
| Glucose 60 min OGTT (mmol/L) | XX | 8.5 (6.9–9.7) | 0.997 | 8.3 (6.8–9.7) | 0.396 | 9 (7.1–10) | 0.007 | 8.2 (6.7–9.6) | 0.236 | 8.1 (6.8–11.1) | 0.808 | 8.3 (6.9–9.5) | 0.724 |
| Xx+xx | 8.3 (6.7–10) | 8.8 (6.7–12) | 7.3 (5.9–8.4) | 8.7 (6.9–11.5) | 8.6 (6.7–9.7) | 8.5 (6.5–10.5) | |||||||
| Glucose 90 min OGTT (mmol/L) | XX | 7.6 (5.7–9.4) | 0.862 | 7.4 (5.9–8.9) | 0.227 | 7.8 (6.6–9.1) | 0.032 | 7.4 (5.8–9.1) | 0.457 | 7.8 (6–9.1) | 0.896 | 7.7 (6.7–9) | 0.849 |
| Xx+xx | 7.6 (6.2–8.7) | 8.8 (6.4–9.6) | 6.2 (5.7–8.1) | 7.9 (6.6–9) | 7.4 (5.8–9.1) | 7.5 (5.8–9.4) | |||||||
| Glucose 120 min OGTT (mmol/L) | XX | 6.9 (5.5–7.9) | 0.396 | 6.5 (5.5–7.7) | 0.048 | 6.6 (5.5–7.9) | 0.742 | 6.9 (5.5–7.8) | 0.995 | 6.6 (5.3–7.7) | 0.708 | 7.2 (5.9–8.2) | 0.091 |
| Xx+xx | 6.4 (5.5–7.8) | 7.6 (6.2–9.3) | 6.4 (5.5–7.8) | 6.6 (5.5–7.9) | 6.9 (5.6–8) | 6.3 (5.4–7.5) | |||||||
| Insulin 0 min OGTT (mU/L) | XX | 12.5 (9.2–21.1) | 0.303 | 11.8 (7–19.7) | 0.116 | 12.2 (7.1–20.1) | 0.611 | 11.4 (6.9–19.6) | 0.076 | 12.6 (7.7–20.2) | 0.829 | 12 (6.1–20.8) | 0.520 |
| Xx+xx | 12.4 (7–19.1) | 14.3 (11.3–24.2) | 13.4 (9.3–20) | 14.2 (10.6–22.5) | 11.8 (7.5–19.8) | 12.4 (8.6–20) | |||||||
| Insulin 30 min OGTT (mU/L) | XX | 64 (40.8–99.9) | 0.090 | 67.8 (44.8–101.3) | 0.420 | 82.7 (45.3–104) | 0.305 | 70.2 (45.2–102.9) | 0.728 | 73.2 (43.4–112) | 0.886 | 81.3 (41.6–113) | 0.755 |
| Xx+xx | 81.3 (47.9–112) | 79.9 (61.8–110) | 60.6 (43.4–110) | 73.2 (44.8–111.5) | 71.1 (49.4–99.2) | 68.1 (46.3–100) | |||||||
| Insulin 60 min OGTT (mU/L) | XX | 80.7 (63.3–138.5) | 0.768 | 94.1 (60.3–129.3) | 0.242 | 102 (72.5–137) | 0.044 | 98.4 (63.2–136.8) | 0.650 | 85.5 (54.2–128) | 0.251 | 104 (75.4–140.3) | 0.268 |
| Xx+xx | 99.2 (62–128.8) | 102 (76.9–147) | 67.4 (54.2–111) | 87.2 (60.8–127.5) | 99.4 (66.8–138) | 85.5 (61.8–127) | |||||||
| Insulin 90 min OGTT (mU/L) | XX | 81.2 (61.3–118.5) | 0.722 | 84.2 (59.2–118.3) | 0.092 | 94.4 (62.4–126) | 0.160 | 90.6 (64.2–128.8) | 0.974 | 79.9 (58.4–122) | 0.647 | 104.1 (59.1–150.5) | 0.470 |
| Xx+xx | 101.6 (64.2–124.5) | 115 (80.3–148) | 80.8 (56.1–115) | 79.9 (60.6–119) | 91.2 (71.4–125) | 85.8 (62.4–119) | |||||||
| Insulin 120 min OGTT (mU/L) | XX | 77.5 (56.4–117.5) | 0.985 | 72.6 (51.6–113.5) | 0.056 | 80 (61.3–128) | 0.138 | 72.6 (49.4–124) | 0.344 | 80 (51.8–122) | 0.678 | 80 (53–146) | 0.694 |
| Xx+xx | 80.3 (48.2–125.5) | 122 (77.8–162) | 65 (44.2–123.5) | 80.3 (55.1–124) | 73.1 (54.4–127.5) | 77.5 (52.9–120) | |||||||
| HOMA IR | XX | 2.8 (2.2–5.1) | 0.313 | 2.8 (1.6–4.9) | 0.104 | 2.8 (1.7–5.1) | 0.664 | 2.7 (1.6–4.8) | 0.079 | 3 (1.9–4.9) | 0.869 | 3 (1.4–4.8) | 0.655 |
| Xx+xx | 2.8 (1.6–4.8) | 3.3 (2.6–6.3) | 3.1 (2.1–4.8) | 3.2 (2.5–5.2) | 2.8 (1.7–5) | 2.8 (2.1–5.1) | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herman, R.; Jensterle, M.; Janež, A.; Goričar, K.; Dolžan, V. Genetic Variability in Antioxidative and Inflammatory Pathways Modifies the Risk for PCOS and Influences Metabolic Profile of the Syndrome. Metabolites 2020, 10, 439. https://doi.org/10.3390/metabo10110439
Herman R, Jensterle M, Janež A, Goričar K, Dolžan V. Genetic Variability in Antioxidative and Inflammatory Pathways Modifies the Risk for PCOS and Influences Metabolic Profile of the Syndrome. Metabolites. 2020; 10(11):439. https://doi.org/10.3390/metabo10110439
Chicago/Turabian StyleHerman, Rok, Mojca Jensterle, Andrej Janež, Katja Goričar, and Vita Dolžan. 2020. "Genetic Variability in Antioxidative and Inflammatory Pathways Modifies the Risk for PCOS and Influences Metabolic Profile of the Syndrome" Metabolites 10, no. 11: 439. https://doi.org/10.3390/metabo10110439
APA StyleHerman, R., Jensterle, M., Janež, A., Goričar, K., & Dolžan, V. (2020). Genetic Variability in Antioxidative and Inflammatory Pathways Modifies the Risk for PCOS and Influences Metabolic Profile of the Syndrome. Metabolites, 10(11), 439. https://doi.org/10.3390/metabo10110439






