Optimizing the Performance of Breast Cancer Classification by Employing the Same Domain Transfer Learning from Hybrid Deep Convolutional Neural Network Model
Abstract
1. Introduction
- A hybrid deep convolutional neural networks (DCNNs) model has been designed based upon an integration of a multi-branch of parallel convolutional layers and residual links.
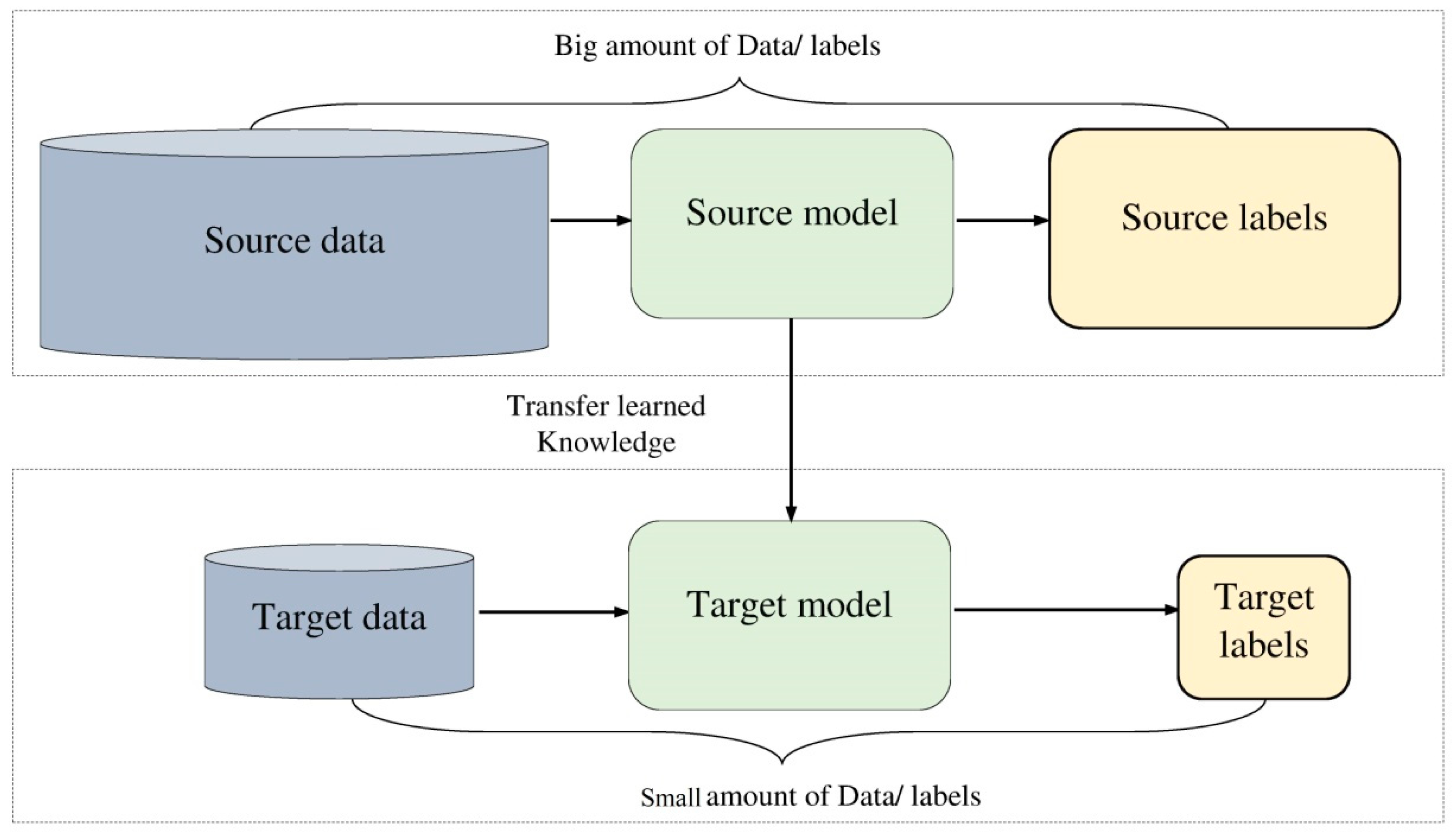
- Transfer learning has been employed to tackle the lack of training data in two ways: transfer learning from the same domain of target task and transfer learning from a domain different of the target task.
- Four different experiments have been utilized to train our proposed model.
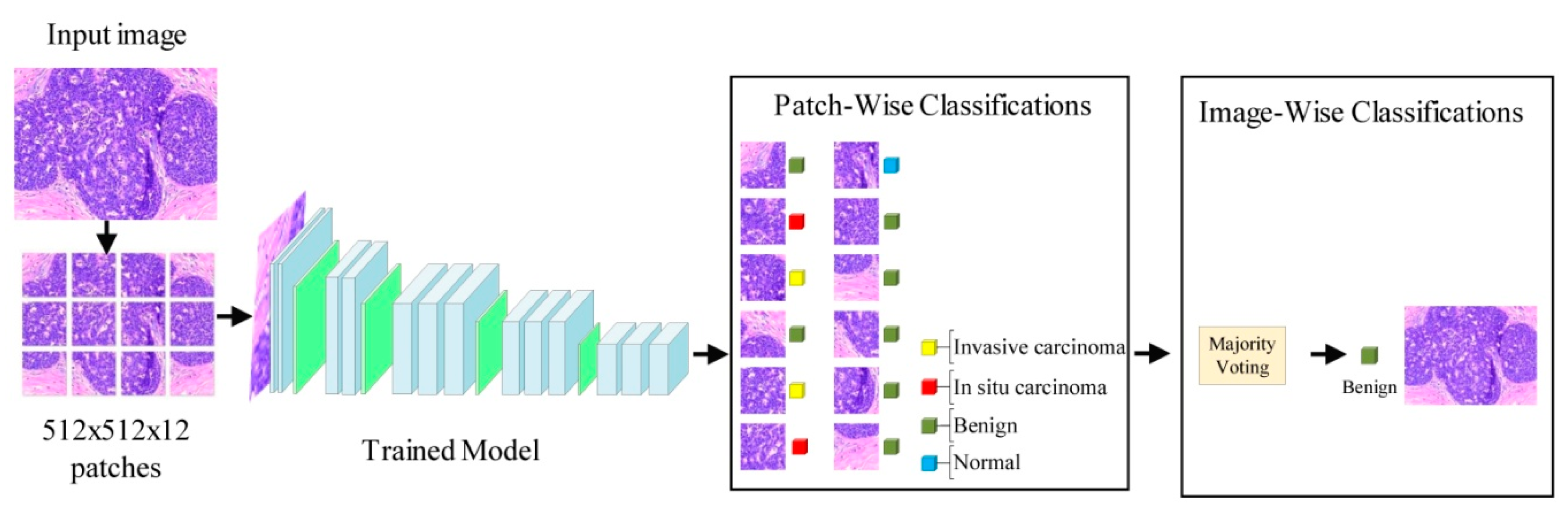
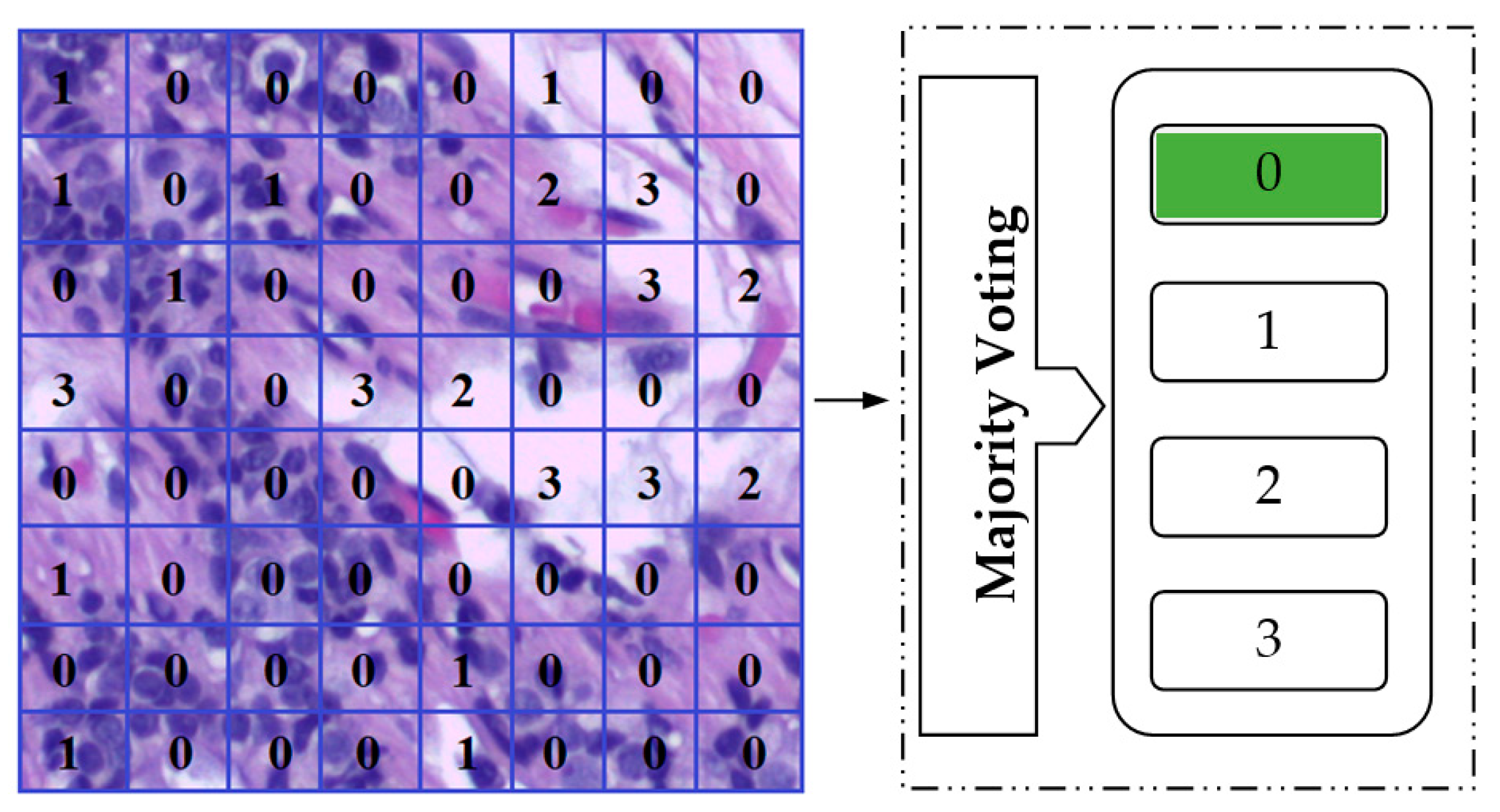
- Two methods of evaluation (image-wise and patch-wise classification modes) have been used.
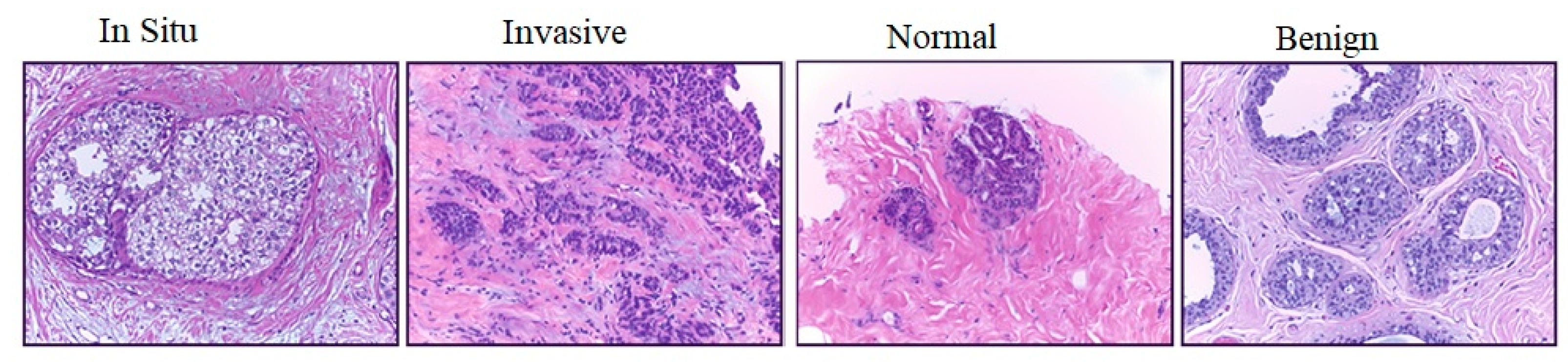
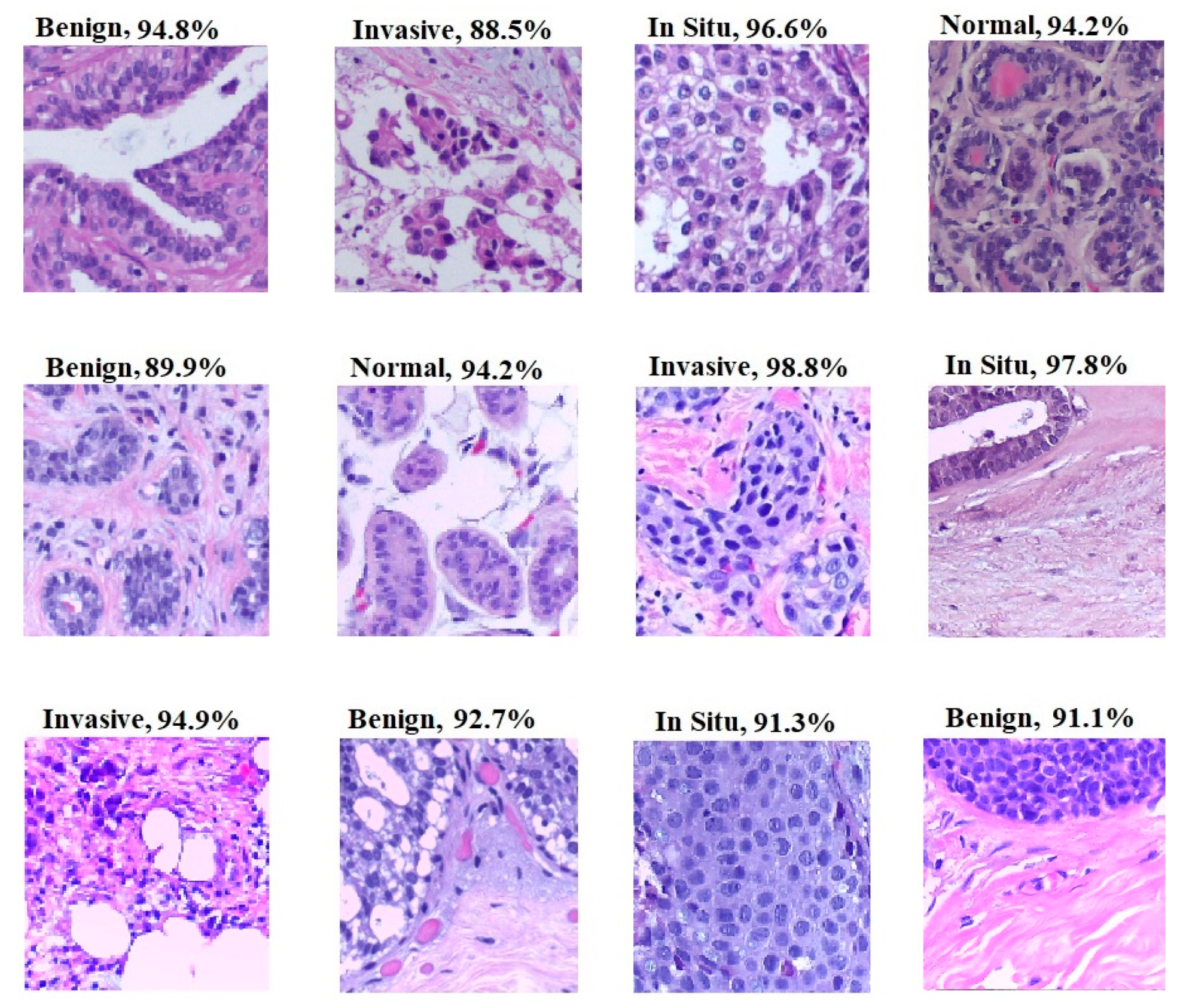
- The proposed model has been utilized to classify hematoxylin–eosin-stained breast biopsy images into four classes, namely invasive carcinoma, in-situ carcinoma, benign tumor and normal tissue. The performance of the breast classification task has been significantly improved in terms of accuracy, and has surpassed the performance of the latest methods by achieving an accuracy of 96.1% in classifying test images of the ICIAR 2018 dataset [16].
- We have empirically proven that transfer learning from the same domain of the target dataset has optimized the performance. This could change the direction of research, especially in the field of medicine with deep learning, since it is hard to collect medical data.
- A concise review of the previous DL architectures and breast cancer classification has been introduced.
2. Literature Review
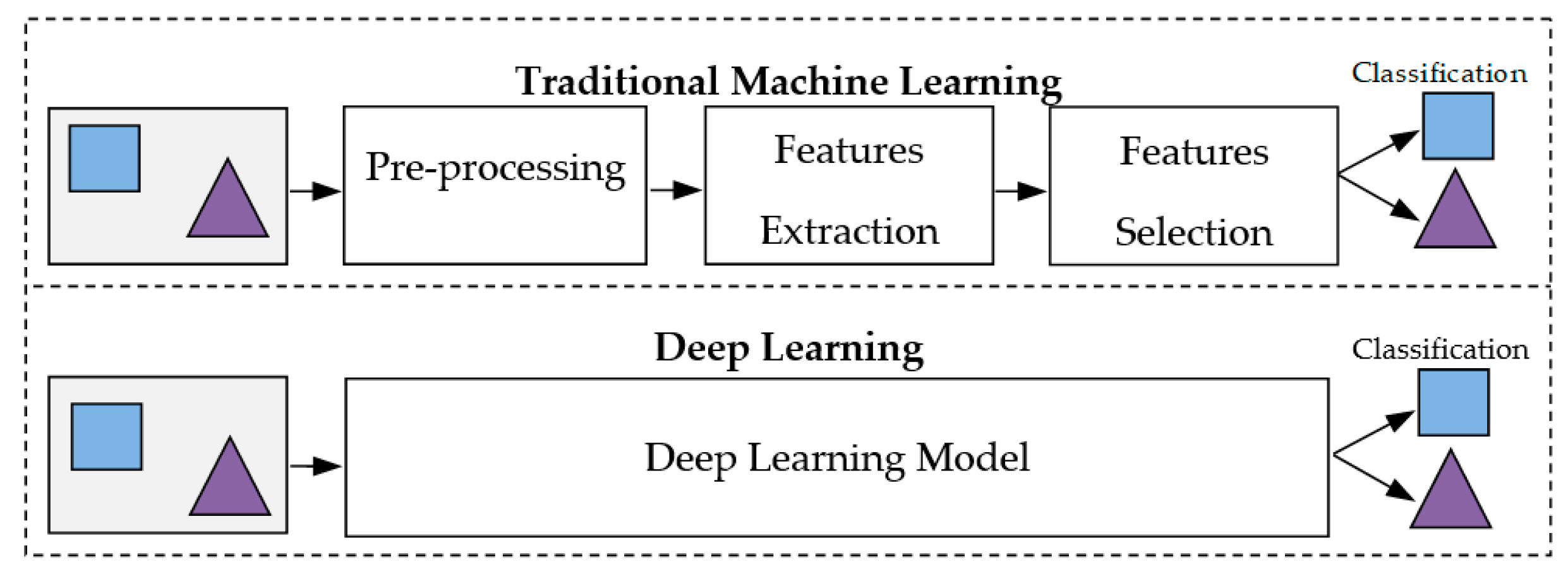
2.1. Deep Learning (DL)
2.2. Application of ML to Breast Cancer Diagnostic
3. Methodology
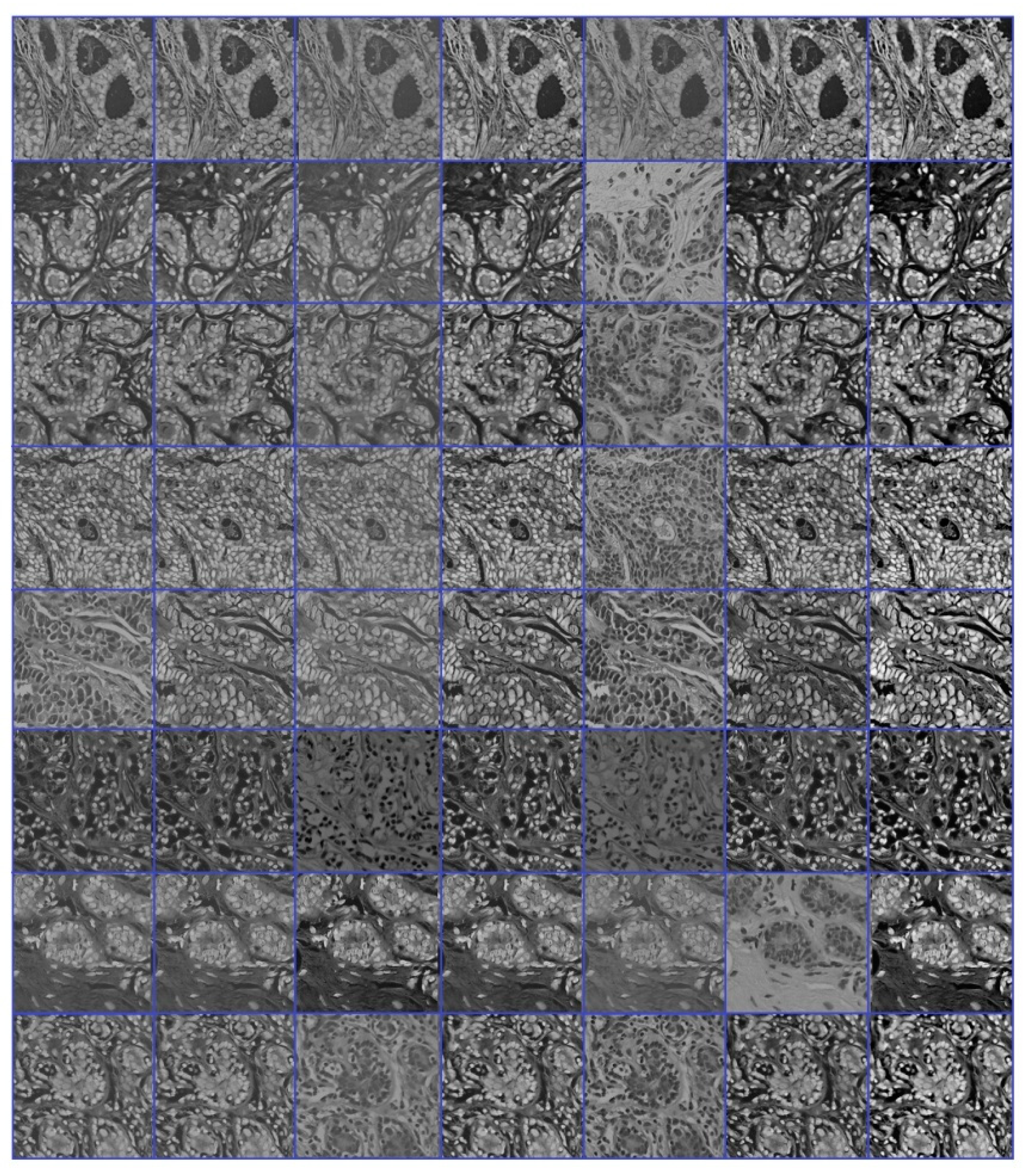
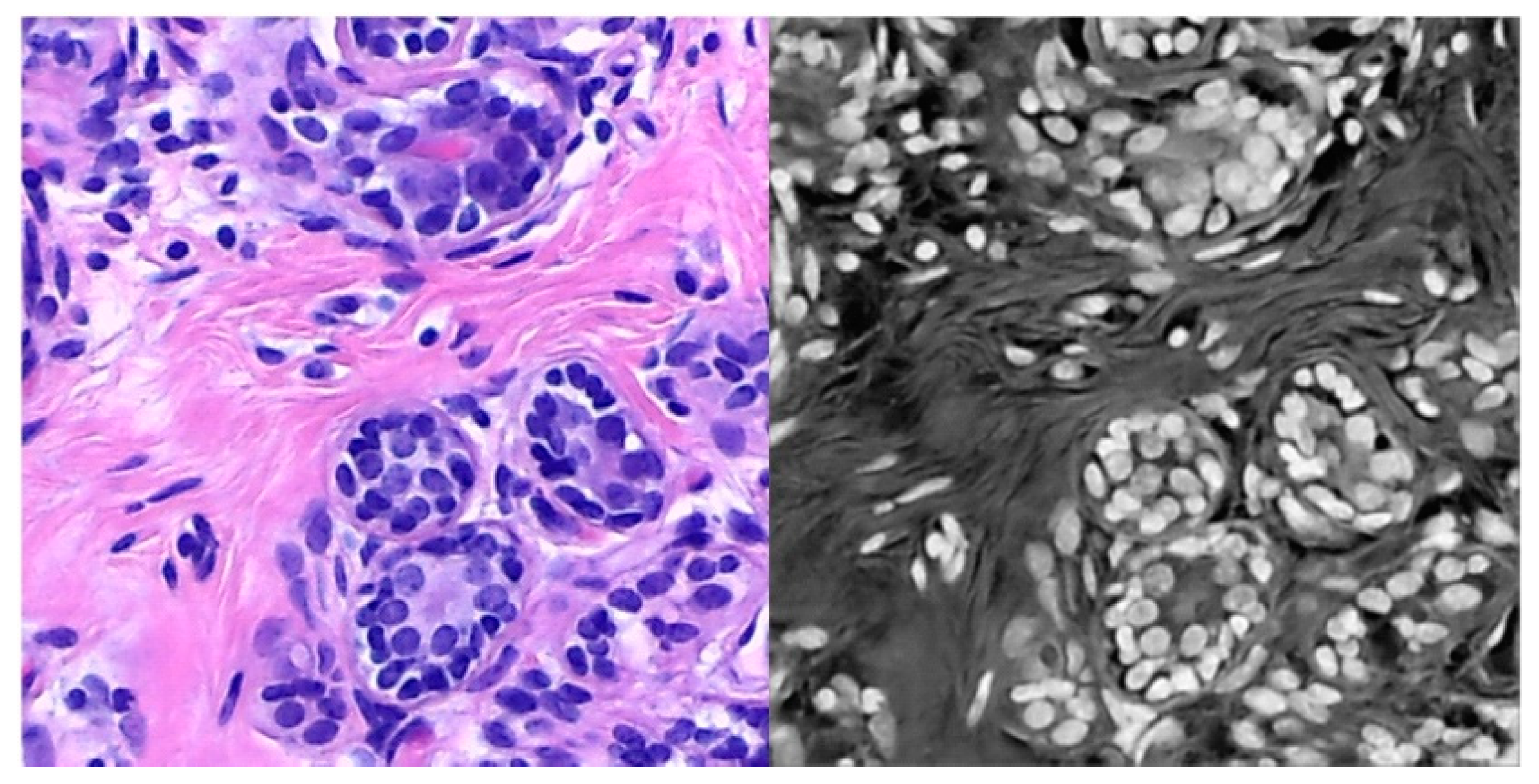
3.1. Datasets
3.1.1. Target Dataset
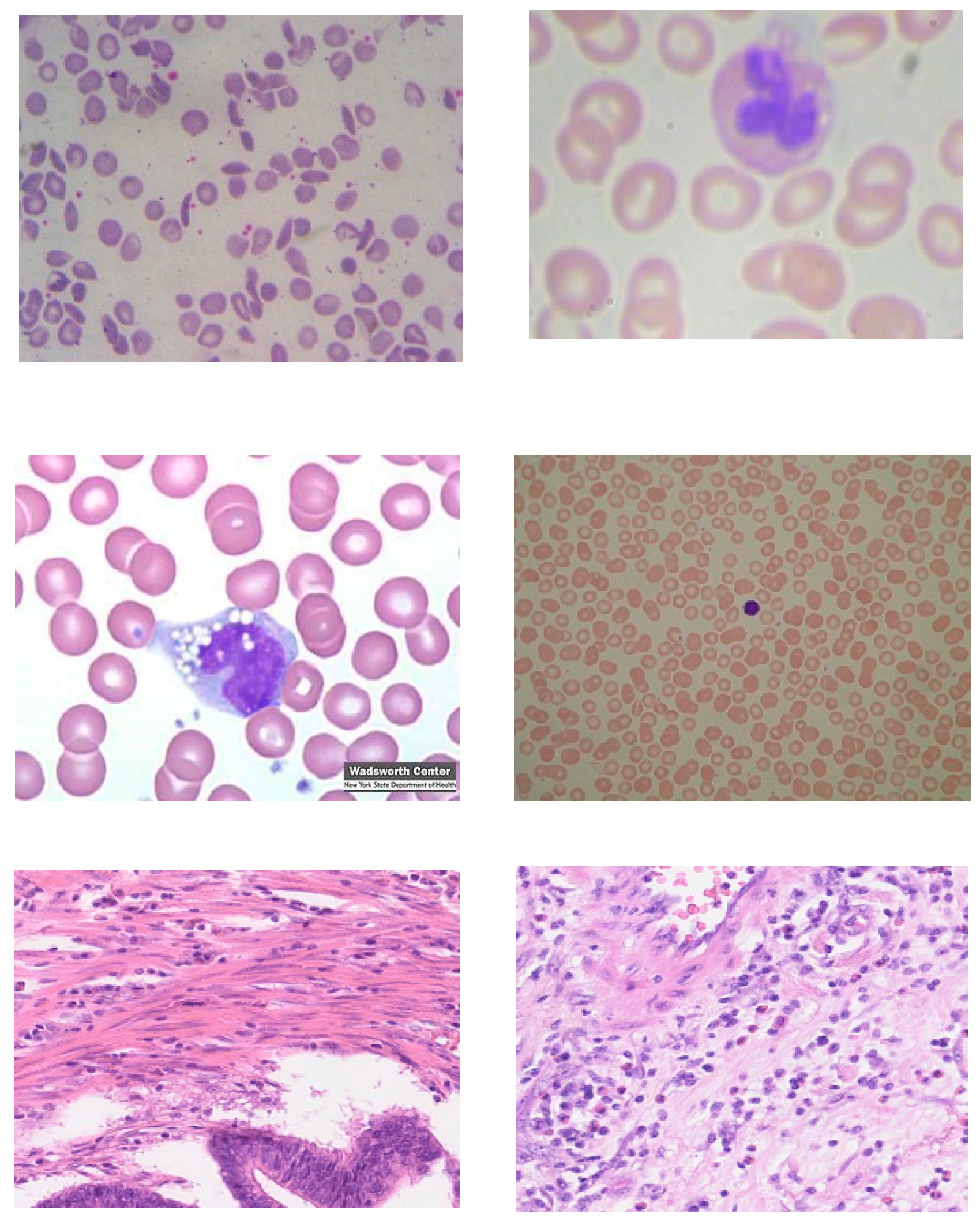
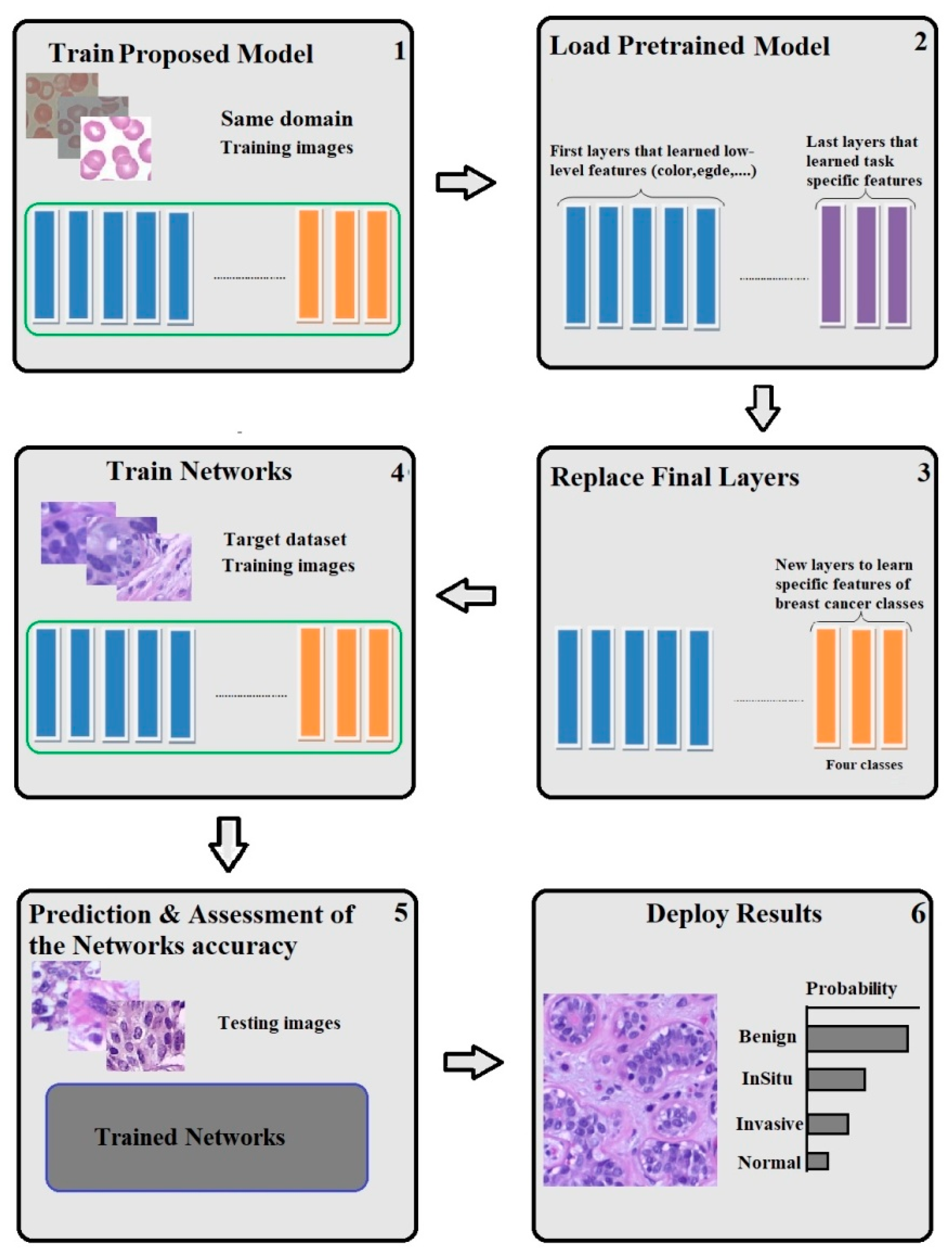
3.1.2. Same Domain Dataset
3.1.3. Different Domain Dataset
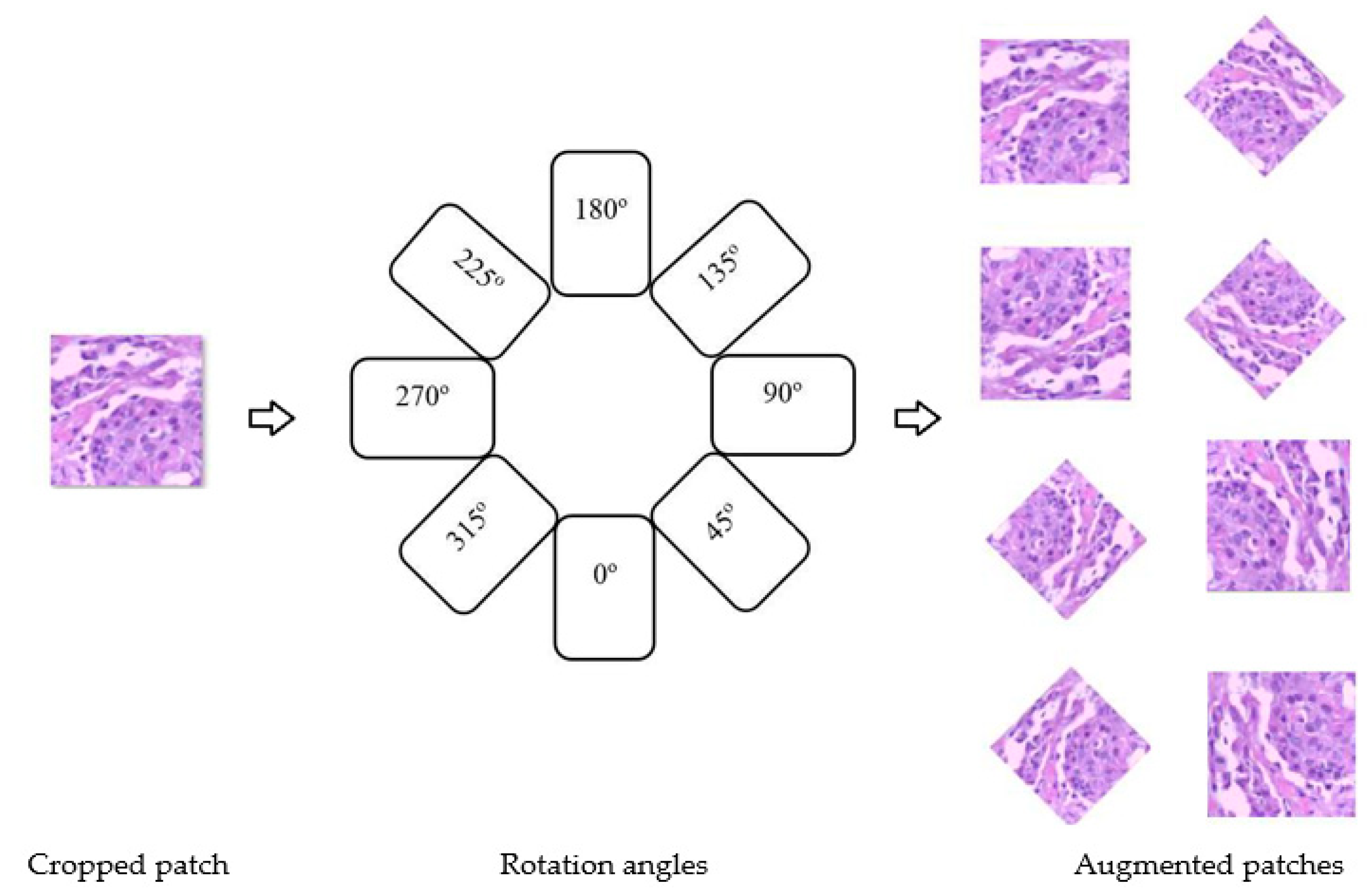
3.2. Image Augmentation
3.3. Transfer Learning
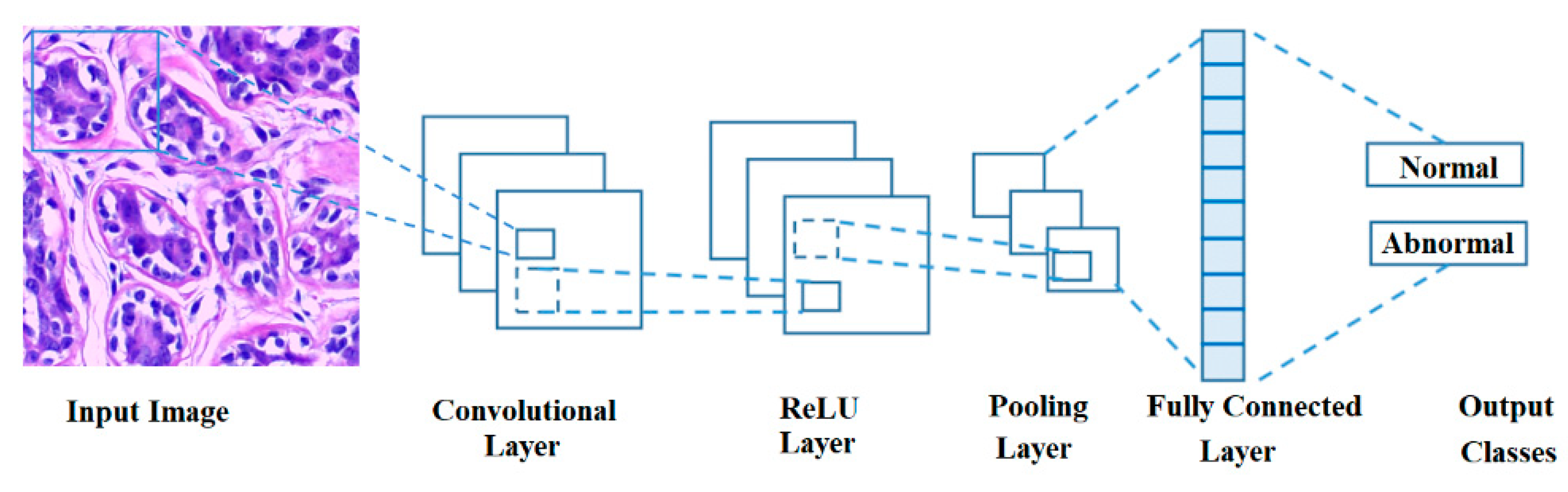
3.4. Convolutional Neural Networks (CNNs)
3.4.1. Convolutional Layer
3.4.2. Pooling Layer
3.4.3. Rectified Linear Unit (ReLU) layer
3.4.4. Fully Connected Layer
3.5. Proposed Model
- Experiment 1: Training only on the target dataset images.
- Experiment 2: Training on target dataset images, plus augmented images of the target dataset. (these augmentation techniques are explained above)
- Experiment 3: Training on transfer learning datasets first, then fine-tune the model and re-train it on the target dataset images.
- (a)
- We train our model from scratch on the different domain dataset first, then fine-tune our model with transferring the learning, and re-train on target dataset images
- (b)
- We train our model from scratch on the same domain dataset first, then fine-tune our model with transferring the learning and re-train on target dataset images.
- Experiment 4: Training on transfer learning dataset plus augmented images of transfer learning dataset first, then fine-tune the model and re-train it on target dataset images plus augmented images of the target dataset. We have applied this experiment on both datasets (different and same domain datasets as explained in experiments 3 a and b).
4. Experimental Results
5. Conclusions and Future Work
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA A Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef]
- U.S. Breast Cancer Statistics. Available online: https://www.breastcancer.org/symptoms/understandfgbc/statistics (accessed on 15 November 2019).
- Fondón, I.; Sarmiento, A.; García, A.I.; Silvestre, M.; Eloy, C.; Polónia, A.; Aguiar, P. Automatic classification of tissue malignancy for breast carcinoma diagnosis. Comput. Biol. Med. 2018, 96, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Breast Cancer Diagnosis; National Breast Cancer Foundation, Inc.: Dallas, TX, USA, 2015.
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. CW Elston & IO Ellis. Histopathology 1991, 19, 403–410: Author commentary. Histopathology 2002, 41, 151. [Google Scholar]
- Rosen, P.P. (Ed.) Rosen’s Breast Pathology, 3rd ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2008. [Google Scholar]
- Gurcan, M.N.; Boucheron, L.; Can, A.; Madabhushi, A.; Rajpoot, N.; Yener, B. Histopathological image analysis: A review. IEEE Rev. Biomed. Eng. 2009, 2, 147. [Google Scholar] [CrossRef] [PubMed]
- Elmore, J.G.; Longton, G.M.; Carney, P.A.; Geller, B.M.; Onega, T.; Tosteson, A.N.; O’Malley, F.P. Diagnostic concordance among pathologists interpreting breast biopsy specimens. J. Am. Med Assoc. 2015, 313, 1122–1132. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Rangayyan, R.M.; Xu, J.; El Naqa, I.; Yang, Y. Computer-aided detection and diagnosis of breast cancer with mammography: Recent advances. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 236–251. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Lin, J.; Xu, L.; Wang, H.; Bai, T.; Pang, Y.; Meen, T.-H. Fusion High-Resolution Network for Diagnosing ChestX-ray Images. Electronics 2020, 9, 190. [Google Scholar] [CrossRef]
- Nurmaini, S.; Darmawahyuni, A.; Sakti Mukti, A.N.; Rachmatullah, M.N.; Firdaus, F.; Tutuko, B. Deep Learning-Based Stacked Denoising and Autoencoder for ECG Heartbeat Classification. Electronics 2020, 9, 135. [Google Scholar] [CrossRef]
- Yang, Z.; Leng, L.; Kim, B.-G. StoolNet for Color Classification of Stool Medical Images. Electronics 2019, 8, 1464. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Fadhel, M.A.; Al-Shamma, O.; Zhang, J.; Duan, Y. Deep Learning Models for Classification of Red Blood Cells in Microscopy Images to Aid in Sickle Cell Anemia Diagnosis. Electronics 2020, 9, 427. [Google Scholar] [CrossRef]
- Abdel-Zaher, A.M.; Eldeib, A.M. Breast cancer classification using deep belief networks. Expert Syst. Appl. 2016, 46, 139–144. [Google Scholar] [CrossRef]
- Al-Zubaidi, L. Deep Learning Based Nuclei Detection for Quantitative Histopathology Image Analysis. Ph.D. Thesis, University of Missouri, Columbia, MO, USA, 2016. [Google Scholar]
- Aresta, G.; Araújo, T.; Kwok, S.; Chennamsetty, S.S.; Safwan, M.; Alex, V.; Marami, B.; Prastawa, M.; Chan, M.; Fernandez, G.; et al. Bach: Grand challenge on breast cancer histology images. Med. Image Anal. 2019, 56, 122–139. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Berg, A.C. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Kang, S.; Park, H.; Park, J.-I. CNN-Based Ternary Classification for Image Steganalysis. Electronics 2019, 8, 1225. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Fadhel, M.A.; Oleiwi, S.R.; Al-Shamma, O.; Zhang, J. DFU_QUTNet: Diabetic foot ulcer classification using novel deep convolutional neural network. Multimed. Tools Appl. 2019, 1–23. [Google Scholar] [CrossRef]
- Fang, B.; Lu, Y.; Zhou, Z.; Li, Z.; Yan, Y.; Yang, L.; Jiao, G.; Li, G. Classification of Genetically Identical Left and Right Irises Using a Convolutional Neural Network. Electronics 2019, 8, 1109. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems 25(NIPS 2012); Curran Associates Inc.: Red Hook, NY, USA, 2012; pp. 1097–1105. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Kumar, A.; Kim, J.; Lyndon, D.; Fulham, M.; Feng, D. An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J. Biomed. Health Inform. 2016, 21, 31–40. [Google Scholar] [CrossRef]
- Nawaz, W.; Ahmed, S.; Tahir, A.; Khan, H.A. Classification of breast cancer histology images using AlexNet. In International Conference Image Analysis and Recognition; Springer: Cham, Switzerland, 2018; pp. 869–876. [Google Scholar]
- Mahbod, A.; Ellinger, I.; Ecker, R.; Smedby, Ö.; Wang, C. Breast cancer histological image classification using fine-tuned deep network fusion. In International Conference Image Analysis and Recognition; Springer: Cham, Switzerland, 2018; pp. 754–762. [Google Scholar]
- Veta, M.; Pluim, J.P.; Van Diest, P.J.; Viergever, M.A. Breast cancer histopathology image analysis: A review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411. [Google Scholar] [CrossRef]
- Kowal, M.; Filipczuk, P. Nuclei segmentation for computer-aided diagnosis of breast cancer. Int. J. Appl. Math. Comput. Sci. 2014, 24, 19–31. [Google Scholar] [CrossRef]
- George, Y.M.; Zayed, H.H.; Roushdy, M.I.; Elbagoury, B.M. Remote computer-aided breast cancer detection and diagnosis system based on cytological images. IEEE Syst. J. 2013, 8, 949–964. [Google Scholar] [CrossRef]
- Belsare, A.D.; Mushrif, M.M.; Pangarkar, M.A.; Meshram, N. Classification of breast cancer histopathology images using texture feature analysis. In Proceedings of the TENCON 2015-2015 IEEE Region 10 Conference, Macao, China, 1–4 November 2015; pp. 1–5. [Google Scholar]
- Brook, A.; El-Yaniv, R.; Isler, E.; Kimmel, R.; Meir, R.; Peleg, D. Breast Cancer Diagnosis from Biopsy Images Using Generic Features and SVMs (No. CS Technion Report CS-2008-07); Computer Science Department, Technion: Haifa, Israel, 2008. [Google Scholar]
- Zhang, B. Breast cancer diagnosis from biopsy images by serial fusion of Random Subspace ensembles. In Proceedings of the 2011 4th International Conference on Biomedical Engineering and Informatics (BMEI), Shanghai, China, 15–17 October 2011; pp. 180–186. [Google Scholar]
- Du, Y.-C.; Muslikhin, M.; Hsieh, T.-H.; Wang, M.-S. Stereo Vision-Based Object Recognition and Manipulation by Regions with Convolutional Neural Network. Electronics 2020, 9, 210. [Google Scholar] [CrossRef]
- Yao, H.; Zhang, X.; Zhou, X.; Liu, S. Parallel Structure Deep Neural Network Using CNN and RNN with an Attention Mechanism for Breast Cancer Histology Image Classification. Cancers 2019, 11, 1901. [Google Scholar] [CrossRef]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Breast cancer histopathological image classification using convolutional neural networks. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 2560–2567. [Google Scholar]
- Cireşan, D.C.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Mitosis detection in breast cancer histology images with deep neural networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention-MICCAI 2013, Nagoya, Japan, 22–26 September 2013; pp. 411–418. [Google Scholar]
- Cruz-Roa, A.; Basavanhally, A.; González, F.; Gilmore, H.; Feldman, M.; Ganesan, S.; Shih, N.; Tomaszewski, J.; Madabhushi, A. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. In Medical Imaging 2014: Digital Pathology; International Society for Optics and Photonics: Bellingham, WA, USA, 2014; p. 904103. [Google Scholar]
- Kassani, S.H.; Kassani, P.H.; Wesolowski, M.J.; Schneider, K.A.; Deters, R. Breast cancer diagnosis with transfer learning and global pooling. arXiv 2019, arXiv:1909.11839. Available online: https://arxiv.org/abs/1909.11839 (accessed on 9 December 2019).
- Wang, Z.; Dong, N.; Dai, W.; Rosario, S.D.; Xing, E.P. Classification of breast cancer histopathological images using convolutional neural networks with hierarchical loss and global pooling. In Proceedings of the International Conference Image Analysis and Recognition, Póvoa de Varzim, Portugal, 27–29 June 2018; pp. 745–753. [Google Scholar]
- Gonzalez-Hidalgo, M.; Guerrero-Pena, F.A.; Herold-Garcia, S.; Jaume-i-Capó, A.; Marrero-Fernández, P.D. Red blood cell cluster separation from digital images for use in sickle cell disease. IEEE J. Biomed. Health Inform. 2014, 19, 1514–1525. [Google Scholar] [CrossRef]
- Parthasarathy, D. WBC-Classification. Available online: https://github.com/dhruvp/wbc-classification/tree/master/Original_Images (accessed on 15 November 2019).
- Wadsworth-Center. White Blood Cell Images. Available online: https://www.wadsworth.org/ (accessed on 15 November 2019).
- Labati, R.D.; Piuri, V.; Scotti, F. All-IDB: The acute lymphoblastic leukemia image database for image processing. In Proceedings of the 2011 18th IEEE International Conference on Image Processing, Brussels, Belgium, 11–14 September 2011; pp. 2045–2048. [Google Scholar]
- Sirinukunwattana, K.; Raza, S.E.A.; Tsang, Y.W.; Snead, D.R.; Cree, I.A.; Rajpoot, N.M. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imaging 2016, 35, 1196–1206. [Google Scholar] [CrossRef]
- Roy, P.; Ghosh, S.; Bhattacharya, S.; Pal, U. Effects of degradations on deep neural network architectures. arXiv 2018, arXiv:1807.10108. Available online: https://arxiv.org/abs/1807.10108 (accessed on 9 December 2019).
- Natural Images. Available online: https://www.kaggle.com/prasunroy/natural-images (accessed on 1 December 2019).
- Animals. Available online: https://www.kaggle.com/alessiocorrado99/animals10#translate.py (accessed on 1 December 2019).
- Collation. Available online: https://www.kaggle.com/mbkinaci/chair-kitchen-knife-saucepan (accessed on 1 December 2019).
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Wang, J.; Perez, L. The effectiveness of data augmentation in image classification using deep learning. arXiv 2017, arXiv:1712.04621. Available online: https://arxiv.org/abs/1712.04621 (accessed on 28 November 2019).
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Baykal, E.; Dogan, H.; Ercin, M.E.; Ersoz, S.; Ekinci, M. Transfer learning with pre-trained deep convolutional neural networks for serous cell classification. Multimed. Tools Appl. 2019. [Google Scholar] [CrossRef]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding transfer learning for medical imaging. In Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019; pp. 3342–3352. [Google Scholar]
- Gil-Martín, M.; Montero, J.M.; San-Segundo, R. Parkinson’s Disease Detection from Drawing Movements Using Convolutional Neural Networks. Electronics 2019, 8, 907. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Qayyum, A.; Anwar, S.M.; Awais, M.; Majid, M. Medical image retrieval using deep convolutional neural network. Neurocomputing 2017, 266, 8–20. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Lv, E.; Wang, X.; Cheng, Y.; Yu, Q. Deep ensemble network based on multi-path fusion. Artif. Intell. Rev. 2019, 52, 151–168. [Google Scholar] [CrossRef]
- Wang, J.; Wei, Z.; Zhang, T.; Zeng, W. Deeply-fused nets. arXiv 2016, arXiv:1605.07716. Available online: https://arxiv.org/abs/1605.07716 (accessed on 10 December 2019).
- Golatkar, A.; Anand, D.; Sethi, A. Classification of Breast Cancer Histology Using Deep Learning; Springer: Cham, Switzerland, 2018; pp. 837–844. [Google Scholar]
- Roy, K.; Banik, D.; Bhattacharjee, D.; Nasipuri, M. Patch-based system for Classification of Breast Histology images using deep learning. Comput. Med. Imaging Graph. 2019, 71, 90–103. [Google Scholar] [CrossRef]
- Ferreira, C.A.; Melo, T.; Sousa, P.; Meyer, M.I.; Shakibapour, E.; Costa, P.; Campilho, A. Classification of Breast Cancer Histology Images through Transfer Learning Using a Pre-Trained Inception ResNet v2; Springer: Cham, Switzerland, 2018; pp. 763–770. [Google Scholar]
- Awan, R.; Koohbanani, N.A.; Shaban, M.; Lisowska, A.; Rajpoot, N. Context-Aware Learning Using Transferable Features for Classification of Breast Cancer Histology Images; Springer: Cham, Switzerland, 2018; pp. 788–795. [Google Scholar]
- Guo, Y.; Dong, H.; Song, F.; Zhu, C.; Liu, J. Breast Cancer Histology Image Classification Based on Deep Neural Networks; Springer: Cham, Switzerland, 2018; pp. 827–836. [Google Scholar]
- Vang, Y.S.; Chen, Z.; Xie, X. Deep Learning Framework for Multi-Class Breast Cancer Histology Image Classification; Springer: Cham, Switzerland, 2018; pp. 914–922. [Google Scholar]
- Sarker, M.I.; Kim, H.; Tarasov, D.; Akhmetzanov, D. Inception Architecture and Residual Connections in Classification of Breast Cancer Histology Images. arXiv 2019, arXiv:1912.04619. Available online: https://arxiv.org/abs/1912.04619 (accessed on 22 December 2019).

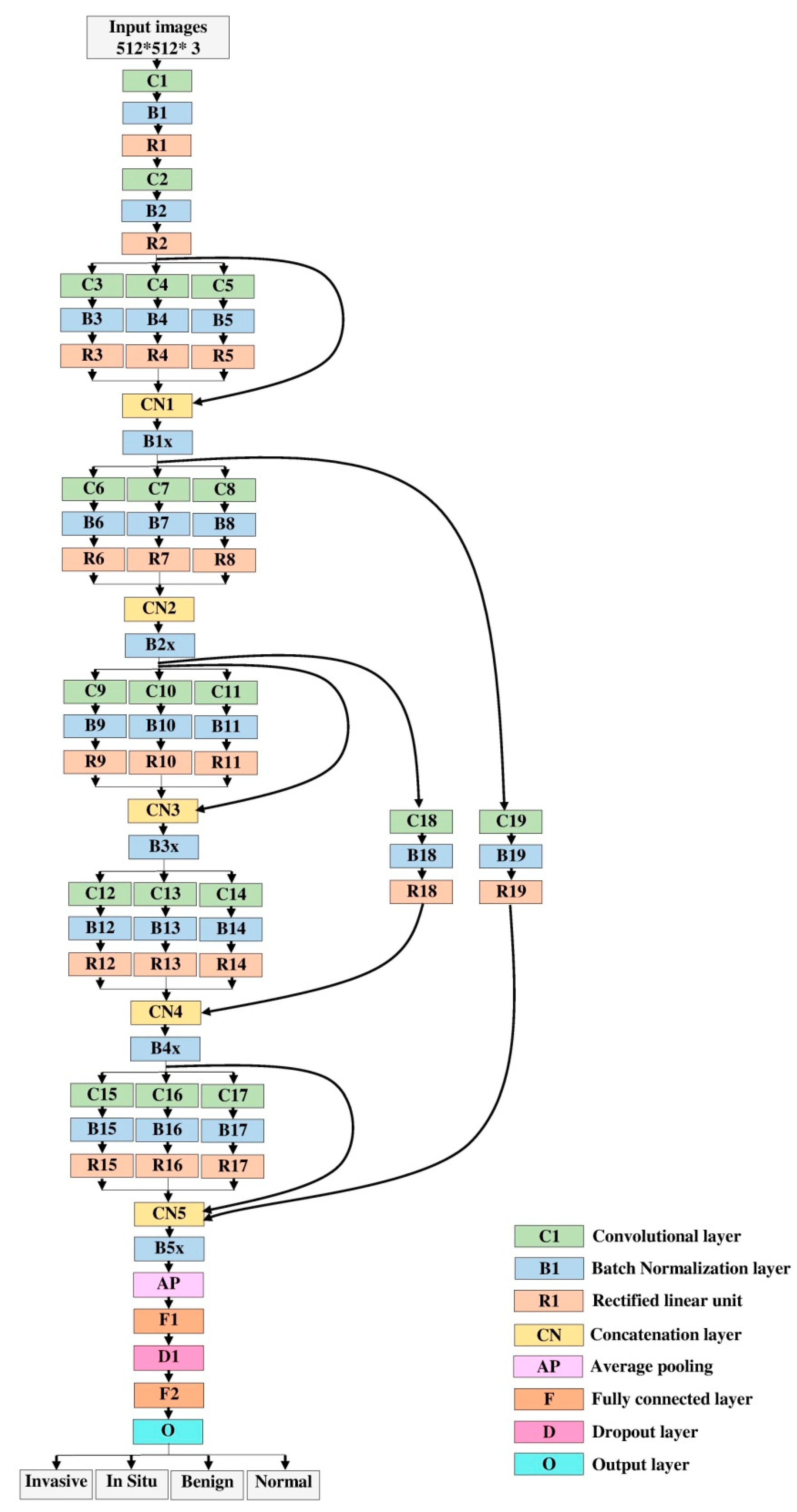
| Name of Layer | Filter Size (FS) & Stride (S) | Activations |
|---|---|---|
| Input layer | - | 512 × 512 × 3 |
| C 1, B 1, R 1 | FS = 5 × 5, S = 1 | 512 × 512 × 16 |
| C 2, B 2, R 2 | FS = 7 × 7, S = 2 | 256 × 256 × 16 |
| C 3, B 3, R 3 | FS = 1 × 1, S = 1 | 256 × 256 × 16 |
| C 4, B 4, R 4 | FS = 3 × 3, S = 1 | 256 × 256 × 16 |
| C 5, B 5, R 5 | FS = 5 × 5, S = 1 | 256 × 256 × 16 |
| CN 1 | Four inputs | 256 × 256 × 64 |
| B1x | Batch Normalization Layer | 256 × 256 × 64 |
| C 6, B 6, R 6 | FS = 1 × 1, S = 2 | 128 × 128 × 32 |
| C 7, B 7, R 7 | FS = 3 × 3, S = 2 | 128 × 128 × 32 |
| C 8, B 8, R 8 | FS = 5 × 5, S = 2 | 128 × 128 × 32 |
| CN 2 | Three inputs | 128 × 128 × 96 |
| B2x | Batch Normalization Layer | 128 × 128 × 96 |
| C 9, B 9, R 9 | FS = 1 × 1, S = 1 | 128 × 128 × 32 |
| C 10, B 10, R 10 | FS = 3 × 3, S = 1 | 128 × 128 × 32 |
| C 11, B 11, R 11 | FS = 5 × 5, S = 1 | 128 × 128 × 32 |
| CN 3 | Four inputs | 128 × 128 × 192 |
| B3x | Batch Normalization Layer | 128 × 128 × 192 |
| C 12, B 12, R 12 | FS = 1 × 1, S = 2 | 64 × 64 × 64 |
| C 13, B 13, R 13 | FS = 3 × 3, S = 2 | 64 × 64 × 64 |
| C 14, B 14, R 14 | FS = 5 × 5, S = 2 | 64 × 64 × 64 |
| CN4 | Four inputs | 64 × 64 × 208 |
| B4x | Batch Normalization Layer | 64 × 64 × 208 |
| C 15, B 15, R 15 | FS = 1 × 1, S = 1 | 64 × 64 × 128 |
| C 16, B 16, R 16 | FS = 3 × 3, S = 1 | 64 × 64 × 128 |
| C 17, B 17, R 17 | FS = 5 × 5, S = 1 | 64 × 64 × 128 |
| CN 5 | Five inputs | 64 × 64 × 608 |
| B5x | Batch Normalization Layer | 64 × 64 × 608 |
| C 18, B 18, R 18 | FS = 3 × 3, S = 2 | 64 × 64 × 16 |
| C 19, B 19, R 19 | FS = 5 × 5, S = 4 | 64 × 64 × 16 |
| AP | Size = 7 × 7, S = 4 | 15 × 15 × 608 |
| F1 | 400 FC | 1 × 1 × 400 |
| D1 | learning rate: 0.5 | 1 × 1 × 400 |
| F2 | 4 FC | 1 × 1 × 4 |
| O (Softmax function) | Invasive, In situ, Benign, Normal | 1 × 1 × 4 |
| Experiment | Patch-Wise (%) | Image-Wise (%) | |
|---|---|---|---|
| Experiment 1 | 76.5 | 80.3 | |
| Experiment 2 | 82.9 | 88.1 | |
| Experiment 3 | (a) Different domain dataset | 77.9 | 81.2 |
| (b) Same domain dataset | 87.8 | 94.1 | |
| Experiment 4 | (a) Different domain dataset | 79.5 | 84.2 |
| (b) Same domain dataset | 90.5 | 97.4 | |
| Method | Patch-Wise (%) | Image-Wise (%) |
|---|---|---|
| Nawaz, W., et al. [28] | 75.73 | - |
| Golatkar, A., et al. [64] | 79 | 85 |
| Mahbod A., et al. [29] | - | 88.5 |
| Roy, K., et al. [65] | 77.4 | 90 |
| Wang, Z., et al. [42] | 87 | 92 |
| Ferreira, C. A., et al. [66] | - | 93 |
| Our model with Experiment 4 (same domain transfer Learning) | 90.5 | 97.4 |
| Experiment | Image-Wise (%) | |
|---|---|---|
| Experiment 1 | 77.4 | |
| Experiment 2 | 86.1 | |
| Experiment 3 | (a) Different domain dataset | 80.2 |
| (b) Same domain dataset | 93.8 | |
| Experiment 4 | (a) Different domain dataset | 82.4 |
| (b) Same domain dataset | 96.1 | |
| Method | Image-Wise (%) |
|---|---|
| Nawaz W., et al. [28] | 81.25 |
| Awan R, et al. [67] | 83.33 |
| Guo Y., et al. [68] | 87.5 |
| Vang Y.S., et al. [69] | 87.5 |
| Sarker M., I et al. [70] | 89 |
| Ferreira C.A., et al. [66] | 90 |
| Kassani S. H., et al. [41] | 92.5 |
| Wang, Z., et al. [42] | 93 |
| Our model with Experiment 4 (same domain-transfer Learning) | 96.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alzubaidi, L.; Al-Shamma, O.; Fadhel, M.A.; Farhan, L.; Zhang, J.; Duan, Y. Optimizing the Performance of Breast Cancer Classification by Employing the Same Domain Transfer Learning from Hybrid Deep Convolutional Neural Network Model. Electronics 2020, 9, 445. https://doi.org/10.3390/electronics9030445
Alzubaidi L, Al-Shamma O, Fadhel MA, Farhan L, Zhang J, Duan Y. Optimizing the Performance of Breast Cancer Classification by Employing the Same Domain Transfer Learning from Hybrid Deep Convolutional Neural Network Model. Electronics. 2020; 9(3):445. https://doi.org/10.3390/electronics9030445
Chicago/Turabian StyleAlzubaidi, Laith, Omran Al-Shamma, Mohammed A. Fadhel, Laith Farhan, Jinglan Zhang, and Ye Duan. 2020. "Optimizing the Performance of Breast Cancer Classification by Employing the Same Domain Transfer Learning from Hybrid Deep Convolutional Neural Network Model" Electronics 9, no. 3: 445. https://doi.org/10.3390/electronics9030445
APA StyleAlzubaidi, L., Al-Shamma, O., Fadhel, M. A., Farhan, L., Zhang, J., & Duan, Y. (2020). Optimizing the Performance of Breast Cancer Classification by Employing the Same Domain Transfer Learning from Hybrid Deep Convolutional Neural Network Model. Electronics, 9(3), 445. https://doi.org/10.3390/electronics9030445










