Enhancing Boundary Precision and Long-Range Dependency Modeling in Medical Imaging via Unified Attention Framework
Abstract
1. Introduction
- A hierarchical attention mechanism is introduced to achieve adaptive fusion of local and global features;
- A fine-grained feature enhancement module is designed to improve the detection and segmentation of small-scale lesions;
- A cross-scale consistency constraint is proposed to guarantee coordination and robustness of segmentation results across scales.
2. Related Work
2.1. Medical Image Segmentation Methods
2.2. Applications of Attention Mechanisms in Medical Imaging
2.3. Hierarchical and Multi-Scale Feature Fusion Methods
3. Materials and Method
3.1. Data Collection
3.2. Data Preprocessing and Augmentation
3.3. Proposed Method
3.3.1. Overall
3.3.2. Local Detail Attention Module
3.3.3. Global Context Attention Module
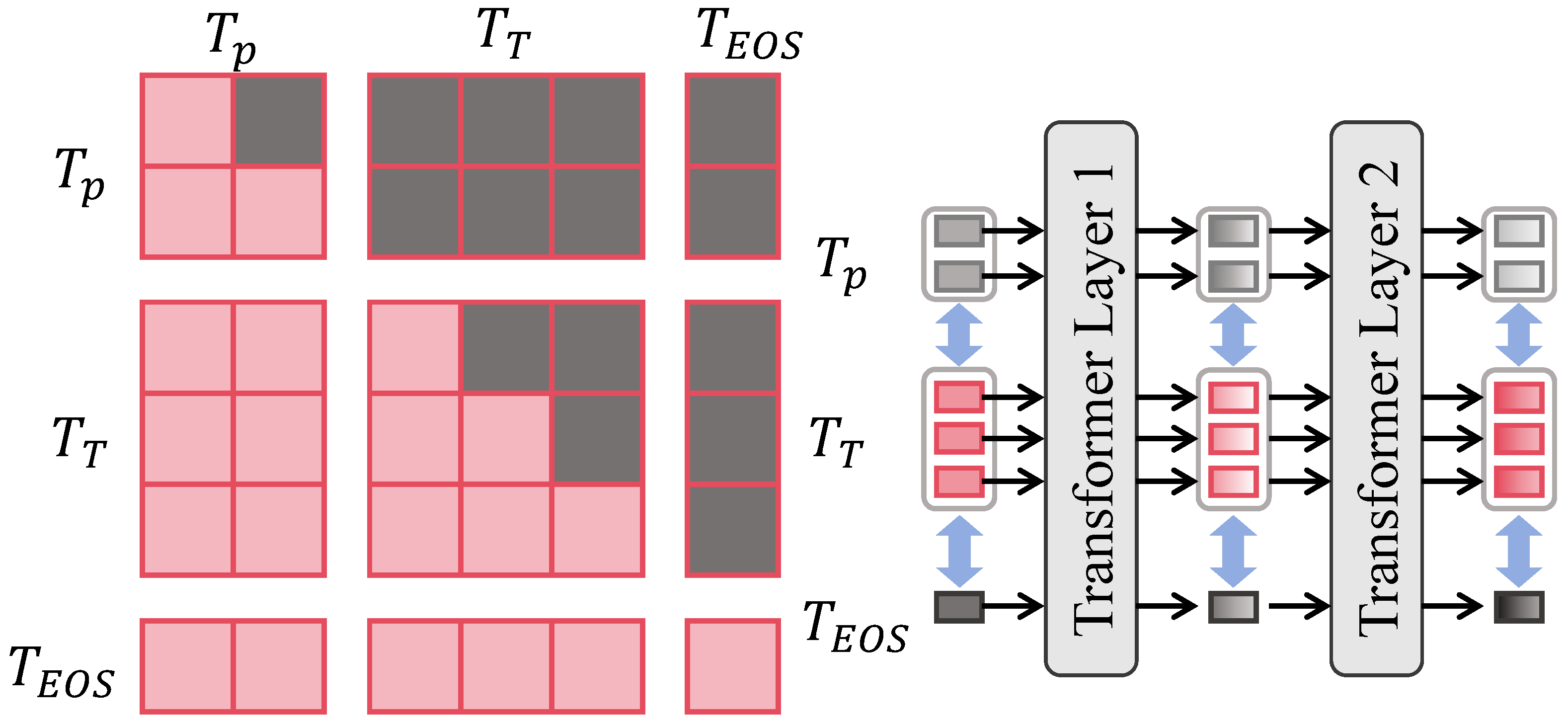
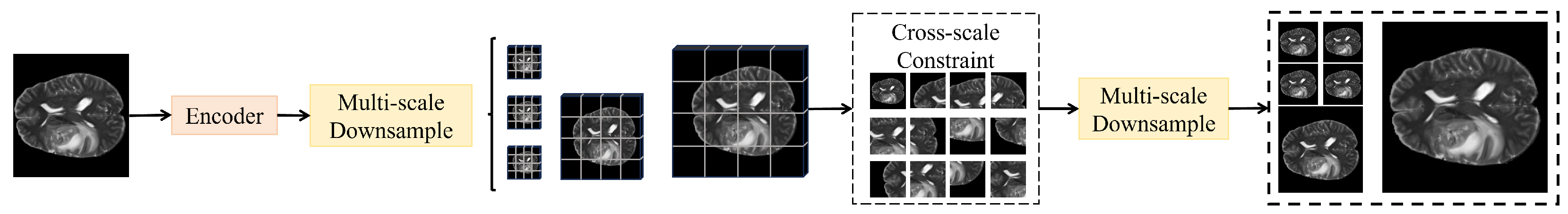
3.3.4. Cross-Scale Consistency Constraint Module
3.4. Experimental Setup
3.4.1. Configurations
3.4.2. Evaluation Metrics
3.4.3. Baseline Models
4. Results and Discussion
4.1. Overall Performance of Different Methods Across All Datasets
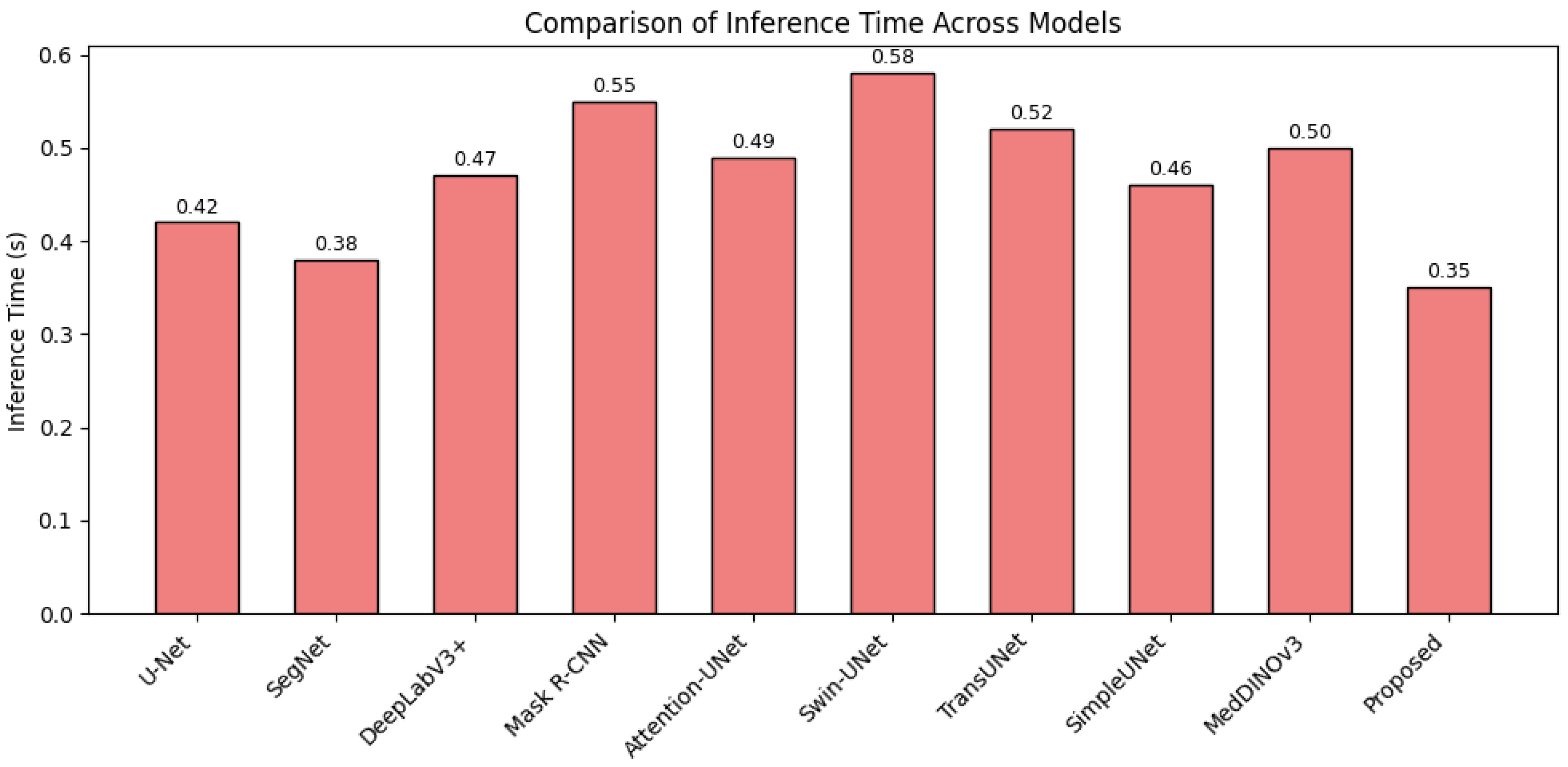
4.2. Detail Analysis
4.3. Ablation Study
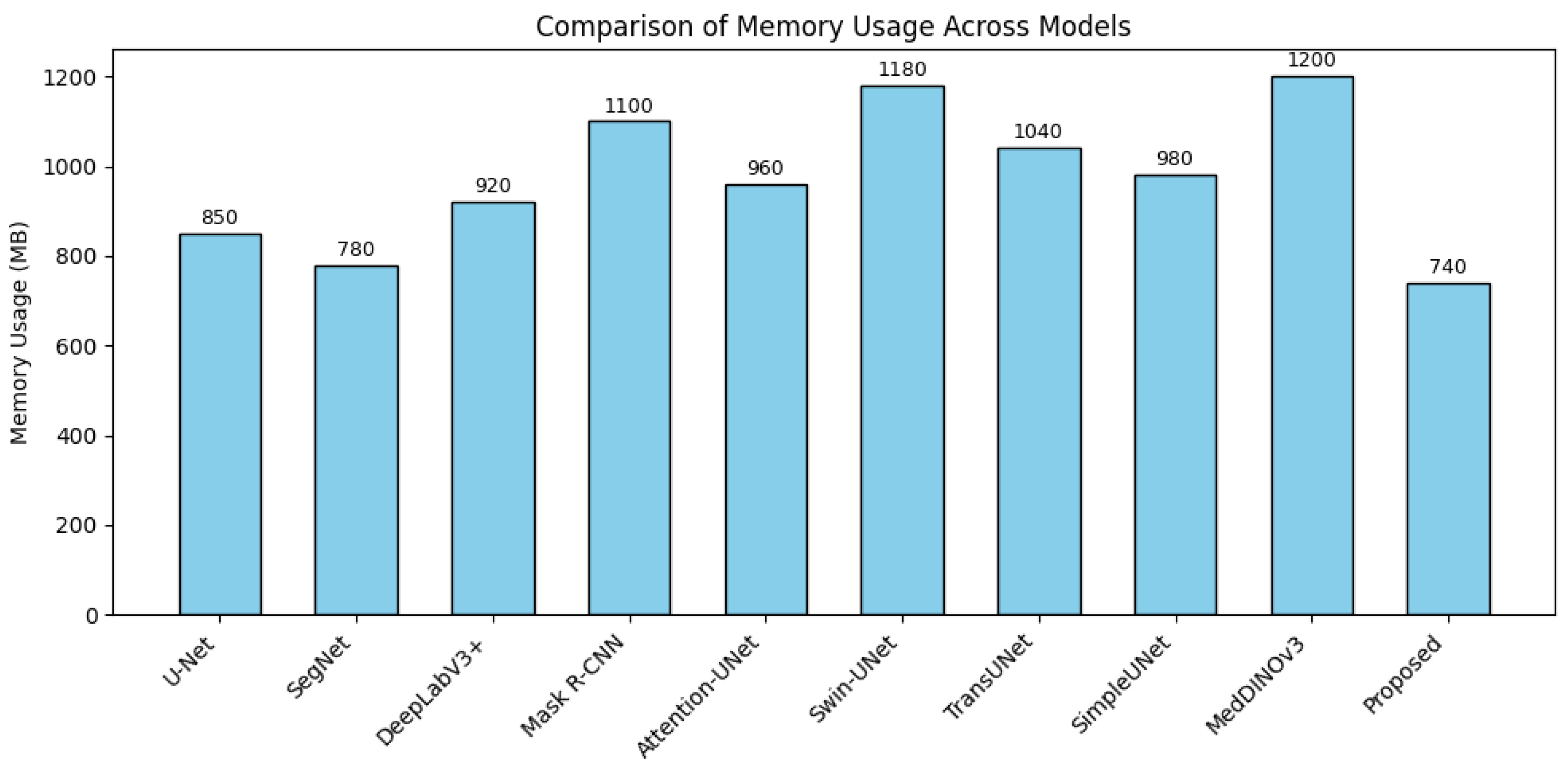
4.4. Discussion
4.5. Limitation and Future Work
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, Y.; Shen, Z.; Jiao, R. Segment anything model for medical image segmentation: Current applications and future directions. Comput. Biol. Med. 2024, 171, 108238. [Google Scholar] [CrossRef]
- Obuchowicz, R.; Strzelecki, M.; Piórkowski, A. Clinical applications of artificial intelligence in medical imaging and image processing—A review. Cancers 2024, 16, 1870. [Google Scholar] [CrossRef]
- Berghout, T. The neural frontier of future medical imaging: A review of deep learning for brain tumor detection. J. Imaging 2024, 11, 2. [Google Scholar] [CrossRef]
- Aparnaa, R.; Dinesh, R. Multi-disease diagnosis using medical images. In Proceedings of the 2024 2nd International Conference on Artificial Intelligence and Machine Learning Applications Theme: Healthcare and Internet of Things (AIMLA), Namakkal, India, 15–16 March 2024; pp. 1–6. [Google Scholar]
- Li, X.; Zhang, L.; Yang, J.; Teng, F. Role of artificial intelligence in medical image analysis: A review of current trends and future directions. J. Med Biol. Eng. 2024, 44, 231–243. [Google Scholar] [CrossRef]
- Li, H.; Bu, Q.; Shi, X.; Xu, X.; Li, J. Non-invasive medical imaging technology for the diagnosis of burn depth. Int. Wound J. 2024, 21, e14681. [Google Scholar] [CrossRef]
- Baştuğ, B.T.; Başol, H.G. Radiology’s role in dermatology: A closer look at two years of data. Eur. Res. J. 2025, 11, 395–403. [Google Scholar] [CrossRef]
- Vavekanand, R. A deep learning approach for medical image segmentation integrating magnetic resonance imaging to enhance brain tumor recognition. SSRN 2024, 4827019, in press. [Google Scholar] [CrossRef]
- Xu, Y.; Quan, R.; Xu, W.; Huang, Y.; Chen, X.; Liu, F. Advances in medical image segmentation: A comprehensive review of traditional, deep learning and hybrid approaches. Bioengineering 2024, 11, 1034. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, R.R.; Houssein, E.H.; Hussien, A.G.; Singh, B.; Emam, M.M. An enhanced chameleon swarm algorithm for global optimization and multi-level thresholding medical image segmentation. Neural Comput. Appl. 2024, 36, 8775–8823. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, S.; Ren, Y.; Zhang, Y.; Fu, J.; Fan, D.; Lin, J.; Wang, Q. Atrous Pyramid GAN Segmentation Network for Fish Images with High Performance. Electronics 2022, 11, 911. [Google Scholar] [CrossRef]
- Azad, R.; Aghdam, E.K.; Rauland, A.; Jia, Y.; Avval, A.H.; Bozorgpour, A.; Karimijafarbigloo, S.; Cohen, J.P.; Adeli, E.; Merhof, D. Medical image segmentation review: The success of u-net. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 10076–10095. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, X.; Wang, S.; Wang, J.; Melo, S.N. MRCA-UNet: A Multiscale Recombined Channel Attention U-Net Model for Medical Image Segmentation. Symmetry 2025, 17, 892. [Google Scholar] [CrossRef]
- Huang, L.; Miron, A.; Hone, K.; Li, Y. Segmenting medical images: From UNet to res-UNet and nnUNet. In Proceedings of the 2024 IEEE 37th International Symposium on Computer-Based Medical Systems (CBMS), Guadalajara, Mexico, 26–28 June 2024; pp. 483–489. [Google Scholar]
- Chen, B.; Li, Y.; Liu, J.; Yang, F.; Zhang, L. MSMHSA-DeepLab V3+: An Effective Multi-Scale, Multi-Head Self-Attention Network for Dual-Modality Cardiac Medical Image Segmentation. J. Imaging 2024, 10, 135. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Yi, C.; Xiong, Q.; Jiang, S. SEA-NET: Medical image segmentation network based on spiral squeeze-and-excitation and attention modules. BMC Med. Imaging 2024, 24, 17. [Google Scholar] [CrossRef]
- Zheng, F.; Chen, X.; Liu, W.; Li, H.; Lei, Y.; He, J.; Pun, C.M.; Zhou, S. Smaformer: Synergistic multi-attention transformer for medical image segmentation. In Proceedings of the 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Lisbon, Portugal, 3–6 December 2024; pp. 4048–4053. [Google Scholar]
- Zhang, C.; Sun, S.; Hu, W.; Zhao, P. FDR-TransUNet: A novel encoder-decoder architecture with vision transformer for improved medical image segmentation. Comput. Biol. Med. 2024, 169, 107858. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, X.; Li, R.; Zhou, P. Swin-HAUnet: A Swin-Hierarchical Attention Unet For Enhanced Medical Image Segmentation. In Proceedings of the Chinese Conference on Pattern Recognition and Computer Vision (PRCV), Urumqi, China, 18–20 October 2024; Springer: Berlin/Heidelberg, Germany, 2024; pp. 371–385. [Google Scholar]
- Ghamsarian, N.; Wolf, S.; Zinkernagel, M.; Schoeffmann, K.; Sznitman, R. Deeppyramid+: Medical image segmentation using pyramid view fusion and deformable pyramid reception. Int. J. Comput. Assist. Radiol. Surg. 2024, 19, 851–859. [Google Scholar] [CrossRef]
- Hikmah, N.F.; Hajjanto, A.D.; Surbakti, A.F.A.; Prakosa, N.A.; Asmaria, T.; Sardjono, T.A. Brain tumor detection using a MobileNetV2-SSD model with modified feature pyramid network levels. Int. J. Electr. Comput. Eng. 2024, 14, 3995–4004. [Google Scholar] [CrossRef]
- Han, X.; Li, T.; Bai, C.; Yang, H. Integrating prior knowledge into a bibranch pyramid network for medical image segmentation. Image Vis. Comput. 2024, 143, 104945. [Google Scholar] [CrossRef]
- Chen, A.; Wei, Y.; Le, H.; Zhang, Y. Learning by teaching with ChatGPT: The effect of teachable ChatGPT agent on programming education. Br. J. Educ. Technol. 2024, in press. [CrossRef]
- Kumar, R.R.; Priyadarshi, R. Denoising and segmentation in medical image analysis: A comprehensive review on machine learning and deep learning approaches. Multimed. Tools Appl. 2025, 84, 10817–10875. [Google Scholar] [CrossRef]
- Zhang, Y.; Mao, Y.; Lu, X.; Zou, X.; Huang, H.; Li, X.; Li, J.; Zhang, H. From single to universal: Tiny lesion detection in medical imaging. Artif. Intell. Rev. 2024, 57, 192. [Google Scholar] [CrossRef]
- Krithika Alias AnbuDevi, M.; Suganthi, K. Review of semantic segmentation of medical images using modified architectures of UNET. Diagnostics 2022, 12, 3064. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Maji, D.; Sigedar, P.; Singh, M. Attention Res-UNet with Guided Decoder for semantic segmentation of brain tumors. Biomed. Signal Process. Control 2022, 71, 103077. [Google Scholar] [CrossRef]
- Ali, S.; Khurram, R.; Rehman, K.u.; Yasin, A.; Shaukat, Z.; Sakhawat, Z.; Mujtaba, G. An improved 3D U-Net-based deep learning system for brain tumor segmentation using multi-modal MRI. Multimed. Tools Appl. 2024, 83, 85027–85046. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 11534–11542. [Google Scholar]
- Chen, J.; Mei, J.; Li, X.; Lu, Y.; Yu, Q.; Wei, Q.; Luo, X.; Xie, Y.; Adeli, E.; Wang, Y.; et al. TransUNet: Rethinking the U-Net architecture design for medical image segmentation through the lens of transformers. Med. Image Anal. 2024, 97, 103280. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 205–218. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Gao, Y.; Zhou, M.; Metaxas, D.N. UTNet: A hybrid transformer architecture for medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 61–71. [Google Scholar]
- Tian, Q.; Wang, Z.; Cui, X. Improved Unet brain tumor image segmentation based on GSConv module and ECA attention mechanism. arXiv 2024, arXiv:2409.13626. [Google Scholar] [CrossRef]
- Zhao, P.; Li, Z.; You, Z.; Chen, Z.; Huang, T.; Guo, K.; Li, D. SE-U-lite: Milling tool wear segmentation based on lightweight U-net model with squeeze-and-excitation module. IEEE Trans. Instrum. Meas. 2024, 73, 1–8. [Google Scholar] [CrossRef]
- Matlala, B.; van der Haar, D.; Vadapalli, H. Automated Gross Tumor Volume Segmentation in Meningioma Using Squeeze and Excitation Residual U-Net for Enhanced Radiotherapy Planning. In Proceedings of the International Conference on AI in Healthcare, Basel, Switzerland, 10–11 November 2025; Springer: Berlin/Heidelberg, Germany, 2025; pp. 57–67. [Google Scholar]
- Wu, Y.; Lin, Y.; Xu, T.; Meng, X.; Liu, H.; Kang, T. Multi-Scale Feature Integration and Spatial Attention for Accurate Lesion Segmentation. In Proceedings of the 6th International Conference on Electronic Communication and Artificial Intelligence (ICECAI), Chengdu, China, 20–22 June 2025. [Google Scholar]
- Chen, Y.; Zou, B.; Guo, Z.; Huang, Y.; Huang, Y.; Qin, F.; Li, Q.; Wang, C. Scunet++: Swin-unet and cnn bottleneck hybrid architecture with multi-fusion dense skip connection for pulmonary embolism ct image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2024; pp. 7759–7767. [Google Scholar]
- Chen, G.; Zhou, L.; Zhang, J.; Yin, X.; Cui, L.; Dai, Y. ESKNet: An enhanced adaptive selection kernel convolution for ultrasound breast tumors segmentation. Expert Syst. Appl. 2024, 246, 123265. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Liu, J.; Zhao, D.; Shen, J.; Geng, P.; Zhang, Y.; Yang, J.; Zhang, Z. HRD-Net: High resolution segmentation network with adaptive learning ability of retinal vessel features. Comput. Biol. Med. 2024, 173, 108295. [Google Scholar] [CrossRef]
- Dai, H.; Xie, W.; Xia, E. SK-Unet++: An improved Unet++ network with adaptive receptive fields for automatic segmentation of ultrasound thyroid nodule images. Med. Phys. 2024, 51, 1798–1811. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Nguyen, N.M.; Huynh, T.Q.; Su, A.K. An enhanced UNet3+ model for accurate identification of COVID-19 in CT images. Digit. Signal Process. 2025, 163, 105205. [Google Scholar] [CrossRef]
- De Verdier, M.C.; Saluja, R.; Gagnon, L.; LaBella, D.; Baid, U.; Tahon, N.H.; Foltyn-Dumitru, M.; Zhang, J.; Alafif, M.; Baig, S.; et al. The 2024 brain tumor segmentation (brats) challenge: Glioma segmentation on post-treatment mri. arXiv 2024, arXiv:2405.18368. [Google Scholar]
- Armato III, S.G.; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Kalpathy-Cramer, J.; Zhao, B.; Goldgof, D.; Gu, Y.; Wang, X.; Yang, H.; Tan, Y.; Gillies, R.; Napel, S. A comparison of lung nodule segmentation algorithms: Methods and results from a multi-institutional study. J. Digit. Imaging 2016, 29, 476–487. [Google Scholar] [CrossRef]
- Yeap, P.L.; Wong, Y.M.; Ong, A.L.K.; Tuan, J.K.L.; Pang, E.P.P.; Park, S.Y.; Lee, J.C.L.; Tan, H.Q. Predicting dice similarity coefficient of deformably registered contours using Siamese neural network. Phys. Med. Biol. 2023, 68, 155016. [Google Scholar] [CrossRef]
- He, Y.; Zhang, N.; Ge, X.; Li, S.; Yang, L.; Kong, M.; Guo, Y.; Lv, C. Passion Fruit Disease Detection Using Sparse Parallel Attention Mechanism and Optical Sensing. Agriculture 2025, 15, 733. [Google Scholar] [CrossRef]
- Zhou, C.; Ge, X.; Chang, Y.; Wang, M.; Shi, Z.; Ji, M.; Wu, T.; Lv, C. A Multimodal Parallel Transformer Framework for Apple Disease Detection and Severity Classification with Lightweight Optimization. Agronomy 2025, 15, 1246. [Google Scholar] [CrossRef]
- Li, L.; Ma, H.; Zhang, X.; Zhao, X.; Lv, M.; Jia, Z. Synthetic aperture radar image change detection based on principal component analysis and two-level clustering. Remote Sens. 2024, 16, 1861. [Google Scholar] [CrossRef]
- Li, L.; Ma, H.; Jia, Z. Multiscale geometric analysis fusion-based unsupervised change detection in remote sensing images via FLICM model. Entropy 2022, 24, 291. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Yu, X.; Chen, Y.; He, G.; Zeng, Q.; Qin, Y.; Liang, M.; Luo, D.; Liao, Y.; Ren, Z.; Kang, C.; et al. Simple is what you need for efficient and accurate medical image segmentation. arXiv 2025, arXiv:2506.13415. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.; Lai, Y.; Hu, M.; Yang, X. MedDINOv3: How to adapt vision foundation models for medical image segmentation? arXiv 2025, arXiv:2509.02379. [Google Scholar] [CrossRef]
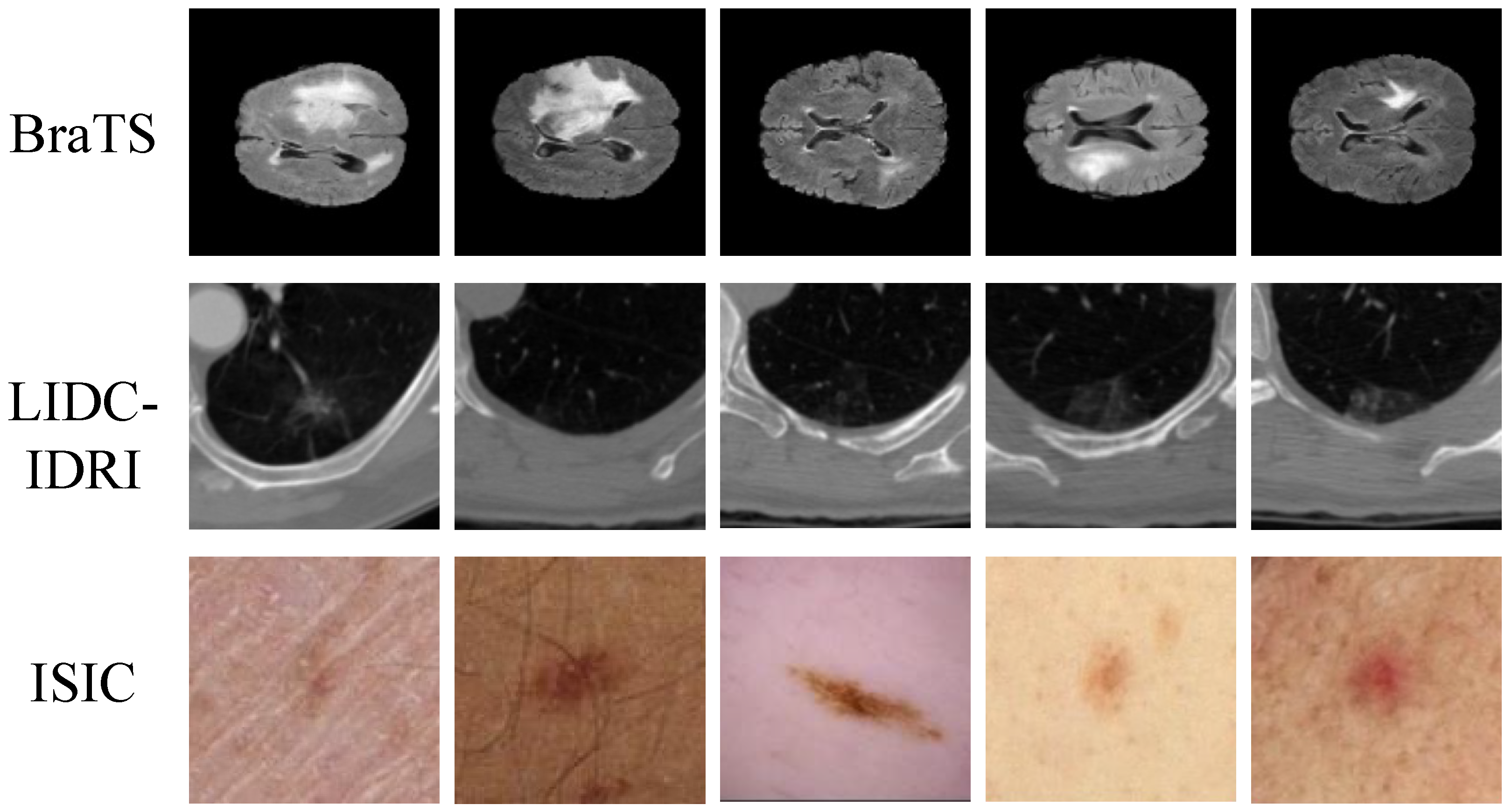



| Dataset | Modality | Collection Time | Resolution/Thickness | Number of Cases |
|---|---|---|---|---|
| BraTS | MRI (T1, T1CE, T2, FLAIR) | 2015–2021 | 1500 | |
| LIDC-IDRI | CT | 2004–2008 | 1 mm slice thickness | 1018 |
| ISIC | Dermoscopy | 2016–2020 | – | 2500 |
| Stage | Operation/Module | Input Tensor | Output Tensor |
|---|---|---|---|
| Input | Preprocessed image | — | |
| Encoder E | Conv + Patch embedding (Stage 1) | I | |
| Hierarchical down-sampling + Transformer blocks (Stage 2–4) | , where , , | ||
| Local Detail Attention Branch | Channel–spatial joint gating + boundary-sensitive residuals | (same shape as inputs) | |
| Global Context Attention Branch | Windowed self-attention + shifted windows + position encoding | (same shape as inputs) | |
| Dynamic Cross-Scale Fusion | Projection + adaptive pixel-wise weighting + concatenation | ||
| Decoder | Progressive up-sampling + skip connections | Z | (multi-scale predictions) |
| Cross-Scale Consistency | Bidirectional alignment among | Consistent feature maps | |
| Classification Head | Global pooling + fully connected layer | Z | Class probabilities |
| Output | Segmentation masks + lesion class probabilities | — |
| Method | Dice | IoU | Precision | Recall | F1 |
|---|---|---|---|---|---|
| U-Net [27] | 0.837 | 0.725 | 0.854 | 0.819 | 0.837 |
| SegNet [58] | 0.819 | 0.708 | 0.841 | 0.799 | 0.820 |
| DeepLabV3+ [43] | 0.853 | 0.744 | 0.872 | 0.835 | 0.853 |
| Mask R-CNN [59] | 0.836 | 0.723 | 0.861 | 0.809 | 0.836 |
| Attention-UNet [60] | 0.861 | 0.757 | 0.873 | 0.848 | 0.861 |
| Swin-UNet [33] | 0.868 | 0.761 | 0.877 | 0.854 | 0.868 |
| TransUNet [32] | 0.872 | 0.765 | 0.880 | 0.858 | 0.872 |
| SimpleUNet [61] | 0.876 | 0.770 | 0.886 | 0.862 | 0.876 |
| MedDINOv3 [62] | 0.879 | 0.774 | 0.889 | 0.867 | 0.879 |
| Proposed | 0.886 | 0.781 | 0.898 | 0.875 | 0.886 |
| Dataset | Method | Dice | IoU | Precision | Recall | F1 |
|---|---|---|---|---|---|---|
| BraTS | U-Net | 0.883 | 0.789 | 0.895 | 0.872 | 0.883 |
| SegNet | 0.861 | 0.758 | 0.880 | 0.845 | 0.862 | |
| DeepLabV3+ | 0.892 | 0.804 | 0.908 | 0.878 | 0.893 | |
| Mask R-CNN | 0.865 | 0.764 | 0.901 | 0.835 | 0.867 | |
| Attention-UNet | 0.889 | 0.799 | 0.907 | 0.874 | 0.889 | |
| Swin-UNet | 0.895 | 0.811 | 0.910 | 0.881 | 0.895 | |
| TransUNet | 0.904 | 0.823 | 0.914 | 0.895 | 0.904 | |
| SimpleUNet | 0.909 | 0.832 | 0.919 | 0.902 | 0.909 | |
| MedDINOv3 | 0.915 | 0.841 | 0.925 | 0.906 | 0.915 | |
| Proposed | 0.922 | 0.853 | 0.930 | 0.915 | 0.922 | |
| LIDC-IDRI | U-Net | 0.751 | 0.606 | 0.774 | 0.732 | 0.752 |
| SegNet | 0.740 | 0.591 | 0.768 | 0.718 | 0.741 | |
| DeepLabV3+ | 0.783 | 0.644 | 0.806 | 0.764 | 0.784 | |
| Mask R-CNN | 0.776 | 0.635 | 0.812 | 0.744 | 0.776 | |
| Attention-UNet | 0.789 | 0.649 | 0.816 | 0.759 | 0.789 | |
| Swin-UNet | 0.792 | 0.654 | 0.818 | 0.766 | 0.792 | |
| TransUNet | 0.795 | 0.660 | 0.812 | 0.779 | 0.795 | |
| SimpleUNet | 0.805 | 0.673 | 0.828 | 0.786 | 0.805 | |
| MedDINOv3 | 0.814 | 0.683 | 0.835 | 0.794 | 0.814 | |
| Proposed | 0.822 | 0.697 | 0.840 | 0.807 | 0.822 | |
| ISIC | U-Net | 0.876 | 0.781 | 0.892 | 0.862 | 0.876 |
| SegNet | 0.855 | 0.748 | 0.876 | 0.836 | 0.855 | |
| DeepLabV3+ | 0.885 | 0.790 | 0.902 | 0.868 | 0.885 | |
| Mask R-CNN | 0.867 | 0.770 | 0.901 | 0.835 | 0.867 | |
| Attention-UNet | 0.883 | 0.788 | 0.904 | 0.861 | 0.883 | |
| Swin-UNet | 0.889 | 0.797 | 0.907 | 0.872 | 0.889 | |
| TransUNet | 0.898 | 0.809 | 0.910 | 0.886 | 0.898 | |
| SimpleUNet | 0.905 | 0.821 | 0.915 | 0.892 | 0.905 | |
| MedDINOv3 | 0.909 | 0.830 | 0.918 | 0.898 | 0.909 | |
| Proposed | 0.914 | 0.842 | 0.922 | 0.906 | 0.914 | |
| Self-Collected | U-Net | 0.792 | 0.649 | 0.818 | 0.775 | 0.792 |
| SegNet | 0.773 | 0.627 | 0.806 | 0.758 | 0.773 | |
| DeepLabV3+ | 0.812 | 0.674 | 0.836 | 0.792 | 0.812 | |
| Mask R-CNN | 0.804 | 0.662 | 0.831 | 0.780 | 0.804 | |
| Attention-UNet | 0.821 | 0.683 | 0.842 | 0.801 | 0.821 | |
| Swin-UNet | 0.829 | 0.692 | 0.849 | 0.808 | 0.829 | |
| TransUNet | 0.835 | 0.701 | 0.855 | 0.816 | 0.835 | |
| SimpleUNet | 0.842 | 0.713 | 0.863 | 0.823 | 0.842 | |
| MedDINOv3 | 0.848 | 0.721 | 0.870 | 0.828 | 0.848 | |
| Proposed | 0.872 | 0.752 | 0.889 | 0.848 | 0.872 |
| Dataset | Configuration | Dice | IoU | Precision | Recall | F1 |
|---|---|---|---|---|---|---|
| BraTS | Baseline (U-Net backbone) | 0.883 | 0.789 | 0.895 | 0.872 | 0.883 |
| + LDA | 0.897 | 0.806 | 0.907 | 0.886 | 0.897 | |
| + LDA + GCA | 0.910 | 0.829 | 0.918 | 0.902 | 0.910 | |
| + LDA + GCA + CS (Proposed) | 0.923 | 0.852 | 0.931 | 0.916 | 0.923 | |
| LIDC-IDRI | Baseline (U-Net backbone) | 0.751 | 0.606 | 0.774 | 0.731 | 0.751 |
| + LDA | 0.770 | 0.632 | 0.796 | 0.751 | 0.770 | |
| + LDA + GCA | 0.800 | 0.669 | 0.820 | 0.781 | 0.800 | |
| + LDA + GCA + CS (Proposed) | 0.823 | 0.696 | 0.841 | 0.809 | 0.823 | |
| ISIC | Baseline (U-Net backbone) | 0.877 | 0.782 | 0.893 | 0.863 | 0.877 |
| + LDA | 0.889 | 0.799 | 0.904 | 0.876 | 0.889 | |
| + LDA + GCA | 0.903 | 0.819 | 0.915 | 0.890 | 0.903 | |
| + LDA + GCA + CS (Proposed) | 0.915 | 0.841 | 0.923 | 0.907 | 0.915 | |
| Self-Collected | Baseline (U-Net backbone) | 0.813 | 0.675 | 0.835 | 0.791 | 0.813 |
| + LDA | 0.831 | 0.700 | 0.850 | 0.808 | 0.831 | |
| + LDA + GCA | 0.857 | 0.733 | 0.874 | 0.834 | 0.857 | |
| + LDA + GCA + CS (Proposed) | 0.871 | 0.753 | 0.888 | 0.849 | 0.871 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Zhu, Y.; Ma, H.; Li, B.; Xiao, L.; Wu, X.; Li, M. Enhancing Boundary Precision and Long-Range Dependency Modeling in Medical Imaging via Unified Attention Framework. Electronics 2025, 14, 4335. https://doi.org/10.3390/electronics14214335
Zhu Y, Zhu Y, Ma H, Li B, Xiao L, Wu X, Li M. Enhancing Boundary Precision and Long-Range Dependency Modeling in Medical Imaging via Unified Attention Framework. Electronics. 2025; 14(21):4335. https://doi.org/10.3390/electronics14214335
Chicago/Turabian StyleZhu, Yi, Yawen Zhu, Hongtao Ma, Bin Li, Luyao Xiao, Xiaxu Wu, and Manzhou Li. 2025. "Enhancing Boundary Precision and Long-Range Dependency Modeling in Medical Imaging via Unified Attention Framework" Electronics 14, no. 21: 4335. https://doi.org/10.3390/electronics14214335
APA StyleZhu, Y., Zhu, Y., Ma, H., Li, B., Xiao, L., Wu, X., & Li, M. (2025). Enhancing Boundary Precision and Long-Range Dependency Modeling in Medical Imaging via Unified Attention Framework. Electronics, 14(21), 4335. https://doi.org/10.3390/electronics14214335





