High-Level Synthesis of Multiclass SVM Using Code Refactoring to Classify Brain Cancer from Hyperspectral Images
Abstract
1. Introduction
2. Materials and Methods
2.1. Zynq-7000 SoC Device from Xilinx
2.2. SDSoC Development Environment by Xilinx
2.3. Support Vector Machine Classifier
SVM Multiclass Classifier
- (1)
- Variables declaration and initialization. Here, the inputs that represents the previously trained model of the algorithm (support vectors, the bias, and the sigmoid function parameters) as well as the samples to be classified are declared and initialized.
- (2)
- Distances computation. In this step, the distances between the samples (i.e., the pixel) and the established hyperplane are computed.
- (3)
- Binary probability computation. This step has the goal of estimating the binary probability of a certain pixel to belong to the two classes under study in the one-vs-one method, taking into account the distances computed in the previous step.
- (4)
- Multiclass probability computation. This final step aims to obtain the multiclass probabilities for each pixel performing a for loop that iteratively refines the probabilities for each pixel associated to a certain class obtained in the previous step. The value of each probability is incrementally modified on the assumption that the difference with the value of the previous iteration is under a certain threshold or if the maximum error is reached (the user establishes both parameters). As soon as one of these two situations is confirmed, the multiclass probabilities of the pixel are computed, and the final classification map is generated.
2.4. In Vivo HS Human Brain Cancer Database
3. Code Refactoring
3.1. Use of Directives and Memory Allocation
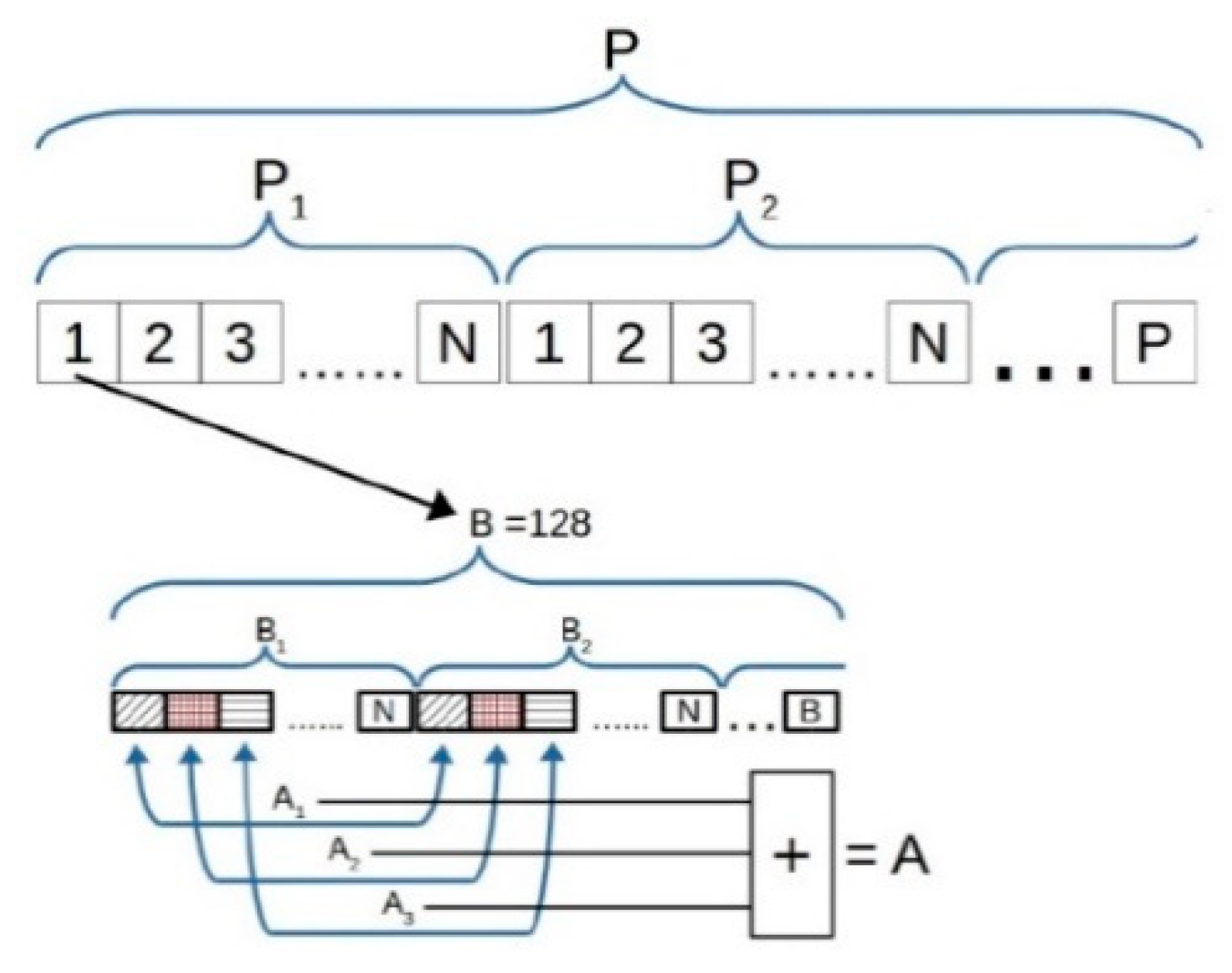
3.2. Improvement in Data Transfer
3.3. Improvement in Data Processing
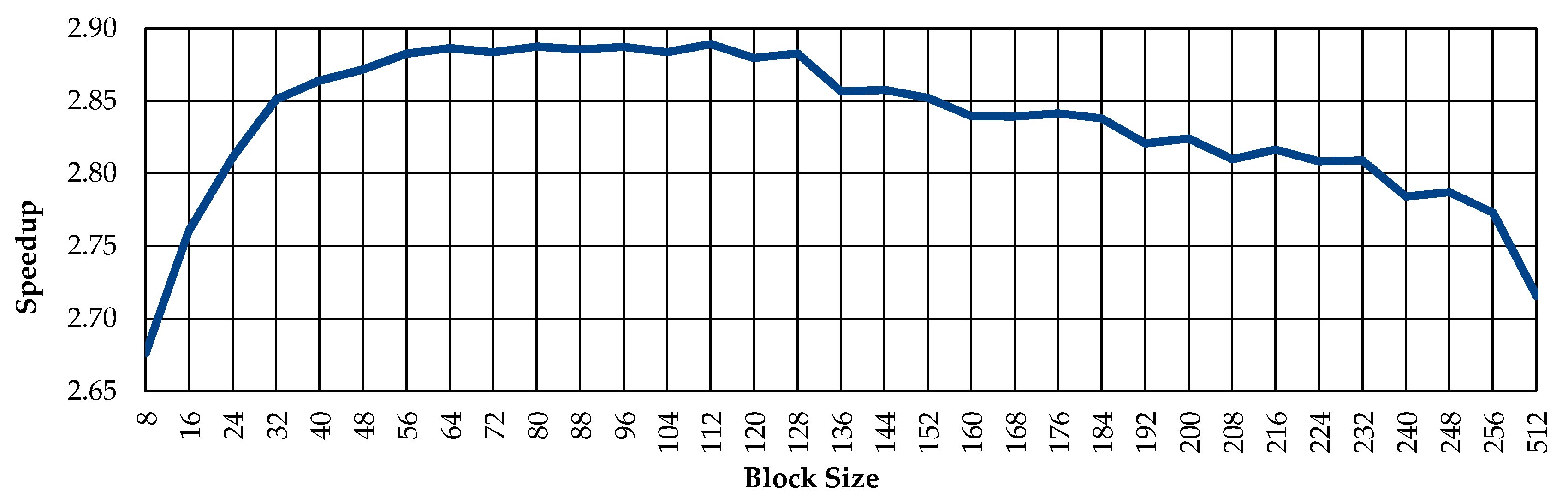
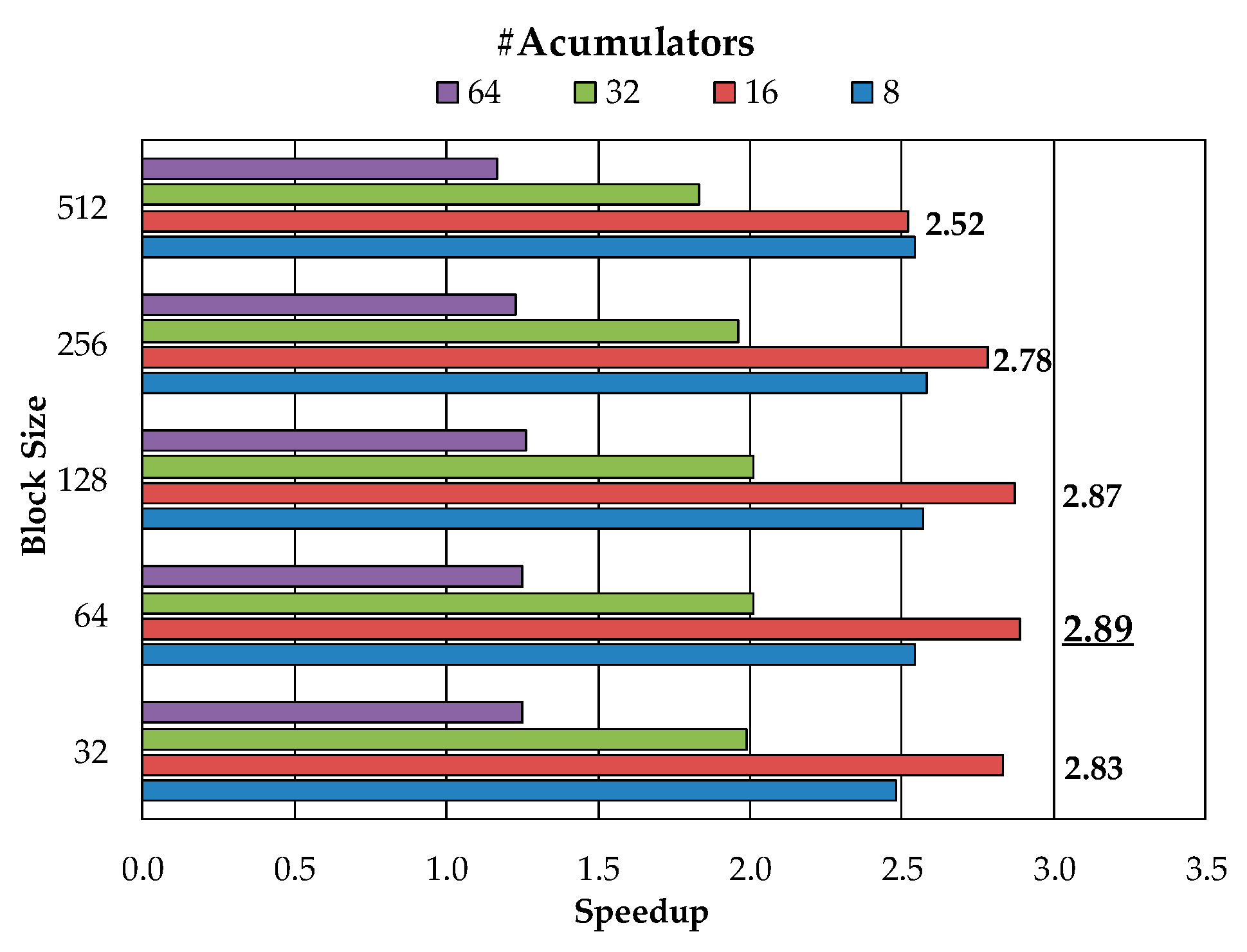
3.4. Including Redundant Data inside Accelerated Function
3.5. Data Type Reduction
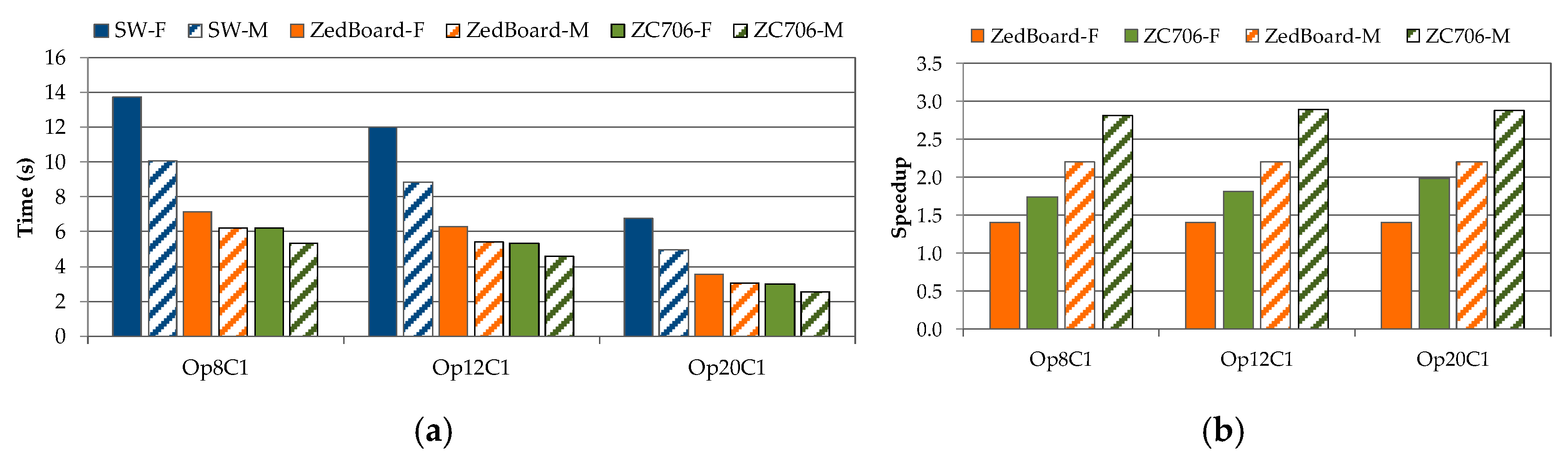
4. Experimental Results and Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Coussy, P.; Gajski, D.D.; Meredith, M.; Takach, A. An Introduction to High-Level Synthesis. IEEE Des. Test Comput. 2009, 26, 8–17. [Google Scholar] [CrossRef]
- Nane, R.; Sima, V.-M.; Pilato, C.; Choi, J.; Fort, B.; Canis, A.; Chen, Y.T.; Hsiao, H.; Brown, S.; Ferrandi, F.; et al. A Survey and Evaluation of FPGA High-Level Synthesis Tools. IEEE Trans. Comput. Des. Integr. Circuits Syst. 2016, 35, 1591–1604. [Google Scholar] [CrossRef]
- Saha, R.; Banik, P.P.; Kim, K.-D.D. HLS Based Approach to Develop an Implementable HDR Algorithm. Electronics 2018, 7, 332. [Google Scholar] [CrossRef]
- Liu, Z.; Chow, P.; Xu, J.; Jiang, J.; Dou, Y.; Zhou, J. A Uniform Architecture Design for Accelerating 2D and 3D CNNs on FPGAs. Electronics 2019, 8, 65. [Google Scholar] [CrossRef]
- Kyrkou, C.; Theocharides, T. A parallel hardware architecture for real-time object detection with support vector machines. IEEE Trans. Comput. 2012, 61, 831–842. [Google Scholar] [CrossRef]
- Jallad, A.H.M.; Mohammed, L.B. Hardware support vector machine (SVM) for satellite on-board applications. In Proceedings of the 2014 NASA/ESA Conference on Adaptive Hardware and Systems (AHS 2014), Leicester, UK, 14–18 July 2014; IEEE Computer Society: Washington, DC, USA, 2014; pp. 256–261. [Google Scholar]
- Anguita, D.; Carlino, L.; Ghio, A.; Ridella, S. A FPGA core generator for embedded classification systems. J. Circuits Syst. Comput. 2011, 20, 263–282. [Google Scholar] [CrossRef]
- Hussain, H.M.; Benkrid, K.; Seker, H. Reconfiguration-based implementation of SVM classifier on FPGA for Classifying Microarray data. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society EMBS, Osaka, Japan, 3–7 July 2013; pp. 3058–3061. [Google Scholar]
- Pan, X.; Yang, H.; Li, L.; Liu, Z.; Hou, L. FPGA implementation of SVM decision function based on hardware-friendly kernel. In Proceedings of the 2013 International Conference on Computational and Information Sciences (ICCIS 2013), Shiyang, China, 21–23 June 2013; pp. 133–136. [Google Scholar]
- Papadonikolakis, M.; Bouganis, C.S. A novel FPGA-based SVM classifier. In Proceedings of the 2010 International Conference on Field-Programmable Technology (FPT’10), Beijing, China, 8–10 December 2010; pp. 283–286. [Google Scholar]
- Vranjković, V.S.; Struharik, R.J.R.; Novak, L.A. Reconfigurable hardware for machine learning applications. J. Circuits Syst. Comput. 2015, 24, 1550064. [Google Scholar] [CrossRef]
- Afifi, S.M.; Gholamhosseini, H.; Sinha, R. Hardware Implementations of SVM on FPGA: A State-of-the-Art Review of Current Practice. Int. J. Innov. Sci. Eng. Technol. 2015, 2, 733–752. [Google Scholar]
- Afifi, S.; GholamHosseini, H.; Sinha, R. A low-cost FPGA-based SVM classifier for melanoma detection. In Proceedings of the IECBES 2016-IEEE-EMBS Conference on Biomedical Engineering and Sciences, Kuala Lumpur, Malaysia, 4–8 December 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 631–636. [Google Scholar]
- Ning, M.; Shaojun, W.; Yeyong, P.; Yu, P. Implementation of LS-SVM with HLS on Zynq. In Proceedings of the 2014 International Conference on Field-Programmable Technology (FPT), Shanghai, China, 10–12 December 2014; IEEE: Piscataway, NJ, USA, 2014; pp. 346–349. [Google Scholar]
- Kamruzzaman, M.; Sun, D.-W. Introduction to Hyperspectral Imaging Technology. In Computer Vision Technology for Food Quality Evaluation, 2nd ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 111–139. [Google Scholar]
- Starr, C.; Evers, C.A.; Starr, L. Biology: Concepts and Applications without Physiology; Cengage Learning: Boston, MA, USA, 2010; ISBN 9780538739252. [Google Scholar]
- Manolakis, D.; Shaw, G. Detection algorithms for hyperspectral imaging applications. IEEE Signal Process. Mag. 2002, 19, 29–43. [Google Scholar] [CrossRef]
- Govender, M.; Chetty, K.; Bulcock, H. A review of hyperspectral remote sensing and its application in vegetation and water resource studies. Water SA 2009, 33, 145–152. [Google Scholar] [CrossRef]
- van der Meer, F.D.; van der Werff, H.M.A.; van Ruitenbeek, F.J.A.; Hecker, C.A.; Bakker, W.H.; Noomen, M.F.; van der Meijde, M.; Carranza, E.J.M.; de Smeth, J.B.; Woldai, T. Multi-and hyperspectral geologic remote sensing: A review. Int. J. Appl. Earth Obs. Geoinf. 2012, 14, 112–128. [Google Scholar] [CrossRef]
- de Carvalho Rocha, W.F.; Sabin, G.P.; Março, P.H.; Poppi, R.J. Quantitative analysis of piroxicam polymorphs pharmaceutical mixtures by hyperspectral imaging and chemometrics. Chemom. Intell. Lab. Syst. 2011, 106, 198–204. [Google Scholar] [CrossRef]
- de Moura França, L.; Pimentel, M.F.; da Silva Simões, S.; Grangeiro, S.; Prats-Montalbán, J.M.; Ferrer, A. NIR hyperspectral imaging to evaluate degradation in captopril commercial tablets. Eur. J. Pharm. Biopharm. 2016, 104, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Edelman, G.J.; Gaston, E.; van Leeuwen, T.G.; Cullen, P.J.; Aalders, M.C.G. Hyperspectral imaging for non-contact analysis of forensic traces. Forensic Sci. Int. 2012, 223, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.S.; Pimentel, M.F.; Honorato, R.S.; Pasquini, C.; Prats-Montalbán, J.M.; Ferrer, A. Near infrared hyperspectral imaging for forensic analysis of document forgery. Analyst 2014, 139, 5176–5184. [Google Scholar] [CrossRef] [PubMed]
- Fernández de la Ossa, M.Á.; Amigo, J.M.; García-Ruiz, C. Detection of residues from explosive manipulation by near infrared hyperspectral imaging: A promising forensic tool. Forensic Sci. Int. 2014, 242, 228–235. [Google Scholar] [CrossRef]
- Wu, D.; Sun, D.-W. Advanced applications of hyperspectral imaging technology for food quality and safety analysis and assessment: A review—Part II: Applications. Innov. Food Sci. Emerg. Technol. 2013, 19, 15–28. [Google Scholar] [CrossRef]
- Feng, Y.-Z.; Sun, D.-W. Application of Hyperspectral Imaging in Food Safety Inspection and Control: A Review. Crit. Rev. Food Sci. Nutr. 2012, 52, 1039–1058. [Google Scholar] [CrossRef]
- Lorente, D.; Aleixos, N.; Gomez-Sanchis, J.; Cubero, S.; Garcia-Navarrete, O.L.; Blasco, J.; Gómez-Sanchis, J.; Cubero, S.; García-Navarrete, O.L.; Blasco, J. Recent Advances and Applications of Hyperspectral Imaging for Fruit and Vegetable Quality Assessment. Food Bioprocess Technol. 2011, 5, 1121–1142. [Google Scholar] [CrossRef]
- Lu, G.; Fei, B. Medical hyperspectral imaging: A review. J. Biomed. Opt. 2014, 19, 10901. [Google Scholar] [CrossRef]
- Calin, M.A.; Parasca, S.V.; Savastru, D.; Manea, D. Hyperspectral imaging in the medical field: Present and future. Appl. Spectrosc. Rev. 2014, 49, 435–447. [Google Scholar] [CrossRef]
- Li, Q.; He, X.; Wang, Y.; Liu, H.; Xu, D.; Guo, F. Review of spectral imaging technology in biomedical engineering: Achievements and challenges. J. Biomed. Opt. 2013, 18, 100901. [Google Scholar] [CrossRef] [PubMed]
- Halicek, M.; Fabelo, H.; Ortega, S.; Callico, G.M.; Fei, B. In-Vivo and Ex-Vivo Tissue Analysis through Hyperspectral Imaging Techniques: Revealing the Invisible Features of Cancer. Cancers 2019, 11, 756. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Kosugi, Y. Hyperspectral imaging: A new modality in surgery. In Recent Advances in Biomedical Engineering; IntechOpen: London, UK, 2009. [Google Scholar]
- Baltussen, E.J.M.; Kok, E.N.D.; Brouwer de Koning, S.G.; Sanders, J.; Aalbers, A.G.J.; Kok, N.F.M.; Beets, G.L.; Flohil, C.C.; Bruin, S.C.; Kuhlmann, K.F.D.; et al. Hyperspectral imaging for tissue classification, a way toward smart laparoscopic colorectal surgery. J. Biomed. Opt. 2019, 24, 016002. [Google Scholar] [CrossRef] [PubMed]
- Pourreza-Shahri, R.; Saki, F.; Kehtarnavaz, N.; Leboulluec, P.; Liu, H. Classification of ex-vivo breast cancer positive margins measured by hyperspectral imaging. In Proceedings of the 2013 IEEE International Conference on Image Processing (ICIP 2013), Melbourne, VIC, Australia, 15–18 September 2013; pp. 1408–1412. [Google Scholar]
- Liu, Z.; Wang, H.; Li, Q. Tongue tumor detection in medical hyperspectral images. Sensors 2012, 12, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Kosugi, Y.; Kojima, K.; Tanaka, N. Detection and Analysis of the Intestinal Ischemia Using Visible and Invisible Hyperspectral Imaging. IEEE Trans. Biomed. Eng. 2010, 57, 2011–2017. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Halig, L.V.; Schuster, D.M.; Osunkoya, A.; Master, V.; Nieh, P.T.; Chen, G.Z.; Fei, B. Hyperspectral imaging and quantitative analysis for prostate cancer detection. J. Biomed. Opt. 2012, 17, 0760051. [Google Scholar] [CrossRef]
- Akbari, H.; Uto, K.; Kosugi, Y.; Kojima, K.; Tanaka, N. Cancer detection using infrared hyperspectral imaging. Cancer Sci. 2011, 102, 852–857. [Google Scholar] [CrossRef]
- Halicek, M.; Lu, G.; Little, J.V.; Wang, X.; Patel, M.; Griffith, C.C.; El-Deiry, M.W.; Chen, A.Y.; Fei, B. Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging. J. Biomed. Opt. 2017, 22, 060503. [Google Scholar] [CrossRef]
- Halicek, M.; Fabelo, H.; Ortega, S.; Little, J.V.; Wang, X.; Chen, A.Y.; Callicó, G.M.; Myers, L.; Sumer, B.; Fei, B. Cancer detection using hyperspectral imaging and evaluation of the superficial tumor margin variance with depth. In Medical Imaging 2019: Image-Guided Procedures, Robotic Interventions, and Modeling; Fei, B., Linte, C.A., Eds.; SPIE: Bellingham, WA, USA, 2019; Volume 10951, p. 45. [Google Scholar]
- APU, A.P.U. Zynq-7000 All Programmable SoC Overview. Available online: https://cdn.hackaday.io/files/19354828041536/ds190-Zynq-7000-Overview.pdf (accessed on 27 October 2019).
- Zedboard.org ZedBoard (Zynq Evaluation and Development) Hardware User’s Guide. Available online: http://www.zedboard.org/sites/default/files/documentations/ZedBoard_HW_UG_v2_2.pdf (accessed on 27 October 2019).
- Xilinx Documentation ZC706 Evaluation Board for the Zynq-7000 XC7Z045 SoC-User Guide. Available online: https://www.xilinx.com/support/documentation/boards_and_kits/zc706/ug954-zc706-eval-board-xc7z045-ap-soc.pdf (accessed on 27 October 2019).
- Cacciotti, M.; Camus, V.; Schlachter, J.; Pezzotta, A.; Enz, C. Hardware Acceleration of HDR-Image Tone Mapping on an FPGA-CPU Platform Through High-Level Synthesis. In Proceedings of the 2018 31st IEEE International System-on-Chip Conference (SOCC), Arlington, VA, USA, 4–7 September 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 158–162. [Google Scholar]
- Kowalczyk, M.; Przewlocka, D.; Krvjak, T. Real-Time Implementation of Contextual Image Processing Operations for 4K Video Stream in Zynq UltraScale+ MPSoC. In Proceedings of the Conference on Design and Architectures for Signal and Image Processing (DASIP), Porto, Portugal, 10–12 October 2018; pp. 37–42. [Google Scholar]
- Xilinx Documentation SDSoC Environment User Guide UG1027. Available online: https://www.xilinx.com/support/documentation/sw_manuals/xilinx2017_4/ug1027-sdsoc-user-guide.pdf (accessed on 27 October 2019).
- Nethercote, N.; Seward, J. Valgrind: A framework for heavyweight dynamic binary instrumentation. ACM Sigplan Not. 2007, 42, 89–100. [Google Scholar] [CrossRef]
- Graham, S.L.; Kessler, P.B.; Mckusick, M.K. Gprof: A call graph execution profiler. ACM Sigplan Not. 1982, 17, 120–126. [Google Scholar] [CrossRef]
- VAPNIK, V. Estimation of Dependences Based on Empirical Data; Springer: Cham, Switzerland, 2006; ISBN 9780387342399. [Google Scholar]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Pittsburgh, PA, USA, 27–29 July 1992; pp. 144–152. [Google Scholar]
- Hsu, C.-W.; Lin, C.-J. A comparison of methods for multiclass support vector machines. IEEE Trans. Neural Netw. 2002, 13, 415–425. [Google Scholar] [PubMed]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Bruzzone, L. Kernel-based methods for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2005, 43, 1351–1362. [Google Scholar] [CrossRef]
- Fabelo, H.; Ortega, S.; Lazcano, R.; Madroñal, D.M.; Callicó, G.; Juárez, E.; Salvador, R.; Bulters, D.; Bulstrode, H.; Szolna, A.; et al. An Intraoperative Visualization System Using Hyperspectral Imaging to Aid in Brain Tumor Delineation. Sensors 2018, 18, 430. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Fabelo, H.; Ortega, S.; Szolna, A.; Bulters, D.; Pineiro, J.F.; Kabwama, S.; J-O’Shanahan, A.; Bulstrode, H.; Bisshopp, S.; Kiran, B.R.; et al. In-Vivo Hyperspectral Human Brain Image Database for Brain Cancer Detection. IEEE Access 2019, 7, 39098–39116. [Google Scholar] [CrossRef]
- Fabelo, H.; Ortega, S.; Ravi, D.; Kiran, B.R.; Sosa, C.; Bulters, D.; Callicó, G.M.; Bulstrode, H.; Szolna, A.; Piñeiro, J.F.; et al. Spatio-spectral classification of hyperspectral images for brain cancer detection during surgical operations. PLoS ONE 2018, 13, e0193721. [Google Scholar] [CrossRef]
- Kelly, C.; Siddiqui, F.M.; Bardak, B.; Woods, R. Histogram of oriented gradients front end processing: An FPGA based processor approach. In Proceedings of the IEEE Workshop on Signal Processing Systems (SiPS: Design and Implementation), Belfast, UK, 20–22 October 2014; IEEE: Piscataway, NJ, USA, 2014. [Google Scholar]
- Mahmoodi, D.; Soleimani, A.; Khosravi, H.; Taghizadeh, M. FPGA Simulation of Linear and Nonlinear Support Vector Machine. J. Softw. Eng. Appl. 2011, 4, 320–328. [Google Scholar] [CrossRef]
- Patil, R.A.; Gupta, G.; Sahula, V.; Mandal, A.S. Power aware hardware prototyping of multiclass SVM classifier through reconfiguration. In Proceedings of the IEEE International Conference on VLSI Design, Hyderabad, India, 7–11 January 2012; pp. 62–67. [Google Scholar]
- Cutajar, M.; Gatt, E.; Grech, I.; Casha, O.; Micallef, J. Hardware-based support vector machine for phoneme classification. In Proceedings of the IEEE EuroCon 2013, Zagreb, Croatia, 1–4 July 2013; pp. 1701–1708. [Google Scholar]
- Mandal, B.; Sarma, M.P.; Sarma, K.K.; Mastorakis, N. Implementation of Systolic Array Based SVM Classifier Using Multiplierless Kernel. In Proceedings of the 16th International Conference on Automatic Control, Modelling & Simulation (ACMOS’14), Brasov, Romania, 26–28 June 2014; ISBN 9789604743834. [Google Scholar]
- Khosravi, H.; Kabir, E. Introducing a very large dataset of handwritten Farsi digits and a study on their varieties. Pattern Recognit. Lett. 2007, 28, 1133–1141. [Google Scholar] [CrossRef]
- Lucey, P.; Cohn, J.F.; Kanade, T.; Saragih, J.; Ambadar, Z.; Matthews, I. The extended Cohn-Kanade dataset (CK+): A complete dataset for action unit and emotion-specified expression. In Proceedings of the 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition-Workshops (CVPRW 2010), San Francisco, CA, USA, 13–18 June 2010; pp. 94–101. [Google Scholar]
- Garofolo, J.S.; Lamel, L.F.; Fisher, W.M.; Fiscus, J.G.; Pallett, D.S. DARPA TIMIT Acoustic-Phonetic Continous Speech Corpus CD-ROM. NIST Speech Disc 1-1.1; NASA STI/Recon Technical Report N: Gaithersburg, MD, USA, 1993; Volume 93.



| ZedBoard (ZC7020) | ZC706 (ZC7045) | |||
|---|---|---|---|---|
| Type | F | M | F | M |
| Dynamic Power (W) | 2.42 | 1.89 | 2.61 | 1.91 |
| Static Power (W) | 0.17 | 0.15 | 0.22 | 0.21 |
| Total (W) | 2.59 | 2.04 | 2.84 | 2.13 |
| Reference Method | [59] | [60] | [61] | [62] | Proposed (M Version) | |
|---|---|---|---|---|---|---|
| Device | Xilinx Virtex-4 | Xilinx Virtex-6 | Xilinx Virtex-II | Xilinx Virtex-7 | ZC7020 (ZedBoard) | ZC7045 (ZC706) |
| Tool | System Generator | Xilinx ISE | n/a | Xilinx XPE 14.1 | SDSOC 2018.2 | SDSOC 2018.2 |
| Clock rate (MHz) | 202.84 | n/a | 42.012 | n/a | 200 | 200 |
| Speedup factor | n/a | n/a | 2.53 | n/a | 2.20 | 2.86 |
| Power (W) | n/a | 2.02 | n/a | 1.70 | 2.04 | 2.13 |
| Slice Registers (%) | 5.00 | 0.15 | 21.00 | 11.00 | n/a | n/a |
| Slice LUTs (%) | 2.00 | 0.35 | 20.00 | 11.00 | n/a | n/a |
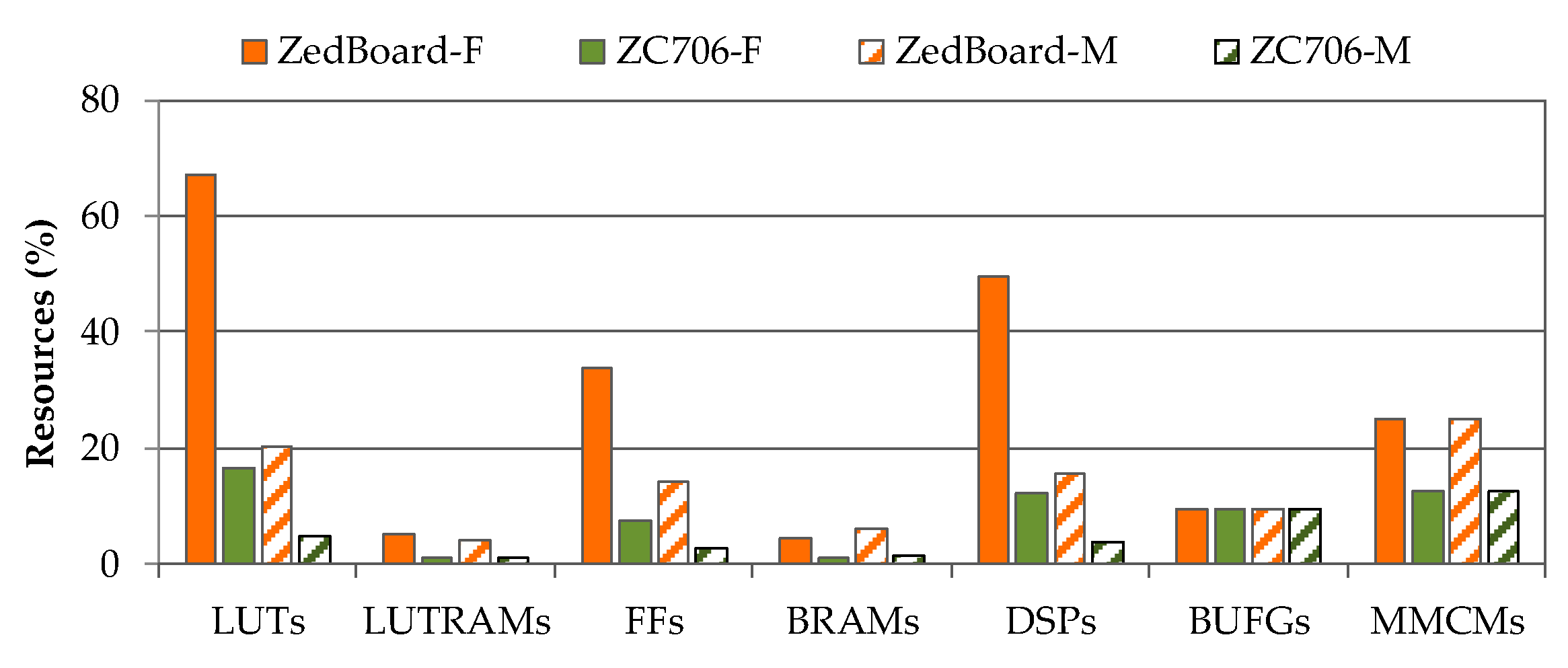
| LUTs (%) | n/a | n/a | n/a | n/a | 20.22 | 4.84 |
| LUTRAM (%) | n/a | n/a | n/a | n/a | 4.30 | 1.00 |
| FF (%) | 4.00 | 32.00 | 2.00 | 100.00 | 14.18 | 2.76 |
| IOBs (%) | 37.00 | 37.00 | 20.00 | 4.00 | n/a | n/a |
| DSP (%) | 14.00 | 0.91 | n/a | 0.00 | 15.45 | 3.78 |
| BUFG (%) | 3.00 | 3.00 | n/a | n/a | 9.38 | 9.38 |
| BRAM (%) | n/a | n/a | n/a | n/a | 6.07 | 1.56 |
| MMCM (%) | n/a | n/a | n/a | n/a | 25.00 | 12.50 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baez, A.; Fabelo, H.; Ortega, S.; Florimbi, G.; Torti, E.; Hernandez, A.; Leporati, F.; Danese, G.; M. Callico, G.; Sarmiento, R. High-Level Synthesis of Multiclass SVM Using Code Refactoring to Classify Brain Cancer from Hyperspectral Images. Electronics 2019, 8, 1494. https://doi.org/10.3390/electronics8121494
Baez A, Fabelo H, Ortega S, Florimbi G, Torti E, Hernandez A, Leporati F, Danese G, M. Callico G, Sarmiento R. High-Level Synthesis of Multiclass SVM Using Code Refactoring to Classify Brain Cancer from Hyperspectral Images. Electronics. 2019; 8(12):1494. https://doi.org/10.3390/electronics8121494
Chicago/Turabian StyleBaez, Abelardo, Himar Fabelo, Samuel Ortega, Giordana Florimbi, Emanuele Torti, Abian Hernandez, Francesco Leporati, Giovanni Danese, Gustavo M. Callico, and Roberto Sarmiento. 2019. "High-Level Synthesis of Multiclass SVM Using Code Refactoring to Classify Brain Cancer from Hyperspectral Images" Electronics 8, no. 12: 1494. https://doi.org/10.3390/electronics8121494
APA StyleBaez, A., Fabelo, H., Ortega, S., Florimbi, G., Torti, E., Hernandez, A., Leporati, F., Danese, G., M. Callico, G., & Sarmiento, R. (2019). High-Level Synthesis of Multiclass SVM Using Code Refactoring to Classify Brain Cancer from Hyperspectral Images. Electronics, 8(12), 1494. https://doi.org/10.3390/electronics8121494










