Metabolomics Study of Serum from a Chronic Alcohol-Fed Rat Model Following Administration of Defatted Tenebrio molitor Larva Fermentation Extract
Abstract
1. Introduction
2. Results
2.1. Metabolic Profiling Analysis and Univariate Analysis
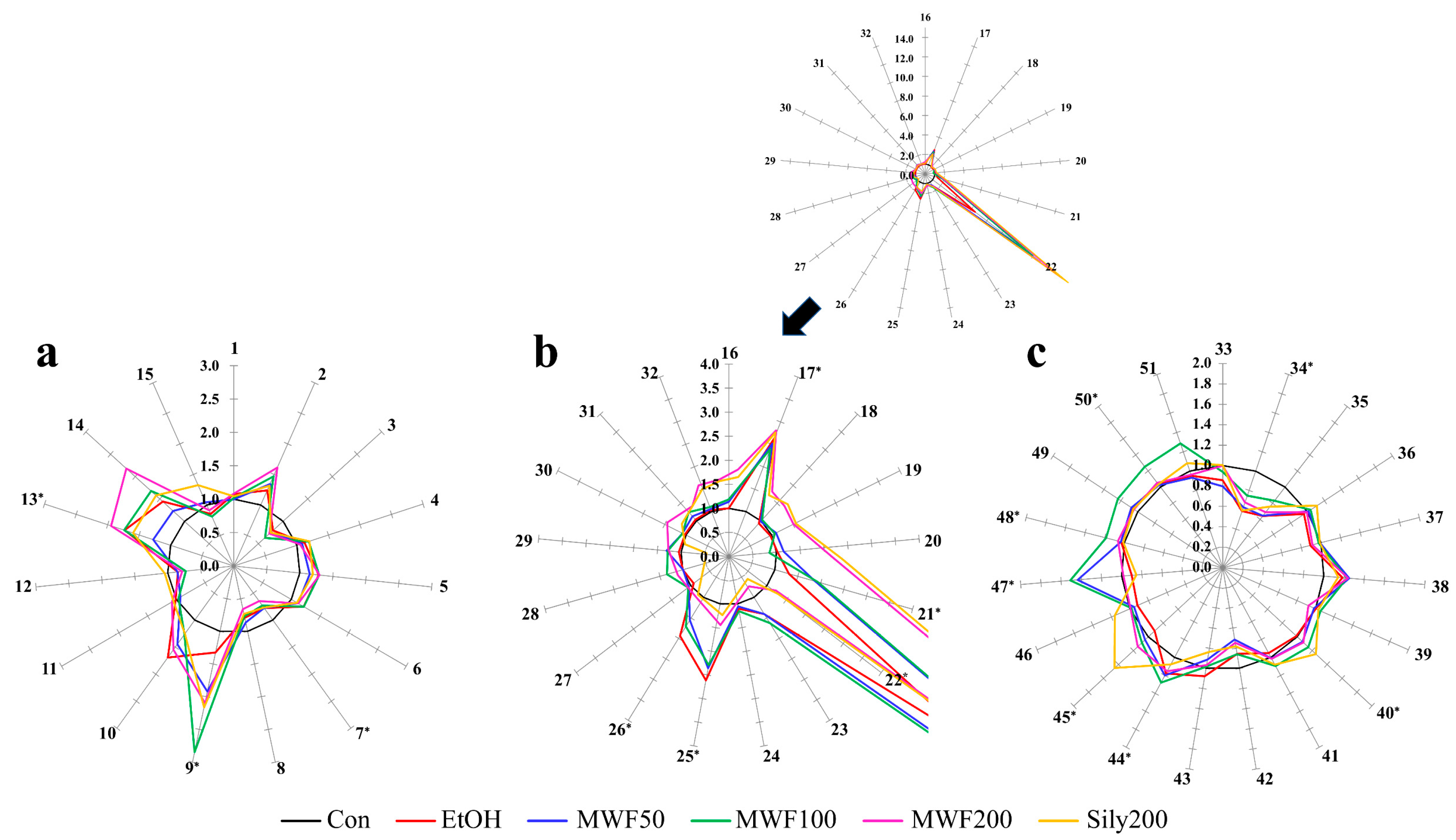
2.2. Star Pattern Recognition Analysis
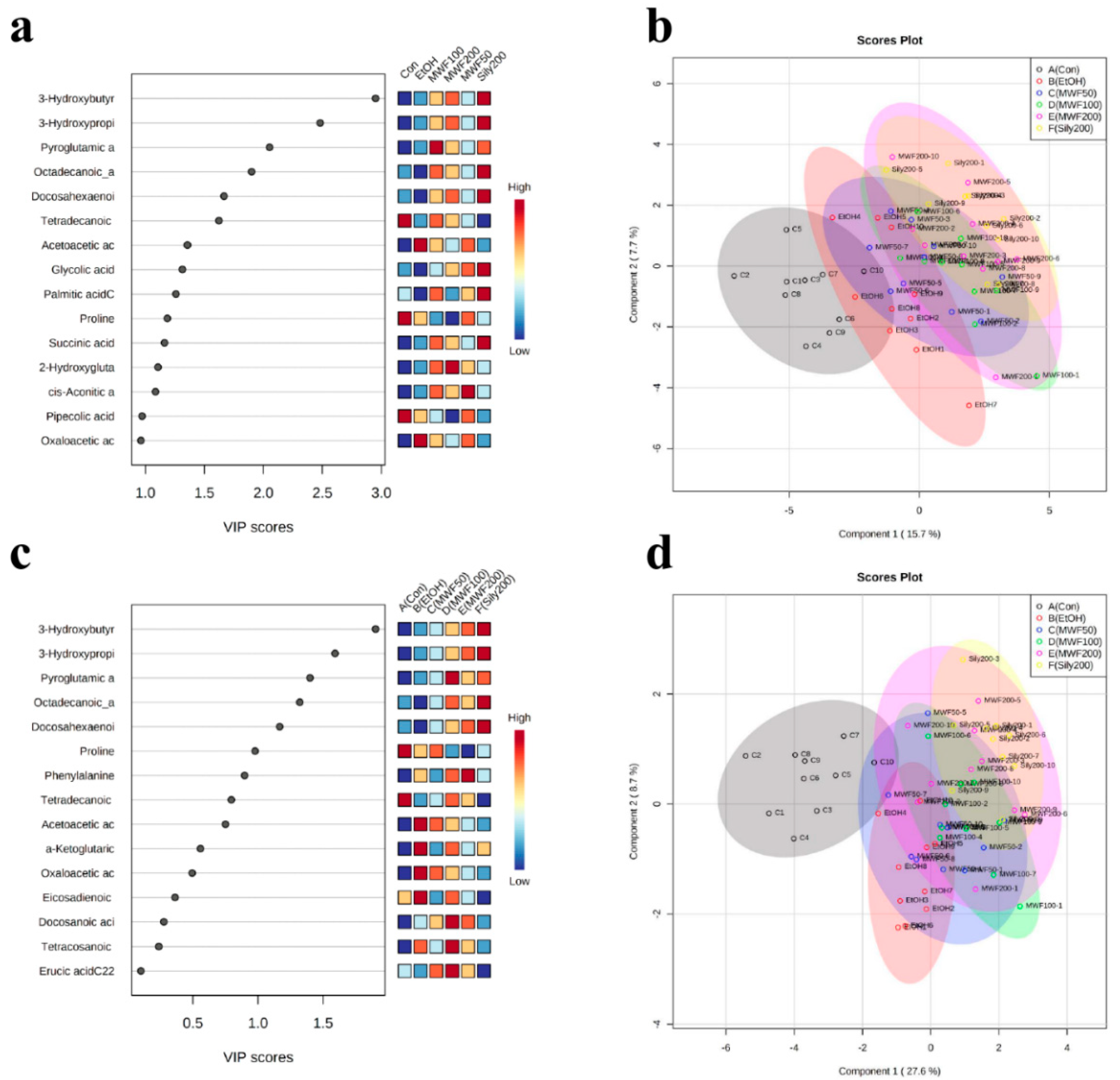
2.3. Multivariate Statistical Analysis
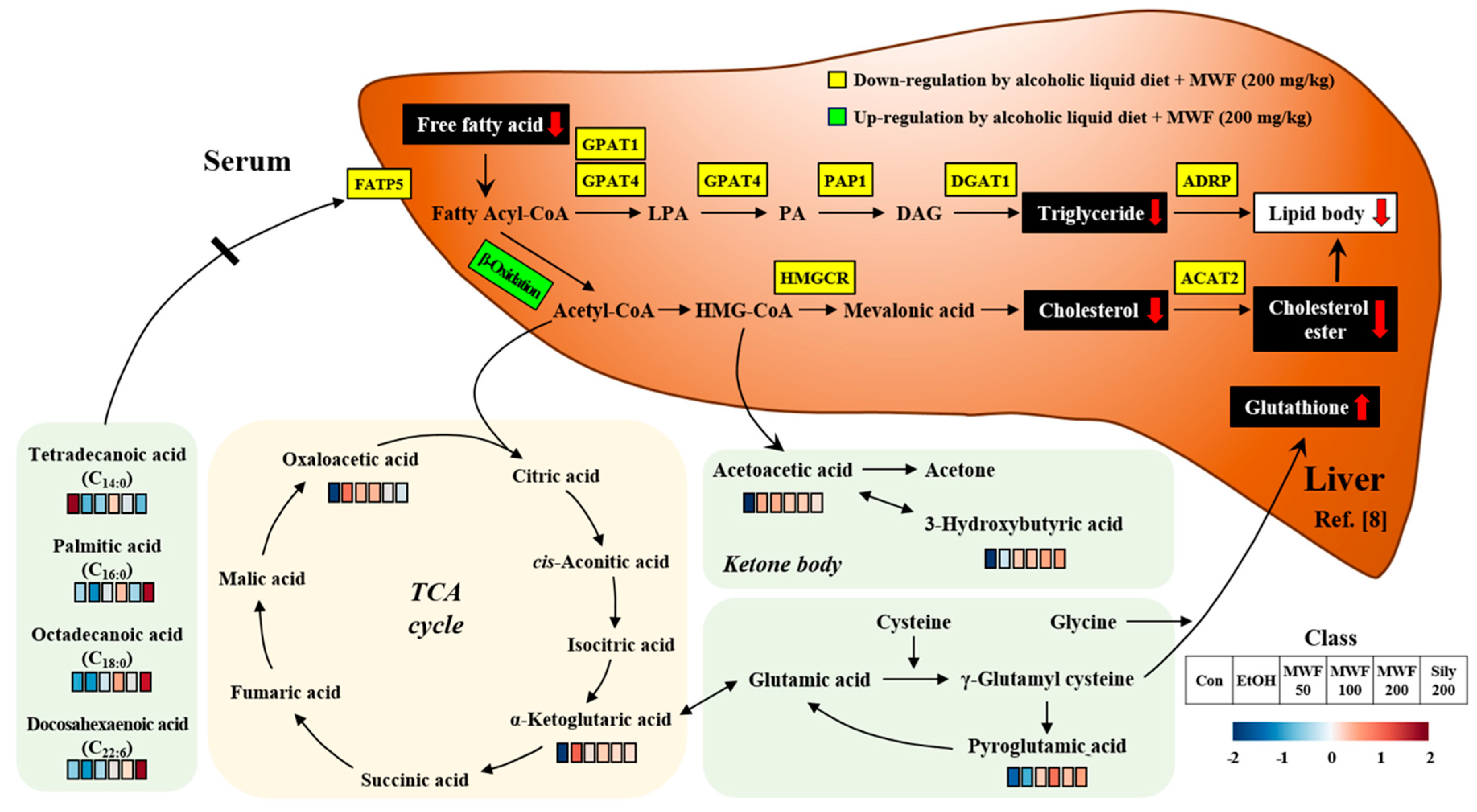
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Preparation of Serum from ALD Rat Model
4.3. Gas Chromatography-Tandem Mass Spectrometry (GC-MS/MS)
4.4. Sample Preparation for Serum AA Profiling Analysis
4.5. Sample Preparation for Serum OA Profiling Analysis
4.6. Sample Preparation for Serum FFA Profiling Analysis
4.7. Star Pattern Recognition Analysis and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sousa, P.; Borges, S.; Pintado, M. Enzymatic Hydrolysis of Insect Alphitobius diaperinus towards the Development of Bioactive Peptide Hydrolysates. Food Funct. 2020, 11, 3539–3548. [Google Scholar] [CrossRef] [PubMed]
- De Marco, M.; Martínez, S.; Hernandez, F.; Madrid, J.; Gai, F.; Rotolo, L.; Belforti, M.; Bergero, D.; Katz, H.; Dabbou, S.; et al. Nutritional Value of Two Insect Larval Meals (Tenebrio molitor and Hermetia illucens) for Broiler Chickens: Apparent Nutrient Digestibility, Apparent Ileal Amino Acid Digestibility and Apparent Metabolizable Energy. Anim. Feed Sci. Technol. 2015, 209, 211–218. [Google Scholar] [CrossRef]
- Seo, M.; Goo, T.W.; Chung, M.Y.; Baek, M.; Hwang, J.S.; Kim, M.A.; Yun, E.Y. Tenebrio molitor Larvae Inhibit Adipogenesis through AMPK and MAPKs Signaling in 3T3-L1 Adipocytes and Obesity in High-Fat Diet-Induced Obese Mice. Int. J. Mol. Sci. 2017, 18, 518. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.H.; Baek, H.K.; Lee, J.S.; Kim, S.J.; Yi, S.S. Chronic Oral Administration of Tenebrio molitor Extract Exhibits Inhibitory Effect on Glucocorticoid Receptor Overexpression in the Hippocampus of Ovariectomy-Induced Estrogen Deficient Mice. J. Food Sci. 2019, 84, 687–694. [Google Scholar] [CrossRef] [PubMed]
- del Hierro, J.N.; Gutiérrez-Docio, A.; Otero, P.; Reglero, G.; Martin, D. Characterization, Antioxidant Activity, and Inhibitory Effect on Pancreatic Lipase of Extracts from the Edible Insects Acheta domesticus and Tenebrio molitor. Food Chem. 2020, 309, 125742. [Google Scholar] [CrossRef]
- Pessina, F.; Frosini, M.; Marcolongo, P.; Fusi, F.; Saponara, S.; Gamberucci, A.; Valoti, M.; Giustarini, D.; Fiorenzani, P.; Gorelli, B.; et al. Antihypertensive, Cardio-and Neuro-Protective Effects of Tenebrio molitor (Coleoptera: Tenebrionidae) Defatted Larvae in Spontaneously Hypertensive Rats. PLoS ONE 2020, 15, e0233788. [Google Scholar] [CrossRef]
- Cho, H.R.; Lee, S.O. Novel Hepatoprotective Peptides Derived from Protein Hydrolysates of Mealworm (Tenebrio molitor). Food Res. Int. 2020, 133, 109194. [Google Scholar] [CrossRef]
- Choi, R.Y.; Ham, J.R.; Ryu, H.S.; Lee, S.S.; Miguel, M.A.; Paik, M.J.; Ji, M.; Park, K.W.; Kang, K.Y.; Lee, H.I.; et al. Defatted Tenebrio molitor Larva Fermentation Extract Modifies Steatosis, Inflammation and Intestinal Microflora in Chronic Alcohol-Fed Rats. Nutrients 2020, 12, 1426. [Google Scholar] [CrossRef]
- Rehm, J.; Mathers, C.; Popova, S.; Thavorncharoensap, M.; Teerawattananon, Y.; Patra, J. Global Burden of Disease and Injury and Economic Cost Attributable to Alcohol Use and Alcohol-Use Disorders. Lancet 2009, 373, 2223–2233. [Google Scholar] [CrossRef]
- Seitz, H.K.; Bataller, R.; Cortez-Pinto, H.; Gao, B.; Gual, A.; Lackner, C.; Mathurin, P.; Mueller, S.; Szabo, G.; Tsukamoto, H. Alcoholic Liver Disease. Nat. Rev. Dis. Primers 2018, 4, 1–22. [Google Scholar] [CrossRef]
- Tajiri, K.; Shimizu, Y. Branched-Chain Amino Acids in Liver Diseases. World J. Gastroenterol. 2013, 19, 7620–7629. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, L.; Corsetti, G.; Ruocco, C.; Ragni, M.; Rossi, F.; Carruba, M.O.; Valerio, A.; Nisoli, E. A Specific Amino Acid Formula Prevents Alcoholic Liver Disease in Rodents. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 314, G566–G582. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Kim, E.H. Therapeutic Effects of Amino Acids in Liver Diseases: Current Studies and Future Perspectives. J. Cancer Prev. 2019, 24, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Mavrelis, P.G.; Ammon, H.V.; Gleysteen, J.J.; Komorowski, R.A.; Charaf, U.K. Hepatic Free Fatty Acids in Alcoholic Liver Disease and Morbid Obesity. Hepatology 1983, 3, 226–231. [Google Scholar] [CrossRef]
- Clish, C.B. Metabolomics: An Emerging but Powerful Tool for Precision Medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef]
- Ma, T.; Li, Y.; Zhu, Y.; Jiang, S.; Cheng, C.; Peng, Z.; Xu, L. Differential Metabolic Pathways and Metabolites in a C57BL/6J Mouse Model of Alcoholic Liver Disease. Med. Sci. Monit. 2020, 26, e924602. [Google Scholar] [CrossRef]
- Dong, Y.; Qiu, P.; Zhao, L.; Zhang, P.; Huang, X.; Li, C.; Chai, K.; Shou, D. Metabolomics Study of the Hepatoprotective Effect of Phellinus igniarius in Chronic Ethanol-Induced Liver Injury Mice using UPLC-Q/TOF-MS Combined with Ingenuity Pathway Analysis. Phytomedicine 2018, 74, 152697. [Google Scholar] [CrossRef]
- Harada, S.; Takebayashi, T.; Kurihara, A.; Akiyama, M.; Suzuki, A.; Hatakeyama, Y.; Sugiyama, D.; Kuwabara, K.; Takeuchi, A.; Okamura, T.; et al. Metabolomic Profiling Reveals Novel Biomarkers of Alcohol Intake and Alcohol-Induced Liver Injury in Community-Dwelling Men. Phytomedicine 2020, 74, 152697. [Google Scholar]
- Lian, J.S.; Liu, W.; Hao, S.R.; Guo, Y.Z.; Huang, H.J.; Chen, D.Y.; Xie, Q.; Pan, X.P.; Xu, W.; Yuan, W.X.; et al. A Serum Metabonomics Study on the Difference Between Alcohol-and HBV-Induced Liver Cirrhosis by Ultraperformance Liquid Chromatography Coupled to Mass Spectrometry Plus Quadrupole Time-of-Flight Mass Spectrometry. Chin. Med. J. 2011, 124, 1367–1373. [Google Scholar]
- Lee, H.S.; Seo, C.; Kim, Y.A.; Park, M.; Choi, B.; Ji, M.; Lee, S.; Paik, M.J. Metabolomic Study of Polyamines in Rat Urine following Intraperitoneal Injection of γ-Hydroxybutyric Acid. Metabolomics 2019, 15, 58. [Google Scholar] [CrossRef]
- Seo, C.; Hwang, Y.H.; Kim, Y.; Joo, B.S.; Yee, S.T.; Kim, C.M.; Paik, M.J. Metabolomic Study of Aging in Mouse Plasma by Gas Chromatography–Mass Spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016, 1025, 1–6. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar] [PubMed]
- Maezono, K.; Mawatari, K.; Kajiwara, K.; Shinkai, A.; Maki, T. Effect of Alanine on D-Galactosamine-Induced Acute Liver Failure in Rats. Hepatology 1996, 24, 1211–1216. [Google Scholar]
- Freudenberg, A.; Petzke, K.J.; Klaus, S. Dietary L-Leucine and L-Alanine Supplementation Have Similar Acute Effects in the Prevention of High-Fat Diet-Induced Obesity. Amino Acids 2013, 44, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Zeb, A.; Rahman, S.U. Protective Effects of Dietary Glycine and Glutamic Acid toward the Toxic Effects of Oxidized Mustard Oil in Rabbits. Food Funct. 2017, 8, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Park, J.G.; Tak, W.Y.; Park, S.Y.; Kweon, Y.O.; Jang, S.Y.; Lee, Y.R.; Bae, S.H.; Jang, J.Y.; Kim, D.Y.; Lee, J.S.; et al. Effects of Branched-Chain Amino Acids (BCAAs) on the Progression of Advanced Liver Disease: A Korean Nationwide, Multicenter, Retrospective, Observational, Cohort study. Medicine 2017, 96, e6580. [Google Scholar] [CrossRef]
- Puchalska, P.; Crawford, P.A. Multi-Dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef]
- Chen, Y.; Ouyang, X.; Hoque, R.; Garcia-Martinez, I.; Yousaf, M.N.; Tonack, S.; Offermanns, S.; Dubuquoy, L.; Louvet, A.; Mathurin, P.; et al. β-Hydroxybutyrate Protects from Alcohol-Induced Liver Injury via a Hcar2-cAMP Dependent Pathway. J. Hepatol. 2018, 69, 687–696. [Google Scholar] [CrossRef]
- Ajmo, J.M.; Liang, X.; Rogers, C.Q.; Pennock, B.; You, M. Resveratrol Alleviates Alcoholic Fatty Liver in Mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 295, G833–G842. [Google Scholar] [CrossRef]
- Li, Q.; Xie, G.; Zhang, W.; Zhong, W.; Sun, X.; Tan, X.; Sun, X.; Jia, W.; Zhou, Z. Dietary Nicotinic Acid Supplementation Ameliorates Chronic Alcohol-Induced Fatty Liver in Rats. Alcohol. Clin. Exp. Res. 2014, 38, 1982–1992. [Google Scholar] [CrossRef]
- Rodríguez-Gallego, E.; Guirro, M.; Riera-Borrull, M.; Hernandez-Aguilera, A.; Marine-Casado, R.; Fernandez-Arroyo, S.; Beltran-Debon, R.; Sabench, F.; Hernandez, M.; del Castillo, D. Mapping of the Circulating Metabolome Reveals α-Ketoglutarate as a Predictor of Morbid Obesity-Associated Non-Alcoholic Fatty Liver Disease. Int. J. Obes. 2015, 39, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Spector, S.R.; Mayan, H.; Loebstein, R.; Markovits, N.; Priel, E.; Massalha, E.; Shafir, Y.; Gueta, I. Pyroglutamic Acidosis as a Cause for High Anion Gap Metabolic Acidosis: A Prospective Study. Sci. Rep. 2019, 9, 3554. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Okita, Y.; de Toledo, A.; Miyazaki, H.; Hirano, E.; Morinaga, T. Pyroglutamic Acid Stimulates DNA Synthesis in Rat Primary Hepatocytes through the Mitogen-Activated Protein Kinase Pathway. Biosci. Biotechnol. Biochem. 2015, 79, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Guelakis, M.P.; Lee, J.; Rosa, J.G.; Madison, S.A.; Damodaran, A.; Kumari, A.; Huang, N.; Harichian, B. Personal Care Compositions with Glutathione Precursor Comprising 4-Substituted Resorcinols and Amino Acids. USA Patent PCT/CN2017/116999, 28 June 2018. [Google Scholar]
- Di Miceli, M.; Gronier, B. Pharmacology, Systematic Review and Recent Clinical Trials of Metadoxine. Rev. Recent Clin. Trials 2018, 13, 114–125. [Google Scholar] [CrossRef]
- Lim, J.; Li, L.; Jacobs, M.D.; Kistler, J.; Donaldson, P.J. Mapping of Glutathione and Its Precursor Amino Acids Reveals a Role for GLYT2 in Glycine Uptake in the Lens Core. Investig. Ophthalmol. Vis. Sci. 2007, 48, 5142–5151. [Google Scholar] [CrossRef] [PubMed]
- Mayatepek, E. Current Opinion in Pediatric Metabolic Disease. J. Pediat. Sci. 2011, 3, e64. [Google Scholar]
- Bando, K.; Kunimatsu, T.; Sakai, J.; Kimura, J.; Funabashi, H.; Seki, T.; Bamba, T.; Fukusaki, E. GC-MS-Based Metabolomics Reveals Mechanism of Action for Hydrazine Induced Hepatotoxicity in Rats. J. Appl. Toxicol. 2011, 31, 524–535. [Google Scholar] [CrossRef]
- Wu, H.; Feng, F. Untargeted Metabolomic Analysis using LC-TOF/MS and LC-MS/MS for Revealing Metabolic Alterations Linked to Alcohol-Induced Hepatic Steatosis in Rat Serum and Plasma. RSC Adv. 2016, 6, 28279–28288. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, X.; Ma, L.J.; Feng, R.B.; Yan, C.; Su, H.; He, C.; Kang, J.X.; Liu, B.; Wan, J. Omega-3 Polyunsaturated Fatty Acids Ameliorate Ethanol-Induced Adipose Hyperlipolysis: A Mechanism for Hepatoprotective Effect against Alcoholic Liver Disease. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 3190–3201. [Google Scholar] [CrossRef]
- Piquet, M.A.; Roulet, M.; Nogueira, V.; Filippi, C.; Sibille, B.; Hourmand-Ollivier, I.; Pilet, M.; Rouleau, V.; Leverve, X.M. Polyunsaturated Fatty Acid Deficiency Reverses Effects of Alcohol on Mitochondrial Energy Metabolism. J. Hepatol. 2004, 41, 721–729. [Google Scholar] [CrossRef]
- Huang, L.L.; Wan, J.B.; Wang, B.; He, C.W.; Ma, H.; Li, T.W.; Kang, J.X. Suppression of Acute Ethanol-Induced Hepatic Steatosis by Docosahexaenoic Acid is Associated with Downregulation of Stearoyl-CoA Desaturase 1 and Inflammatory Cytokines. Prostaglandins Leukot. Essent. Fatty Acids 2013, 88, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Saini, R.K.; Keum, Y.S. Omega-3 and Omega-6 Polyunsaturated Fatty Acids: Dietary Sources, Metabolism, and Significance-A Review. Life Sci. 2018, 203, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Guan, Y.S. Cautiously using Natural Medicine to Treat Liver Problems. World J. Gastroenterol. 2017, 23, 3388–3395. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Lee, G.; Moon, S.M.; Park, M.J.; Hong, S.K.; Ahn, Y.H.; Kim, K.R.; Paik, M.J. Metabolomic Screening and Star Pattern Recognition by Urinary Amino Acid Profile Analysis from Bladder Cancer Patients. Metabolomics 2010, 6, 202–206. [Google Scholar] [CrossRef]
- Seo, C.; Park, M.; Choi, B.; Lee, S.; Paik, M.J. Metabolomic Analysis of Urinary Organic Acids following Intraperitoneal Injection with γ-Hydroxybutyric Acid in Rats. Metabolomics 2016, 12, 190. [Google Scholar] [CrossRef]
- Seo, C.; Yoon, J.; Rhee, Y.; Kim, J.J.; Nam, S.J.; Lee, W.; Lee, G.; Yee, S.T.; Paik, M.J. Simultaneous Analysis of Seven 2-Hydroxy Fatty Acids as Tert-Butyldimethylsilyl Derivatives in Plasma by Gas Chromatography–Mass Spectrometry. Biomed. Chromatogr. 2015, 29, 156–160. [Google Scholar] [CrossRef]


| No. | Metabolite | Concentration (μg/Serum of 50 μL) | Normalized Value a | p-Value b | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Con | EtOH | MWF 50 | MWF 100 | MWF 200 | Sily 200 | EtOH | MWF 50 | MWF 100 | MWF 200 | Sily 200 | ANOVA | FDR c | ||
| 7 | Proline | 1.65 ± 0.43 | 1.28 ± 0.24 | 1.28 ± 0.28 | 1.20 ± 0.24 | 1.07 ± 0.22 | 1.27 ± 0.23 | 0.78 | 0.78 | 0.73 | 0.65 | 0.77 | 0.005 | 0.019 |
| 9 | Pyroglutamic acid | 0.47 ± 0.13 | 0.62 ± 0.28 | 0.90 ± 0.21 | 1.33 ± 1.14 | 0.98 ± 0.34 | 1.01 ± 0.22 | 1.32 | 1.92 | 2.85 | 2.09 | 2.16 | <0.001 | 0.001 |
| 13 | Phenylalanine | 0.67 ± 0.27 | 1.15 ± 0.35 | 0.85 ± 0.20 | 1.16 ± 0.41 | 1.30 ± 0.27 | 1.07 ± 0.49 | 1.73 | 1.28 | 1.74 | 1.95 | 1.60 | 0.001 | 0.007 |
| 17 | Acetoacetic acid | 0.44 ± 0.27 | 1.18 ± 0.61 | 1.11 ± 0.45 | 1.06 ± 0.55 | 0.97 ± 0.29 | 0.95 ± 0.46 | 2.66 | 2.52 | 2.40 | 2.20 | 2.15 | 0.007 | 0.023 |
| 21 | 3-Hydroxypropionic acid | 0.30 ± 0.14 | 0.39 ± 0.12 | 0.55 ± 0.18 | 0.57 ± 0.14 | 0.59 ± 0.16 | 0.67 ± 0.11 | 1.31 | 1.88 | 1.94 | 2.01 | 2.25 | <0.001 | 0.001 |
| 22 | 3-Hydroxybutyric acid | 0.26 ± 0.17 | 1.67 ± 0.70 | 3.53 ± 2.62 | 3.58 ± 1.60 | 4.59 ± 1.83 | 4.79 ± 1.87 | 6.48 | 13.67 | 13.87 | 17.77 | 18.53 | <0.001 | <0.001 |
| 25 | Oxaloacetic acid | 0.02 ± 0.00 | 0.06 ± 0.01 | 0.05 ± 0.02 | 0.05 ± 0.02 | 0.04 ± 0.02 | 0.04 ± 0.01 | 2.62 | 2.36 | 2.30 | 2.03 | 1.82 | <0.001 | 0.001 |
| 26 | α-Ketoglutaric acid | 0.20 ± 0.04 | 0.39 ± 0.10 | 0.31 ± 0.08 | 0.34 ± 0.12 | 0.33 ± 0.14 | 0.32 ± 0.10 | 1.94 | 1.55 | 1.71 | 1.67 | 1.60 | 0.005 | 0.019 |
| 34 | Tetradecanoic acid (C14:0) | 0.10 ± 0.03 | 0.06 ± 0.01 | 0.06 ± 0.02 | 0.07 ± 0.02 | 0.06 ± 0.02 | 0.06 ± 0.01 | 0.59 | 0.63 | 0.75 | 0.68 | 0.59 | 0.001 | 0.006 |
| 40 | Octadecanoic acid (C18:0) | 7.56 ± 0.62 | 7.48 ± 0.68 | 8.10 ± 1.00 | 8.68 ± 0.94 | 8.17 ± 0.83 | 9.45 ± 1.27 | 0.99 | 1.07 | 1.15 | 1.08 | 1.25 | 0.001 | 0.006 |
| 44 | Eicosanoic acid (C20:0) | 0.03 ± 0.00 | 0.03 ± 0.00 | 0.03 ± 0.01 | 0.04 ± 0.00 | 0.03 ± 0.00 | 0.03 ± 0.00 | 1.18 | 1.20 | 1.28 | 1.15 | 1.08 | <0.001 | 0.001 |
| 45 | Docosahexaenoic acid (DHA, C22:6) | 2.32 ± 0.57 | 2.12 ± 0.34 | 2.41 ± 0.68 | 2.52 ± 0.53 | 2.65 ± 0.65 | 3.36 ± 0.70 | 0.91 | 1.04 | 1.09 | 1.14 | 1.45 | 0.002 | 0.012 |
| 47 | Erucic acid (C22:1) | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.90 | 1.44 | 1.51 | 0.98 | 0.86 | 0.001 | 0.005 |
| 48 | Docosanoic acid (C22:0) | 0.02 ± 0.00 | 0.02 ± 0.00 | 0.02 ± 0.00 | 0.02 ± 0.01 | 0.02 ± 0.00 | 0.02 ± 0.00 | 1.04 | 1.04 | 1.19 | 1.07 | 1.01 | 0.002 | 0.012 |
| 50 | Tetracosanoic acid (C24:0) | 0.02 ± 0.00 | 0.02 ± 0.00 | 0.02 ± 0.00 | 0.02 ± 0.01 | 0.02 ± 0.00 | 0.02 ± 0.00 | 1.04 | 1.03 | 1.25 | 1.05 | 1.04 | 0.003 | 0.013 |
| No. | Metabolite | Unsupervised Learning | Supervised Learning | |
|---|---|---|---|---|
| PCA Loading Score | PLS-DA | |||
| PC1 | PC2 | VIP Score a | ||
| 1 | Alanine | −0.137 | 0.110 | 0.240 |
| 2 | Glycine | −0.043 | 0.314 | 0.557 |
| 3 | α-Aminobutyric acid | −0.025 | −0.033 | 0.850 |
| 4 | Valine | −0.219 | −0.187 | 0.828 |
| 5 | Leucine | −0.247 | 0.021 | 0.356 |
| 6 | Isoleucine | −0.231 | −0.101 | 0.185 |
| 7 | Proline | 0.048 | −0.203 | 0.734 |
| 8 | Pipecolic acid | 0.197 | −0.036 | 0.705 |
| 9 | Pyroglutamic acid | −0.237 | −0.002 | 2.375 |
| 10 | Methionine | −0.011 | 0.079 | 0.491 |
| 11 | Serine | 0.034 | 0.203 | 0.365 |
| 12 | Threonine | 0.001 | −0.171 | 0.064 |
| 13 | Phenylalanine | −0.112 | 0.265 | 0.323 |
| 14 | Aspartic acid | −0.047 | 0.280 | 0.008 |
| 15 | 4-Hydroxyproline | 0.002 | −0.167 | 0.707 |
| 16 | Pyruvic acid | −0.100 | 0.003 | 0.615 |
| 17 | Acetoacetic acid | −0.056 | 0.092 | 0.660 |
| 18 | Lactic acid | −0.178 | −0.001 | 0.223 |
| 19 | Glycolic acid | −0.107 | −0.034 | 1.438 |
| 20 | 2-Hydroxybutyric acid | −0.142 | −0.037 | 0.471 |
| 21 | 3-Hydroxypropionic acid | −0.183 | 0.020 | 2.384 |
| 22 | 3-Hydroxybutyric acid | −0.194 | 0.124 | 2.391 |
| 23 | Succinic acid | −0.235 | 0.047 | 1.114 |
| 24 | Fumaric acid | −0.193 | 0.115 | 0.147 |
| 25 | Oxaloacetic acid | −0.205 | 0.115 | 0.334 |
| 26 | α-Ketoglutaric acid | −0.216 | 0.058 | 0.233 |
| 27 | 4-Hydroxyphenylacetic acid | 0.014 | 0.049 | 0.080 |
| 28 | Malic acid | −0.174 | 0.160 | 0.572 |
| 29 | 2-Hydroxyglutaric acid | −0.146 | 0.078 | 0.988 |
| 30 | cis-Aconitic acid | −0.051 | 0.093 | 1.083 |
| 31 | Citric acid | −0.092 | 0.164 | 0.650 |
| 32 | Isocitric acid | −0.096 | 0.153 | 0.569 |
| 33 | Dodecanoic acid (C12:0) | −0.016 | −0.064 | 0.463 |
| 34 | Tetradecanoic acid (C14:0) | 0.046 | −0.079 | 0.695 |
| 35 | Palmitoleic acid (C16:1) | 0.080 | −0.184 | 0.143 |
| 36 | Palmitic acid (C16:0) | 0.113 | −0.155 | 1.812 |
| 37 | γ-Linolenic acid (γ-C18:3) | −0.180 | −0.249 | 0.935 |
| 38 | Linoleic acid (C18:2) | −0.061 | −0.135 | 0.514 |
| 39 | Oleic acid (C18:1) | −0.115 | −0.097 | 0.522 |
| 40 | Octadecanoic acid (C18:0) | −0.230 | −0.184 | 2.524 |
| 41 | Arachidonic acid (C20:4) | −0.210 | −0.115 | 1.359 |
| 42 | 11-Eicosenic acid (C20:1) | −0.081 | −0.220 | 0.351 |
| 43 | Eicosadienoic acid (C20:2) | −0.099 | −0.240 | 0.859 |
| 44 | Eicosanoic acid (C20:0) | −0.186 | −0.192 | 0.569 |
| 45 | Docosahexaenoic acid (DHA, C22:6) | −0.180 | 0.007 | 1.974 |
| 46 | Docosatetraenoic acid (C22:4) | −0.145 | −0.131 | 1.129 |
| 47 | Erucic acid (C22:1) | −0.103 | −0.163 | 0.835 |
| 48 | Docosanoic acid (C22:0) | −0.093 | −0.038 | 0.550 |
| 49 | Nervonic acid (C24:1) | −0.083 | 0.042 | 0.620 |
| 50 | Tetracosanoic acid (C24:0) | −0.058 | 0.004 | 0.575 |
| 51 | Hexacosanoic acid(C26:0) | 0.030 | 0.015 | 0.532 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, R.-Y.; Ji, M.; Lee, M.-K.; Paik, M.-J. Metabolomics Study of Serum from a Chronic Alcohol-Fed Rat Model Following Administration of Defatted Tenebrio molitor Larva Fermentation Extract. Metabolites 2020, 10, 436. https://doi.org/10.3390/metabo10110436
Choi R-Y, Ji M, Lee M-K, Paik M-J. Metabolomics Study of Serum from a Chronic Alcohol-Fed Rat Model Following Administration of Defatted Tenebrio molitor Larva Fermentation Extract. Metabolites. 2020; 10(11):436. https://doi.org/10.3390/metabo10110436
Chicago/Turabian StyleChoi, Ra-Yeong, Moongi Ji, Mi-Kyung Lee, and Man-Jeong Paik. 2020. "Metabolomics Study of Serum from a Chronic Alcohol-Fed Rat Model Following Administration of Defatted Tenebrio molitor Larva Fermentation Extract" Metabolites 10, no. 11: 436. https://doi.org/10.3390/metabo10110436
APA StyleChoi, R.-Y., Ji, M., Lee, M.-K., & Paik, M.-J. (2020). Metabolomics Study of Serum from a Chronic Alcohol-Fed Rat Model Following Administration of Defatted Tenebrio molitor Larva Fermentation Extract. Metabolites, 10(11), 436. https://doi.org/10.3390/metabo10110436





