Hybrid CNN–Transformer with Fusion Discriminator for Ovarian Tumor Ultrasound Imaging Classification
Abstract
1. Introduction
- 1.
- A novel deep network architecture based on local–global attention fusion is introduced to enhance tumor boundary recognition in CDFI images.
- 2.
- A local enhancement module is designed to highlight fine-grained lesion features.
- 3.
- A global attention mechanism is incorporated to capture long-range dependencies between blood flow signals and lesion regions.
- 4.
- The superiority of the proposed model is validated on multi-center CDFI datasets, achieving substantial improvements over classical networks such as ResNet and DenseNet.
- 5.
- An economic analysis framework is proposed to investigate how the model alleviates insufficient healthcare resources (e.g., shortage of experienced sonographers). Through cost-effectiveness simulation, scalability analysis, and social value evaluation, the framework provides a comprehensive perspective on the economic impact of early diagnosis for both patients and society.
2. Related Work
2.1. Ultrasound Imaging Analysis of Ovarian Tumor Malignancy
2.2. Applications of Deep Learning in Medical Ultrasound
2.3. Attention Mechanisms and Global Modeling Methods
2.4. Medical Artificial Intelligence and Health Economics
3. Materials and Method
3.1. Data Collection
3.2. Data Preprocessing and Augmentation
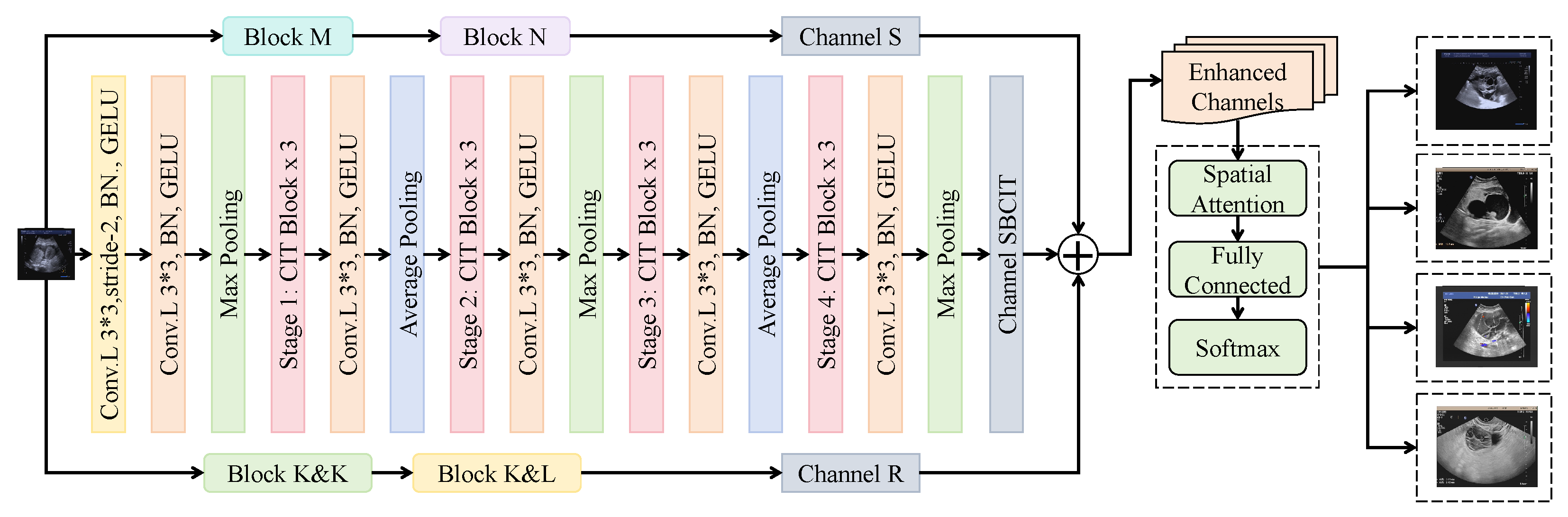
3.3. Proposed Method
3.3.1. Local Enhancement Module
3.3.2. Fusion Discriminator
3.3.3. Global Attention Module
4. Results and Discussion
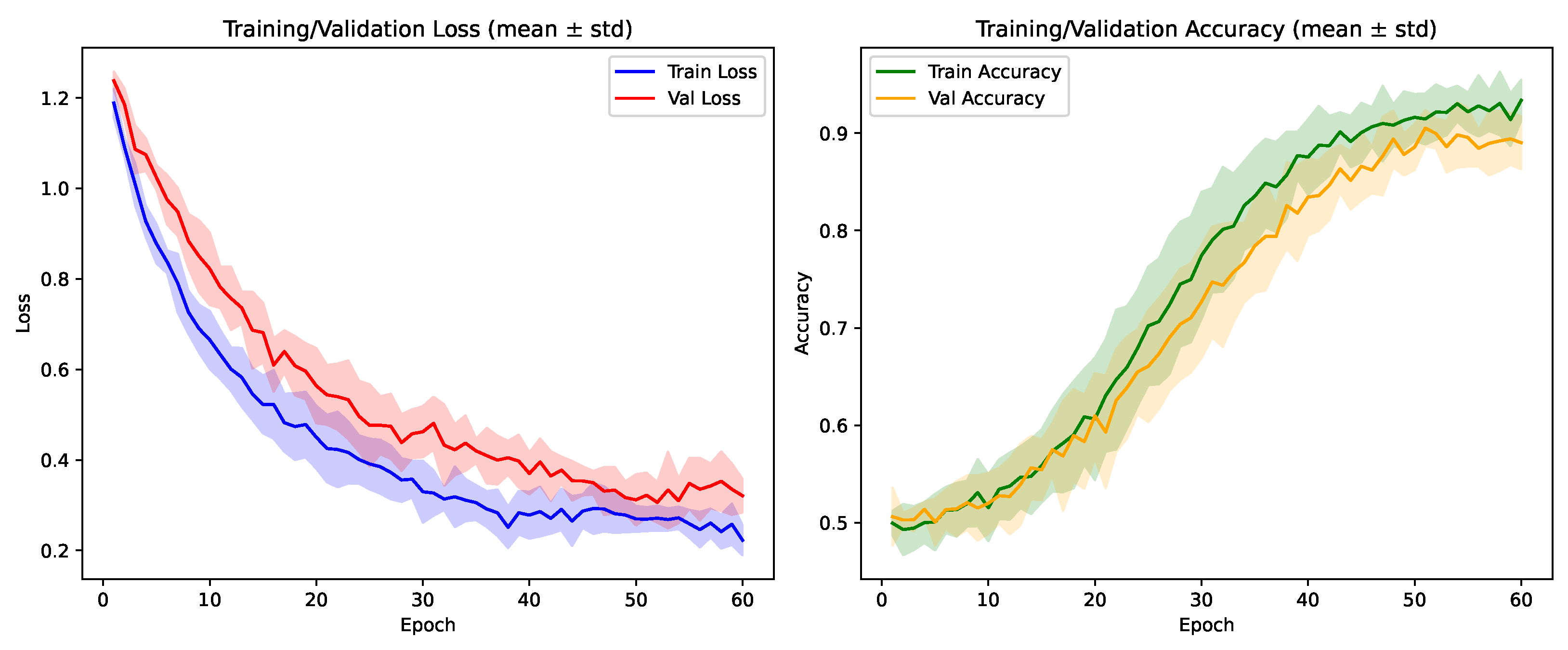
4.1. Experimental Setup
4.1.1. Hardware and Software Environment
4.1.2. Hyperparameters
4.1.3. Baseline Models
4.1.4. Evaluation Metrics
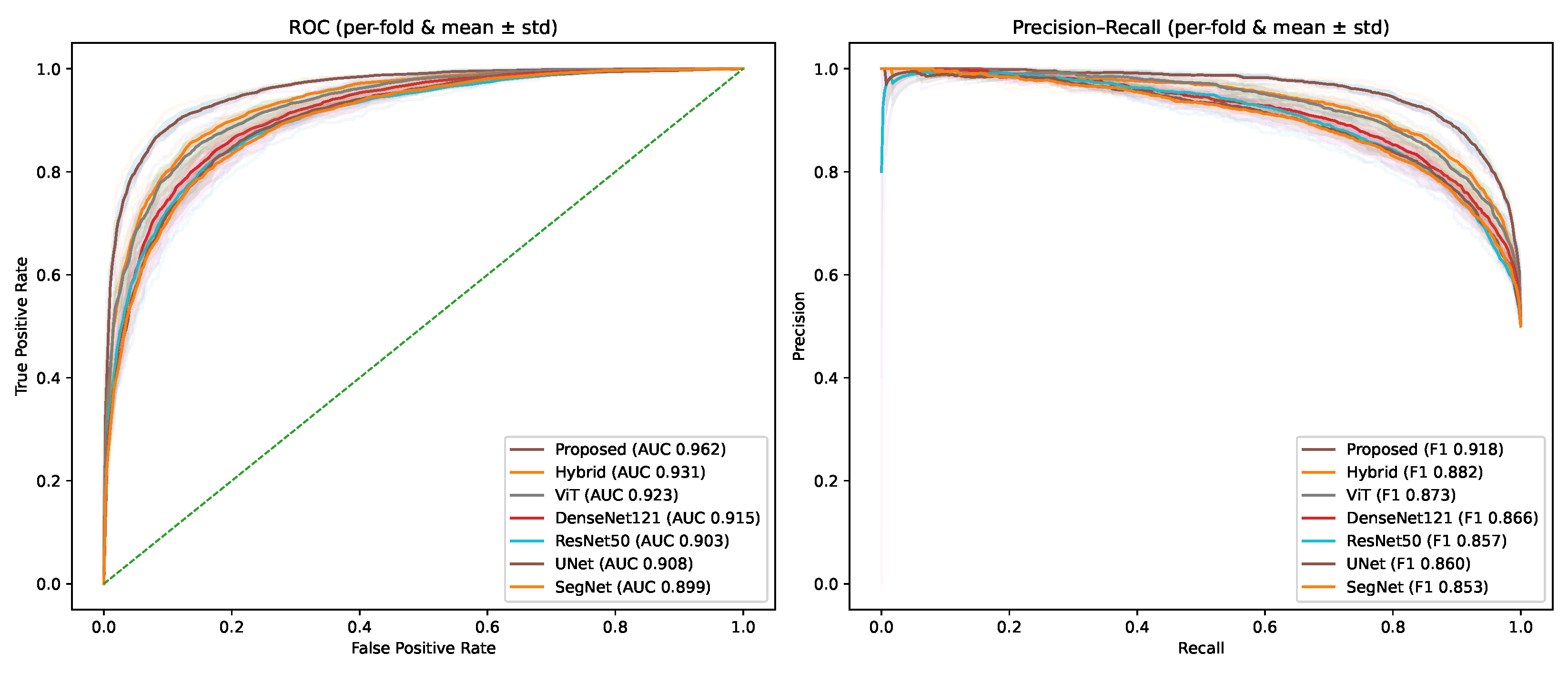
4.2. Performance Comparison of Different Baseline Models and the Proposed Method
4.3. Ablation Study on the Proposed Method
4.4. Cross-Validation Performance of the Proposed Method
4.5. Robustness Evaluation Under Realistic Imaging Perturbations
4.6. Subgroup Evaluation
4.7. Discussion
4.8. Limitation and Future Work
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Prasad, S.; Jha, M.K.; Sahu, S.; Bharat, I.; Sehgal, C. Evaluation of ovarian masses by color Doppler imaging and histopathological correlation. Int. J. Contemp. Med. Surg. Radiol. 2019, 4, 66. [Google Scholar] [CrossRef]
- Lin, X.; Wa, S.; Zhang, Y.; Ma, Q. A dilated segmentation network with the morphological correction method in farming area image Series. Remote Sens. 2022, 14, 1771. [Google Scholar] [CrossRef]
- Zhao, X.; Zhu, Q.; Wu, J. AResNet-ViT: A Hybrid CNN-Transformer Network for Benign and Malignant Breast Nodule Classification in Ultrasound Images. arXiv 2024, arXiv:2407.19316. [Google Scholar]
- Liu, X.; Gao, K.; Liu, B.; Pan, C.; Liang, K.; Yan, L.; Ma, J.; He, F.; Zhang, S.; Pan, S.; et al. Advances in deep learning-based medical image analysis. Health Data Sci. 2021, 2021, 8786793. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, S.; Ren, Y.; Zhang, Y.; Fu, J.; Fan, D.; Lin, J.; Wang, Q. Atrous Pyramid GAN Segmentation Network for Fish Images with High Performance. Electronics 2022, 11, 911. [Google Scholar] [CrossRef]
- Sebia, H.; Guyet, T.; Pereira, M.; Valdebenito, M.; Berry, H.; Vidal, B. Vascular segmentation of functional ultrasound images using deep learning. Comput. Biol. Med. 2025, 194, 110377. [Google Scholar] [CrossRef]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical image analysis using convolutional neural networks: A review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef]
- Qu, X.; Lu, H.; Tang, W.; Wang, S.; Zheng, D.; Hou, Y.; Jiang, J. A VGG attention vision transformer network for benign and malignant classification of breast ultrasound images. Med. Phys. 2022, 49, 5787–5798. [Google Scholar] [CrossRef]
- Sehgal, N. Efficacy of color doppler ultrasonography in differentiation of ovarian masses. J. Mid-Life Health 2019, 10, 22–28. [Google Scholar] [CrossRef]
- Dhir, Y.R.; Roy, A.; Maji, S.; Karim, R. Sonological Accuracy in Defining Various Benign and Malignant Ovarian Neoplasms with Colour Doppler and Histopathological Correlation. Int. J. Acad. Med. Pharm. 2023, 5, 2308–2311. [Google Scholar]
- Deckers, P.J.; Manning, R.; Laursen, T.; Worthy, S.; Kulkarni, S. The Clinical and Economic Impact of the Early Detection and Diagnosis of Cancer. Health L. Pol’y Brief 2020, 14, 1. [Google Scholar]
- Dicle, O. Artificial intelligence in diagnostic ultrasonography. Diagn. Interv. Radiol. 2023, 29, 40. [Google Scholar] [CrossRef] [PubMed]
- Lyu, H.; Fu, H.; Hu, X.; Liu, L. Esnet: Edge-based segmentation network for real-time semantic segmentation in traffic scenes. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; IEEE: New York, NY, USA, 2019; pp. 1855–1859. [Google Scholar]
- Aslam, M.A.; Naveed, A.; Ahmed, N. Hybrid Attention Network for Accurate Breast Tumor Segmentation in Ultrasound Images. arXiv 2025, arXiv:2506.16592. [Google Scholar]
- Sadeghi, M.H.; Sina, S.; Omidi, H.; Farshchitabrizi, A.H.; Alavi, M. Deep learning in ovarian cancer diagnosis: A comprehensive review of various imaging modalities. Pol. J. Radiol. 2024, 89, e30. [Google Scholar] [CrossRef]
- Jung, Y.; Kim, T.; Han, M.R.; Kim, S.; Kim, G.; Lee, S.; Choi, Y.J. Ovarian tumor diagnosis using deep convolutional neural networks and a denoising convolutional autoencoder. Sci. Rep. 2022, 12, 17024. [Google Scholar] [CrossRef]
- Mahale, N.; Kumar, N.; Mahale, A.; Ullal, S.; Fernandes, M.; Prabhu, S. Validity of ultrasound with color Doppler to differentiate between benign and malignant ovarian tumours. Obstet. Gynecol. Sci. 2024, 67, 227–234. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Ma, X. A new strategy for tuning ReLUs: Self-adaptive linear units (SALUs). In Proceedings of the ICMLCA 2021; 2nd International Conference on Machine Learning and Computer Application, Shenyang, China, 17–19 December 2021; VDE: Alzenau, Germany, 2021; pp. 1–8. [Google Scholar]
- Zhao, Y.; Li, X.; Zhou, C.; Peng, H.; Zheng, Z.; Chen, J.; Ding, W. A review of cancer data fusion methods based on deep learning. Inf. Fusion 2024, 108, 102361. [Google Scholar] [CrossRef]
- Chen, G.; Li, L.; Zhang, J.; Dai, Y. Rethinking the unpretentious U-net for medical ultrasound image segmentation. Pattern Recognit. 2023, 142, 109728. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, F.; Liang, H.; Bai, Y.; Wang, Z.; Zhao, L.; Ma, S.; Niu, Q.; Li, F.; Xie, T.; et al. CNN-based automatic segmentations and radiomics feature reliability on contrast-enhanced ultrasound images for renal tumors. Front. Oncol. 2023, 13, 1166988. [Google Scholar] [CrossRef]
- Yi, J.; Kang, H.K.; Kwon, J.H.; Kim, K.S.; Park, M.H.; Seong, Y.K.; Kim, D.W.; Ahn, B.; Ha, K.; Lee, J.; et al. Technology trends and applications of deep learning in ultrasonography: Image quality enhancement, diagnostic support, and improving workflow efficiency. Ultrasonography 2021, 40, 7–22. [Google Scholar] [CrossRef]
- Xiao, H.; Li, L.; Liu, Q.; Zhu, X.; Zhang, Q. Transformers in medical image segmentation: A review. Biomed. Signal Process. Control 2023, 84, 104791. [Google Scholar] [CrossRef]
- Chen, B.; Liu, Y.; Zhang, Z.; Lu, G.; Kong, A.W.K. Transattunet: Multi-level attention-guided u-net with transformer for medical image segmentation. IEEE Trans. Emerg. Top. Comput. Intell. 2023, 8, 55–68. [Google Scholar] [CrossRef]
- Azad, R.; Niggemeier, L.; Hüttemann, M.; Kazerouni, A.; Aghdam, E.K.; Velichko, Y.; Bagci, U.; Merhof, D. Beyond self-attention: Deformable large kernel attention for medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2024; pp. 1287–1297. [Google Scholar]
- Xie, Y.; Yang, B.; Guan, Q.; Zhang, J.; Wu, Q.; Xia, Y. Attention mechanisms in medical image segmentation: A survey. arXiv 2023, arXiv:2305.17937. [Google Scholar]
- Dai, Y.; Gao, Y.; Liu, F. Transmed: Transformers advance multi-modal medical image classification. Diagnostics 2021, 11, 1384. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Jin, Y.; Tao, K.; Xia, K.; Gu, J.; Yu, L.; Du, L.; Chen, C. MTS-Net: Dual-Enhanced Positional Multi-Head Self-Attention for 3D CT Diagnosis of May-Thurner Syndrome. arXiv 2024, arXiv:2406.04680. [Google Scholar]
- Liu, Z.; Lv, Q.; Yang, Z.; Li, Y.; Lee, C.H.; Shen, L. Recent progress in transformer-based medical image analysis. Comput. Biol. Med. 2023, 164, 107268. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.A.; Zaman, M.; Jawad, A.; Santamaria-Pang, A.; Lee, H.H.; Tarapov, I.; See, K.; Imran, M.S.; Roy, A.; Fallah, Y.P.; et al. WaveFormer: A 3D Transformer with Wavelet-Driven Feature Representation for Efficient Medical Image Segmentation. arXiv 2025, arXiv:2503.23764. [Google Scholar]
- Chetia, D.; Dutta, D.; Kalita, S.K. Image Segmentation with transformers: An Overview, Challenges and Future. arXiv 2025, arXiv:2501.09372. [Google Scholar]
- Huo, J.; Ouyang, X.; Ourselin, S.; Sparks, R. Generative medical segmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, Philadelphia, PA, USA, 25 February–4 March 2025; Volume 39, pp. 3851–3859. [Google Scholar]
- Li, J.; Xu, Q.; He, X.; Liu, Z.; Zhang, D.; Wang, R.; Qu, R.; Qiu, G. Cfformer: Cross cnn-transformer channel attention and spatial feature fusion for improved segmentation of low quality medical images. arXiv 2025, arXiv:2501.03629. [Google Scholar]
- Tran, P.N.; Truong Pham, N.; Dang, D.N.M.; Huh, E.N.; Hong, C.S. QTSeg: A Query Token-Based Architecture for Efficient 2D Medical Image Segmentation. arXiv 2024, arXiv:2412.17241. [Google Scholar]
- Al Kuwaiti, A.; Nazer, K.; Al-Reedy, A.; Al-Shehri, S.; Al-Muhanna, A.; Subbarayalu, A.V.; Al Muhanna, D.; Al-Muhanna, F.A. A review of the role of artificial intelligence in healthcare. J. Pers. Med. 2023, 13, 951. [Google Scholar] [CrossRef] [PubMed]
- Obuchowicz, R.; Strzelecki, M.; Piórkowski, A. Clinical applications of artificial intelligence in medical imaging and image processing—A review. Cancers 2024, 16, 1870. [Google Scholar] [CrossRef] [PubMed]
- Alnaggar, O.A.M.F.; Jagadale, B.N.; Saif, M.A.N.; Ghaleb, O.A.; Ahmed, A.A.; Aqlan, H.A.A.; Al-Ariki, H.D.E. Efficient artificial intelligence approaches for medical image processing in healthcare: Comprehensive review, taxonomy, and analysis. Artif. Intell. Rev. 2024, 57, 221. [Google Scholar] [CrossRef]
- Hunter, B.; Hindocha, S.; Lee, R.W. The role of artificial intelligence in early cancer diagnosis. Cancers 2022, 14, 1524. [Google Scholar] [CrossRef]
- Wu, H.; Lu, X.; Wang, H. The application of artificial intelligence in health care resource allocation before and during the COVID-19 pandemic: Scoping review. JMIR AI 2023, 2, e38397. [Google Scholar] [CrossRef]
- Hendrix, N.; Veenstra, D.L.; Cheng, M.; Anderson, N.C.; Verguet, S. Assessing the economic value of clinical artificial intelligence: Challenges and opportunities. Value Health 2022, 25, 331–339. [Google Scholar] [CrossRef]
- Gomez Rossi, J.; Feldberg, B.; Krois, J.; Schwendicke, F. Evaluation of the clinical, technical, and financial aspects of cost-effectiveness analysis of artificial intelligence in medicine: Scoping review and framework of analysis. JMIR Med. Inform. 2022, 10, e33703. [Google Scholar] [CrossRef]
- Reason, T.; Rawlinson, W.; Langham, J.; Gimblett, A.; Malcolm, B.; Klijn, S. Artificial intelligence to automate health economic modelling: A case study to evaluate the potential application of large language models. PharmacoEconomics-Open 2024, 8, 191–203. [Google Scholar] [CrossRef]
- Koonce, B. ResNet 50. In Convolutional Neural Networks with Swift for Tensorflow: Image Recognition and Dataset Categorization; Springer: Berlin/Heidelberg, Germany, 2021; pp. 63–72. [Google Scholar]
- Chhabra, G.S.; Verma, M.; Gupta, K.; Kondekar, A.; Choubey, S.; Choubey, A. Smart helmet using IoT for alcohol detection and location detection system. In Proceedings of the 2022 4th International Conference on Inventive Research in Computing Applications (ICIRCA), Coimbatore, India, 21–23 September 2022; IEEE: New York, NY, USA, 2022; pp. 436–440. [Google Scholar]
- Yuan, L.; Chen, Y.; Wang, T.; Yu, W.; Shi, Y.; Jiang, Z.H.; Tay, F.E.; Feng, J.; Yan, S. Tokens-to-token vit: Training vision transformers from scratch on imagenet. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 558–567. [Google Scholar]
- Wang, Y.; Qiu, Y.; Cheng, P.; Zhang, J. Hybrid CNN-transformer features for visual place recognition. IEEE Trans. Circuits Syst. Video Technol. 2022, 33, 1109–1122. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]




| Category | Surgical Approach | Number of Cases |
|---|---|---|
| Benign ovarian tumors | Umbilical + 1-port laparoscopy | 426 |
| Malignant ovarian tumors | Laparotomy | 394 |
| Model | Accuracy | Sensitivity | Specificity | AUC | F1-Score |
|---|---|---|---|---|---|
| ResNet-50 [43] | 0.861 | 0.842 | 0.876 | 0.903 [0.887–0.918] | 0.857 |
| DenseNet-121 [44] | 0.872 | 0.856 | 0.884 | 0.915 [0.900–0.929] | 0.866 |
| ViT [45] | 0.884 | 0.867 | 0.893 | 0.923 [0.909–0.937] | 0.873 |
| Hybrid CNN–Transformer [46] | 0.891 | 0.874 | 0.902 | 0.931 [0.917–0.944] | 0.882 |
| U-Net [47] | 0.865 | 0.849 | 0.878 | 0.908 [0.892–0.923] | 0.860 |
| SegNet [48] | 0.858 | 0.841 | 0.871 | 0.899 [0.883–0.915] | 0.853 |
| Proposed (GAM + LEM + FD) | 0.923 | 0.911 | 0.934 | 0.962 [0.950–0.973] | 0.918 |
| Variant | Accuracy | Sensitivity | Specificity | AUC | F1-Score | FLOPs (G) |
|---|---|---|---|---|---|---|
| Without GAM | 0.897 | 0.881 | 0.906 | 0.941 | 0.890 | 47.1 |
| Without LEM | 0.889 | 0.870 | 0.902 | 0.935 | 0.884 | 45.8 |
| Without FD | 0.901 | 0.886 | 0.910 | 0.944 | 0.895 | 46.5 |
| GAM + LEM only | 0.913 | 0.898 | 0.922 | 0.951 | 0.907 | 49.2 |
| LEM + FD only | 0.908 | 0.892 | 0.918 | 0.948 | 0.902 | 48.7 |
| GAM + FD only | 0.911 | 0.896 | 0.920 | 0.949 | 0.905 | 48.9 |
| LEM → SE | 0.902 | 0.887 | 0.913 | 0.944 | 0.897 | 45.9 |
| LEM → CBAM | 0.906 | 0.890 | 0.916 | 0.947 | 0.900 | 46.2 |
| FD → Concat+MLP | 0.904 | 0.889 | 0.914 | 0.945 | 0.898 | 46.0 |
| FD → Concat+Self-Attention | 0.910 | 0.894 | 0.919 | 0.949 | 0.904 | 46.4 |
| GAM → Swin window attention | 0.907 | 0.891 | 0.915 | 0.946 | 0.901 | 47.0 |
| GAM → Linformer attention | 0.910 | 0.893 | 0.917 | 0.948 | 0.903 | 46.8 |
| GAM → Deformable attention | 0.912 | 0.895 | 0.919 | 0.949 | 0.905 | 47.3 |
| Full model (GAM + LEM + FD) | 0.923 | 0.911 | 0.934 | 0.962 | 0.918 | 49.5 |
| Fold | Accuracy | Sensitivity | Specificity | AUC | F1-Score |
|---|---|---|---|---|---|
| Fold 1 | 0.918 | 0.905 | 0.929 | 0.958 | 0.914 |
| Fold 2 | 0.922 | 0.910 | 0.933 | 0.960 | 0.917 |
| Fold 3 | 0.926 | 0.913 | 0.937 | 0.964 | 0.920 |
| Fold 4 | 0.919 | 0.907 | 0.931 | 0.959 | 0.915 |
| Fold 5 | 0.925 | 0.912 | 0.936 | 0.963 | 0.919 |
| Average | 0.922 | 0.909 | 0.933 | 0.961 | 0.917 |
| Clean | mAUC (↓AUC) | mSensitivity (↓Sens) | |||||
|---|---|---|---|---|---|---|---|
| Model | AUC/Sens | Rician | Motion | Gibbs | Rician | Motion | Gibbs |
| ResNet-50 | 0.903/0.842 | 0.872 (↓0.031) | 0.881 (↓0.022) | 0.876 (↓0.027) | 0.804 (↓0.038) | 0.812 (↓0.030) | 0.808 (↓0.034) |
| ViT | 0.923/0.867 | 0.901 (↓0.022) | 0.907 (↓0.016) | 0.903 (↓0.020) | 0.836 (↓0.031) | 0.842 (↓0.025) | 0.838 (↓0.029) |
| Hybrid CNN-Transf. | 0.931/0.874 | 0.909 (↓0.022) | 0.914 (↓0.017) | 0.911 (↓0.020) | 0.845 (↓0.029) | 0.850 (↓0.024) | 0.847 (↓0.027) |
| Proposed (GAM + LEM + FD) | 0.962/0.911 | 0.944 (↓0.018) | 0.949 (↓0.013) | 0.946 (↓0.016) | 0.892 (↓0.019) | 0.897 (↓0.014) | 0.894 (↓0.017) |
| Subgroup | ResNet-50 AUC | Hybrid CNN-Transformer AUC | Proposed AUC | Proposed Sensitivity |
|---|---|---|---|---|
| Small lesions (<1 cm) | 0.881 | 0.902 | 0.952 | 0.911 |
| Large lesions (≥1 cm) | 0.907 | 0.931 | 0.968 | 0.932 |
| Age <40 years | 0.889 | 0.915 | 0.956 | 0.919 |
| Age ≥60 years | 0.872 | 0.910 | 0.949 | 0.903 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, D.; He, X.; Zhang, R.; Zhang, Y.; Li, M.; Zhan, Y. Hybrid CNN–Transformer with Fusion Discriminator for Ovarian Tumor Ultrasound Imaging Classification. Electronics 2025, 14, 4040. https://doi.org/10.3390/electronics14204040
Xu D, He X, Zhang R, Zhang Y, Li M, Zhan Y. Hybrid CNN–Transformer with Fusion Discriminator for Ovarian Tumor Ultrasound Imaging Classification. Electronics. 2025; 14(20):4040. https://doi.org/10.3390/electronics14204040
Chicago/Turabian StyleXu, Donglei, Xinyi He, Ruoyun Zhang, Yinuo Zhang, Manzhou Li, and Yan Zhan. 2025. "Hybrid CNN–Transformer with Fusion Discriminator for Ovarian Tumor Ultrasound Imaging Classification" Electronics 14, no. 20: 4040. https://doi.org/10.3390/electronics14204040
APA StyleXu, D., He, X., Zhang, R., Zhang, Y., Li, M., & Zhan, Y. (2025). Hybrid CNN–Transformer with Fusion Discriminator for Ovarian Tumor Ultrasound Imaging Classification. Electronics, 14(20), 4040. https://doi.org/10.3390/electronics14204040





