Evaluating Retrieval-Augmented Generation Variants for Clinical Decision Support: Hallucination Mitigation and Secure On-Premises Deployment
Abstract
1. Introduction
2. Related Work
2.1. Evaluation Metrics and Benchmark Datasets
2.2. Privacy-Preserving and On-Premises Retrieval
2.3. RAG Toolkit Ecosystem
2.4. Applications of RAG in Medical Data Analysis
3. Materials and Methods
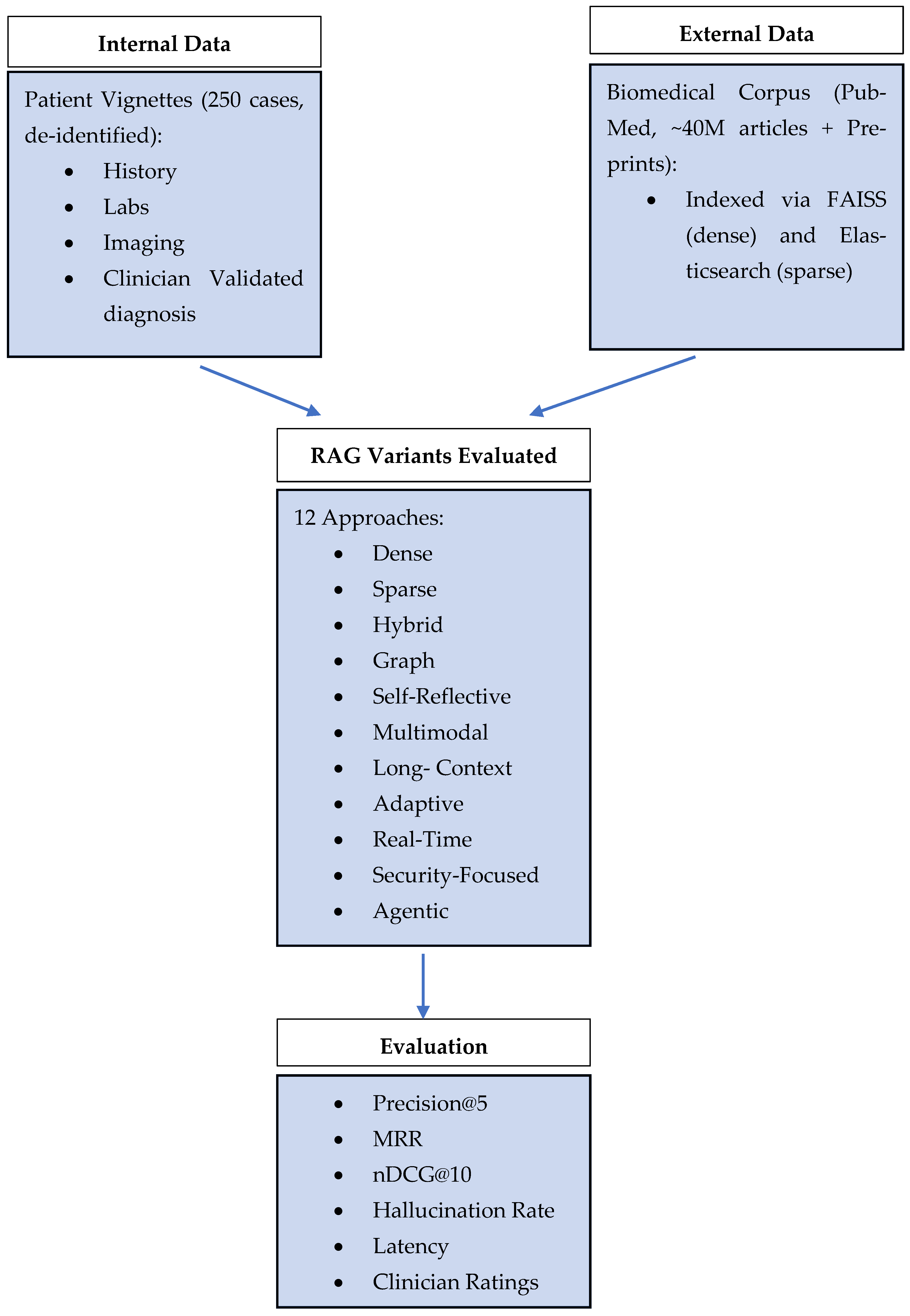
3.1. Data Sources
3.2. Experimental Setup Architecture
3.2.1. Dense Retrieval (Vector Search)
3.2.2. Keyword Search (Sparse Retrieval)
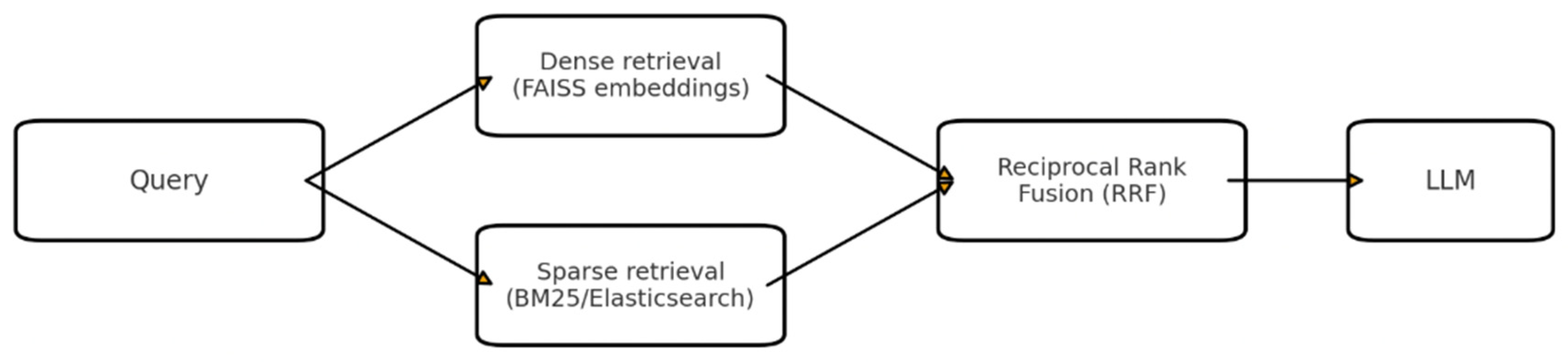
3.2.3. Hybrid Retrieval with Reciprocal Rank Fusion (RRF)
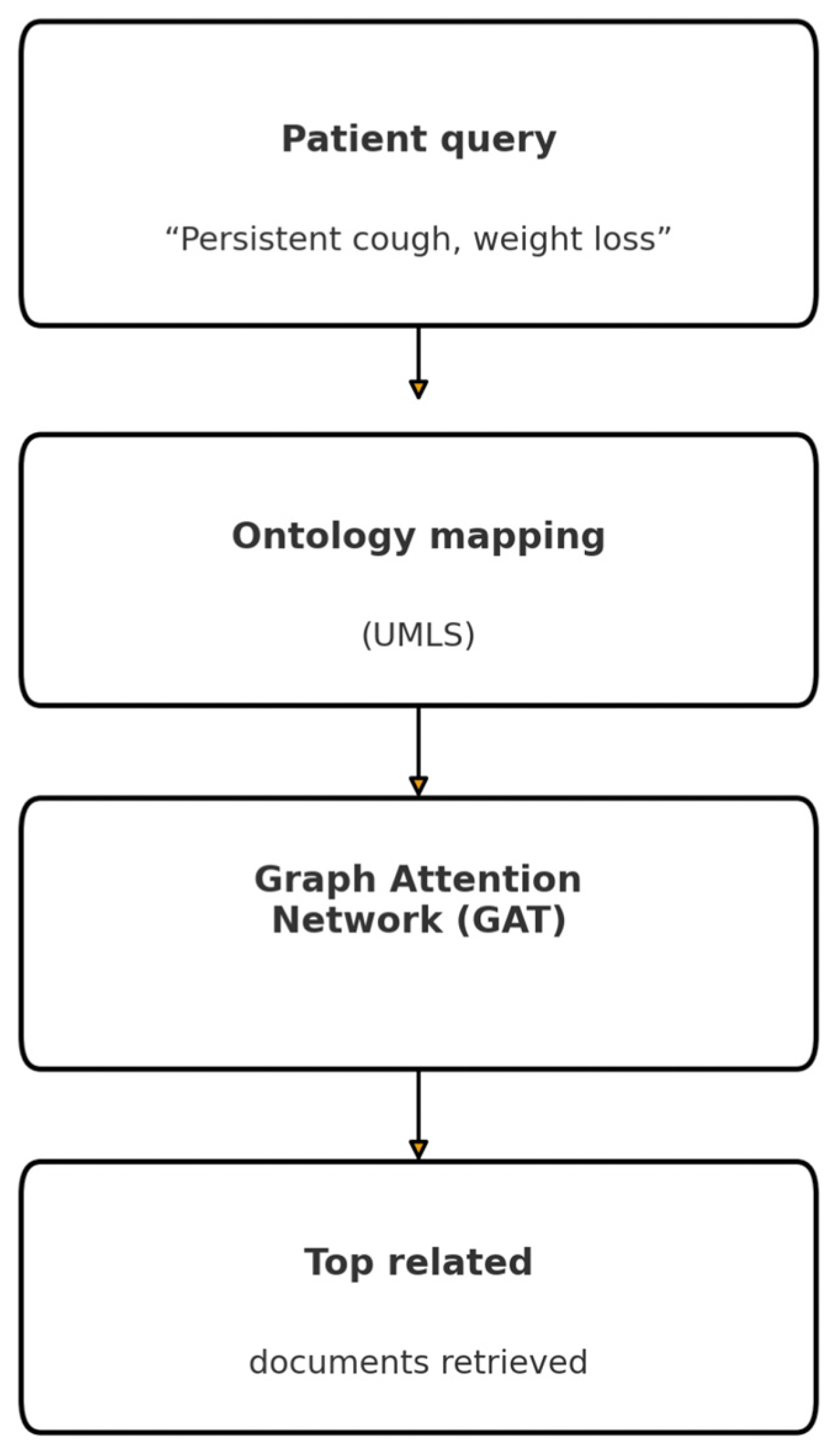
3.2.4. Graph-Based RAG (GraphRAG)
3.2.5. Long-Context Retrieval (Long RAG)
3.2.6. Adaptive Retrieval
3.2.7. Multimodal RAG
3.2.8. Self-Reflective RAG (SELF-RAG)
3.2.9. Real-Time Knowledge Integration (CRAG)
3.2.10. Security-Focused Retrieval (TrustRAG)
3.2.11. Agentic RAG
3.2.12. Haystack Pipeline and Re-Ranking
3.3. Comparison Table for RAG Variants
3.4. Hallucination Reduction Strategies and Evaluation Protocol
3.5. Generalizability to Other Datasets
3.6. Overall Framerwork Architechture
4. Results
4.1. Comparison Table
4.2. Limitations
5. Discussion
6. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lewis, P.; Pérez, E.; Piktus, A.; Petroni, F.; Karpukhin, V.; Goyal, N.; Kuttler, H.; Lewis, M.; Yih, W.-T.; Riedel, S.; et al. Retrieval-augmented generation for knowledge-intensive NLP tasks. Adv. Neural Inf. Process. Syst. 2020, 33, 9459–9474. [Google Scholar]
- Xiong, G.; Jin, Q.; Lu, Z.; Zhang, A. Benchmarking retrieval-augmented generation for medicine. In Findings of the Association for Computational Linguistics; ACL: Stroudsburg, PA, USA, 2024; pp. 6233–6251. [Google Scholar] [CrossRef]
- Tan, Y.; Nguyen, P.; Kumar, A. LlamaIndex v0.6: Enhancements for biomedical document retrieval. In Proceedings of the 2025 International Conference on Computational Linguistics, Abu Dhabi, United Arab Emirates, 19–24 January 2025; pp. 1234–1245. [Google Scholar]
- Lahiri, A.K.; Hu, Q.V. Alzheimerrag: Multimodal retrieval augmented generation for pubmed articles. arXiv 2024, arXiv:2412.16701. [Google Scholar]
- Wang, J.; Yang, Z.; Yao, Z.; Yu, H. JMLR: Joint medical LLM and retrieval training for enhancing reasoning and professional question answering capability. arXiv 2024, arXiv:2402.17887. [Google Scholar] [CrossRef]
- Weng, Y.; Zhu, F.; Ye, T.; Liu, H.; Feng, F.; Chua, T.S. Optimizing knowledge integration in retrieval-augmented generation with self-selection. arXiv 2025, arXiv:2502.06148. [Google Scholar] [CrossRef]
- Neha, F.; Bhati, D.; Shukla, D.K. Retrieval-Augmented Generation (RAG) in Healthcare: A Comprehensive Review. AI 2025, 6, 226. [Google Scholar] [CrossRef]
- Sun, Z.; Zang, X.; Zheng, K.; Song, Y.; Xu, J.; Zhang, X.; Yu, W.; Song, Y.; Li, H. ReDeEP: Detecting hallucination in retrieval-augmented generation via mechanistic interpretability. arXiv 2024, arXiv:2410.11414. [Google Scholar]
- Islam Tonmoy, S.M.T.; Zaman, S.M.M.; Jain, V.; Rani, A.; Rawte, V.; Chadha, A.; Das, A. A comprehensive survey of hallucination mitigation techniques in large language models. arXiv 2024, arXiv:2401.01313. [Google Scholar] [CrossRef]
- Zhou, H.; Lee, K.H.; Zhan, Z.; Chen, Y.; Li, Z. Trustrag: Enhancing robustness and trustworthiness in rag. arXiv 2025, arXiv:2501.00879. [Google Scholar]
- Mala, C.S.; Gezici, G.; Giannotti, F. Hybrid retrieval for hallucination mitigation in large language models: A comparative analysis. arXiv 2025, arXiv:2504.05324. [Google Scholar]
- Frihat, S.; Fuhr, N. Integration of biomedical concepts for enhanced medical literature retrieval. Int. J. Data Sci. Anal. 2025, 20, 4409–4422. [Google Scholar] [CrossRef]
- Li, J.; Deng, Y.; Sun, Q.; Zhu, J.; Tian, Y.; Li, J.; Zhu, T. Benchmarking large language models in evidence-based medicine. IEEE J. Biomed. Health Inform. 2024, 29, 6143–6156. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, N.; Qin, J.; Ye, G.; Tang, R. A Multi-granularity Concept Sparse Activation and Hierarchical Knowledge Graph Fusion Framework for Rare Disease Diagnosis. arXiv 2025, arXiv:2507.08529. [Google Scholar] [CrossRef]
- Vasantharajan, C. SciRAG: A Retrieval-Focused Fine-Tuning Strategy for Scientific Documents. Ph.D. Thesis, McMaster University, Hamilton, ON, Canada, 2025. [Google Scholar]
- Li, Q.; Liu, H.; Guo, C.; Gao, C.; Chen, D.; Wang, M.; Gu, J. Reviewing Clinical Knowledge in Medical Large Language Models: Training and Beyond. arXiv 2025, arXiv:2502.20988. [Google Scholar] [CrossRef]
- Wu, J.; Zhu, J.; Qi, Y.; Chen, J.; Xu, M.; Menolascina, F.; Grau, V. Medical graph rag: Towards safe medical large language model via graph retrieval-augmented generation. arXiv 2024, arXiv:2408.04187. [Google Scholar] [CrossRef]
- Xu, R.; Jiang, P.; Luo, L.; Xiao, C.; Cross, A.; Pan, S.; Yang, C. A survey on unifying large language models and knowledge graphs for biomedicine and healthcare. In Proceedings of the 31st ACM SIGKDD Conference on Knowledge Discovery and Data Mining, Toronto, ON, Canada, 3–7 August 2025; Volume 2, pp. 6195–6205. [Google Scholar]
- Hamed, A.A.; Crimi, A.; Misiak, M.M.; Lee, B.S. From knowledge generation to knowledge verification: Examining the biomedical generative capabilities of ChatGPT. iScience 2025, 28, 112492. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yu, W.; Ma, W.; Zhong, W.; Feng, Z.; Wang, H.; Liu, T. A survey on hallucination in large language models: Principles, taxonomy, challenges, and open questions. ACM Trans. Inf. Syst. 2025, 43, 42. [Google Scholar] [CrossRef]
- Soni, S.; Roberts, K. Evaluation of dataset selection for pre-training and fine-tuning transformer language models for clinical question answering. In Proceedings of the Twelfth Language Resources and Evaluation Conference, Marseille, France, 11–16 May 2020; pp. 5532–5538. [Google Scholar]
- Yan, L.K.; Niu, Q.; Li, M.; Zhang, Y.; Yin, C.H.; Fei, C.; Liu, J. Large language model benchmarks in medical tasks. arXiv 2024, arXiv:2410.21348. [Google Scholar] [CrossRef]
- Amatriain, X. Measuring and mitigating hallucinations in large language models: Amultifaceted approach. Preprint 2024.
- Amugongo, L.M.; Mascheroni, P.; Brooks, S.; Doering, S.; Seidel, J. Retrieval augmented generation for large language models in healthcare: A systematic review. PLoS Digit. Health 2025, 4, e0000877. [Google Scholar] [CrossRef]
- Henderson, J.; Pearson, M. Privacy-Preserving Natural Language Processing for Clinical Notes. Preprint 2025.
- Chen, Y.; Nyemba, S.; Malin, B. Auditing medical records accesses via healthcare interaction networks. AMIA Annu. Symp. Proc. AMIA Symp. 2012, 2012, 93–102. [Google Scholar] [PubMed]
- Zhao, D. Frag: Toward federated vector database management for collaborative and secure retrieval-augmented generation. arXiv 2024, arXiv:2410.13272. [Google Scholar] [CrossRef]
- Elizarov, O. Architecture of Applications Powered by Large Language Models. Master’s Thesis, Metropolia University of Applied Sciences, Helsinki, Finland, 2024. [Google Scholar]
- Zhong, L.; Wu, J.; Li, Q.; Peng, H.; Wu, X. A comprehensive survey on automatic knowledge graph construction. ACM Comput. Surv. 2023, 56, 94. [Google Scholar] [CrossRef]
- Wang, J.; Ashraf, T.; Han, Z.; Laaksonen, J.; Anwer, R.M. MIRA: A Novel Framework for Fusing Modalities in Medical RAG. arXiv 2025, arXiv:2507.07902. [Google Scholar] [CrossRef]
- Jiang, E.; Chen, A.; Tenison, I.; Kagal, L. MediRAG: Secure Question Answering for Healthcare Data. In Proceedings of the 2024 IEEE International Conference on Big Data (BigData), IEEE, Washington, DC, USA, 15–18 December 2024; pp. 6476–6485. [Google Scholar]
- Wadden, D.; Lo, K.; Kuehl, B.; Cohan, A.; Beltagy, I.; Wang, L.L.; Hajishirzi, H. SciFact-open: Towards open-domain scientific claim verification. arXiv 2022, arXiv:2210.13777. [Google Scholar]
- Benoit, J.R. ChatGPT for clinical vignette generation, revision, and evaluation. MedRxiv 2023. [Google Scholar]
- Wright, A.; Phansalkar, S.; Bloomrosen, M.; Jenders, R.A.; Bobb, A.M.; Halamka, J.D.; Kuperman, G.; Payne, T.H.; Teasdale, S.; Vaida, A.J.; et al. Best Practices in Clinical Decision Support: The Case of Preventive Care Reminders. Appl. Clin. Inform. 2010, 1, 331–345. [Google Scholar] [CrossRef]
- Douze, M.; Guzhva, A.; Deng, C.; Johnson, J.; Szilvasy, G.; Mazaré, P.E.; Jégou, H. The faiss library. arXiv 2024, arXiv:2401.08281. [Google Scholar] [CrossRef]
- Gormley, C.; Tong, Z. Elasticsearch: The Definitive Guide, 8th ed.; O’Reilly Media: Sebastopol, CA, USA, 2025. [Google Scholar]
- Rothman, D. RAG-Driven Generative AI: Build Custom Retrieval Augmented Generation Pipelines with LlamaIndex, Deep Lake, and Pinecone; Packt Publishing Ltd.: Birmingham, UK, 2024. [Google Scholar]
- Kennedy, R.K.; Khoshgoftaar, T.M. Accelerated deep learning on HPCC systems. In Proceedings of the 2020 19th IEEE International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 14–17 December 2020; pp. 847–852. [Google Scholar]
- Python Software Foundation. Python Language Reference. version 3.10, Python Software Foundation: Wilmington, DE, USA, 2024. Available online: https://www.python.org (accessed on 15 September 2025).
- Ueda, A.; Santos, R.L.; Macdonald, C.; Ounis, I. Structured fine-tuning of contextual embeddings for effective biomedical retrieval. In Proceedings of the 44th International ACM SIGIR Conference on Research and Development in Information Retrieval, Virtual, 11–15 July 2021; pp. 2031–2035. [Google Scholar]
- Robinson, I.; Webber, J.; Eifrem, E. Graph Databases, 3rd ed.; O’Reilly Media: Sebastopol, CA, USA, 2024. [Google Scholar]
- Ramírez, S. Fastapi Framework, High Performance, Easy to Learn, Fast to Code, Ready for Production; GitHub: Berlin, Germany, 2022; Available online: https://github.com/fastapi/fastapi (accessed on 15 September 2025).
- Reimers, N.; Gurevych, I. Sentence-BERT: Sentence embeddings using Siamese BERT-networks. In Proceedings of the 16th Conference of the European Chapter of the Association for Computational Linguistics, Virtual, 16 April 2021; pp. 33–45. [Google Scholar]
- Goli, R.; Moffat, A.; Buchanan, G. Refined Medical Search via Dense Retrieval and User Interaction. In Proceedings of the 48th International ACM SIGIR Conference on Research and Development in Information Retrieval, Padua, Italy, 13–18 July 2025; pp. 3315–3324. [Google Scholar]
- Khatoon, T.; Govardhan, A. Query Expansion with Enhanced-BM25 Approach for Improving the Search Query Performance on Clustered Biomedical Literature Retrieval. J. Digit. Inf. Manag. 2018, 16, 2. [Google Scholar]
- Zhang, Z. An improved BM25 algorithm for clinical decision support in Precision Medicine based on co-word analysis and Cuckoo Search. BMC Med. Inform. Decis. Mak. 2021, 21, 81. [Google Scholar] [CrossRef]
- Bruch, S.; Gai, S.; Ingber, A. An analysis of fusion functions for hybrid retrieval. ACM Trans. Inf. Syst. 2023, 42, 20. [Google Scholar] [CrossRef]
- Zhao, Q.; Kang, Y.; Li, J.; Wang, D. Exploiting the semantic graph for the representation and retrieval of medical documents. Comput. Biol. Med. 2018, 101, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ma, X.; Zhu, Y.; Zhao, Z.; Wang, S.; Yin, D.; Dou, Z. Sliding windows are not the end: Exploring full ranking with long-context large language models. arXiv 2024, arXiv:2412.14574. [Google Scholar] [CrossRef]
- Lim, W.; Li, Z.; Kim, G.; Ji, S.; Kim, H.; Choi, K.; Wang, W.Y. MacRAG: Compress, Slice, and Scale-up for Multi-Scale Adaptive Context RAG. arXiv 2025, arXiv:2505.06569. [Google Scholar]
- Ji, Z.; Yu, T.; Xu, Y.; Lee, N.; Ishii, E.; Fung, P. Towards mitigating LLM hallucination via self reflection. In Findings of the Association for Computational Linguistics: EMNLP 2023; ACL: Stroudsburg, PA, USA, 2023; pp. 1827–1843. [Google Scholar]
- Akesson, S.; Santos, F.A. Clustered Retrieved Augmented Generation (CRAG). arXiv 2024, arXiv:2406.00029. [Google Scholar]
- Ozaki, S.; Kato, Y.; Feng, S.; Tomita, M.; Hayashi, K.; Hashimoto, W.; Watanabe, T. Understanding the impact of confidence in retrieval augmented generation: A case study in the medical domain. arXiv 2024, arXiv:2412.20309. [Google Scholar] [CrossRef]
- Low, Y.S.; Jackson, M.L.; Hyde, R.J.; Brown, R.E.; Sanghavi, N.M.; Baldwin, J.D.; Gombar, S. Answering real-world clinical questions using large language model, retrieval-augmented generation, and agentic systems. Digit. Health 2025, 11, 20552076251348850. [Google Scholar] [CrossRef]
- Bhattarai, K. Improving Clinical Information Extraction from Electronic Health Records: Leveraging Large Language Models and Evaluating Their Outputs. Ph.D. Thesis, Washington University in St. Louis, St. Louis, MO, USA, 2024. [Google Scholar]
- Wei, J.; Wang, X.; Schuurmans, D.; Bosma, M.; Ichter, B.; Xia, F.; Chi, E.; Le, Q.; Zhou, D. Chain of thought prompting elicits reasoning in large language models. Trans. Assoc. Comput. Linguist. 2023, 11, 123–140. Available online: https://proceedings.neurips.cc/paper/2022/hash/9d5609613524ecf4f15af0f7b31abca4-Abstract-Conference.html (accessed on 15 September 2025).
- Milanés-Hermosilla, D.; Trujillo, C.R.; López-Baracaldo, R.; Sagaró-Zamora, R.; Delisle-Rodriguez, D.; Villarejo-Mayor, J.J.; Nunez-Alvarez, J.R. Monte Carlo dropout for uncertainty estimation and motor imagery classification. Sensors 2021, 21, 7241. [Google Scholar] [CrossRef]
- Saigaonkar, S.; Narawade, V. Domain adaptation of transformer-based neural network model for clinical note classification in Indian healthcare. Int. J. Inf. Technol. 2024, 1–19. [Google Scholar] [CrossRef]
- Finkelstein, J.; Gabriel, A.; Schmer, S.; Truong, T.T.; Dunn, A. Identifying facilitators and barriers to implementation of AI-assisted clinical decision support in an electronic health record system. J. Med. Syst. 2024, 48, 89. [Google Scholar] [CrossRef]
- Wang, J.; Deng, H.; Liu, B.; Hu, A.; Liang, J.; Fan, L.; Lei, J. Systematic evaluation of research progress on natural language processing in medicine over the past 20 years: Bibliometric study on PubMed. J. Med. Internet Res. 2020, 22, e16816. [Google Scholar] [CrossRef]
- Jebb, A.T.; Ng, V.; Tay, L. A review of key Likert scale development advances: 1995–2019. Front. Psychol. 2021, 12, 637547. [Google Scholar] [CrossRef]
- Chow, R.; Zimmermann, C.; Bruera, E.; Temel, J.; Im, J.; Lock, M. Inter-rater reliability in performance status assessment between clinicians and patients: A systematic review and meta-analysis. BMJ Support. Palliat. Care 2020, 10, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.; Culpepper, J.S.; Blanco, R.; Crane, M.; Clarke, C.L.; Lin, J. Efficient and Effective Tail Latency Minimization in Multi-Stage Retrieval Systems. arXiv 2017, arXiv:1704.03970. [Google Scholar] [CrossRef]
- Kharitonova, K.; Pérez-Fernández, D.; Gutiérrez-Hernando, J.; Gutiérrez-Fandiño, A.; Callejas, Z.; Griol, D. Leveraging Retrieval-Augmented Generation for Reliable Medical Question Answering Using Large Language Models. In International Conference on Hybrid Artificial Intelligence Systems; Springer Nature: Cham, Switzerland, 2024; pp. 141–153. [Google Scholar]
- Ahire, P.R.; Hanchate, R.; Kalaiselvi, K. Optimized Data Retrieval and Data Storage for Healthcare Applications. In Predictive Data Modelling for Biomedical Data and Imaging; River Publishers: Gistrup, Denmark, 2024; pp. 107–126. [Google Scholar]
- Paulson, D.; Cannon, B. Auditing and Logging Systems for Privacy Assurance in Medical AI Pipelines. Preprint 2025.
- Rojas, J.C.; Teran, M.; Umscheid, C.A. Clinician trust in artificial intelligence: What is known and how trust can be facilitated. Crit. Care Clin. 2023, 39, 769–782. [Google Scholar] [CrossRef]
- Atf, Z.; Safavi-Naini, S.A.A.; Lewis, P.R.; Mahjoubfar, A.; Naderi, N.; Savage, T.R.; Soroush, A. The challenge of uncertainty quantification of large language models in medicine. arXiv 2025, arXiv:2504.05278. [Google Scholar] [CrossRef]
- Wang, G.; Ran, J.; Tang, R.; Chang, C.Y.; Chuang, Y.N.; Liu, Z.; Hu, X. Assessing and enhancing large language models in rare disease question-answering. arXiv 2024, arXiv:2408.08422. [Google Scholar] [CrossRef]
- Neehal, N.; Wang, B.; Debopadhaya, S.; Dan, S.; Murugesan, K.; Anand, V.; Bennett, K.P. CTBench: A comprehensive benchmark for evaluating language model capabilities in clinical trial design. arXiv 2024, arXiv:2406.17888. [Google Scholar] [CrossRef]
- Ting, L.P.Y.; Zhao, C.; Zeng, Y.H.; Lim, Y.J.; Chuang, K.T. Beyond RAG: Reinforced Reasoning Augmented Generation for Clinical Notes. arXiv 2025, arXiv:2506.05386. [Google Scholar]
- Rector, A.; Minor, K.; Minor, K.; McCormack, J.; Breeden, B.; Nowers, R.; Dorris, J. Validating Pharmacogenomics Generative Artificial Intelligence Query Prompts Using Retrieval-Augmented Generation (RAG). arXiv 2025, arXiv:2507.21453. [Google Scholar] [CrossRef]


| RAG Variant | Clinical Challenges Addressed |
|---|---|
| Dense Retrieval | Captures semantic similarity, useful for rare diseases where terminology varies. |
| Sparse Retrieval (BM25) | Provides rapid responses, valuable in time-sensitive situations like emergency care. |
| Hybrid Retrieval (RRF) | Balances precision and recall by combining dense and sparse retrieval. |
| Graph-Based RAG | Leverages medical ontologies to reason across related conditions and linked entities. |
| Long-Context RAG | Handles extended patient histories and complex longitudinal records. |
| Adaptive Retrieval | Dynamically adjusts strategy when initial confidence is low, reducing unsupported outputs. |
| Self-Reflective RAG | Minimizes hallucinations through iterative critique and refinement of responses. |
| Multimodal RAG | Integrates imaging data with clinical text for real-world diagnostic workflows. |
| Security-Focused RAG (TrustRAG) | Ensures HIPAA/GDPR compliance with encryption, provenance, and audit trails. |
| Agentic RAG | Decomposes complex queries into steps, mirroring clinician reasoning processes. |
| Haystack Pipeline (DPR + BM25 + Cross-Encoder) | Achieves state-of-the-art retrieval accuracy by combining multiple retrieval signals. |
| Study/Variant | Application Area | Advantages | Limitations |
|---|---|---|---|
| Xiong et al. (2024) [2] | Benchmarking RAG for PubMedQA and MedMCQA | Improved factual accuracy (+10% vs. baseline) | Evaluated only on benchmark datasets, not clinical use |
| Wu et al. (2024)—Medical GraphRAG [17] | Medical document retrieval via knowledge graphs | Supports multi-hop reasoning with ontology grounding and cited sources | Requires high-quality, up-to-date KGs; scalability and maintenance overhead |
| Wang et al. (2025)—MIRA: Multimodal Medical RAG [30] | Clinical decision support combining text and imaging | Fuses modalities to ground answers in multimodal evidence; improved diagnostic support in controlled evaluations | Higher compute and latency; performance sensitive to image–text representations |
| Li et al. (2024)—Benchmarking LLMs in Evidence-Based Medicine [13] | Clinical RAG evaluation and evidence-based assessment | Clearer evaluation protocols for EBM tasks; emphasis on citation quality and evidence grounding | Hallucination and evidence-quality gaps persist across models |
| Jiang et al. (2024)—MediRAG: Secure QA for Healthcare Data [31] | Privacy-preserving clinical retrieval and QA | Security-aware RAG design and governance suitable for clinical settings | Additional complexity and latency for secure deployments |
| Wadden et al. (2022)—SciFact-Open (Scientific Claim Verification) [32] | Biomedical fact verification for RAG outputs | Automated claim checking reduces unsupported statements in scientific/biomedical answers | Coverage limited to annotated datasets; domain shift can degrade performance |
| Method | Retrieval Type/Setup | Distinctive Feature | Clinical Relevance |
|---|---|---|---|
| Dense Retrieval | BioMed-RoBERTa encoder → 768-dim embeddings, indexed in FAISS (IVFFlat, 8-bit quantization). | Semantic similarity search. Adaptive n-probe scheduling. | Captures meaning beyond keywords, useful for rare/variably described diseases. |
| Sparse Retrieval (BM25) | Elasticsearch BM25 with dynamic term weighting, phrase boosting, and pseudo-relevance feedback. | Keyword/phrase-based search. | Rapid response; suitable for emergency or keyword-heavy cases. |
| Hybrid Retrieval (RRF) | Combines dense + sparse scores via Reciprocal Rank Fusion with adaptive weighting. | Balances precision and recall. | Ensures coverage without sacrificing ranking quality. |
| Graph-Based RAG | UMLS graph (~3 M nodes, 15 M relations) with Graph Attention Network scoring. | Traverses medical ontology relations. | Useful for reasoning across linked conditions and biomedical concepts. |
| Long-Context RAG | Dual-phase retrieval: sparse window selection + dense re-ranking. | Handles long passages with coherence scoring. | Effective for patients with complex or longitudinal histories. |
| Adaptive Retrieval | Confidence thresholds; fallback re-queries with expanded terms. | Dynamically adjusts strategy when confidence is low. | Reduces unsupported outputs and improves reliability in uncertain cases. |
| Self-Reflective RAG (SELF-RAG) | Three-step loop: generate → check citations → refine. | Iterative self-critique and refinement. | Minimizes hallucinations; most reliable for safety-critical workflows. |
| Multimodal RAG | Vision Transformer (ViT) embeddings + text embeddings fused via MLP. | Integrates imaging + text. | Supports clinical workflows involving radiology alongside text. |
| Security-Focused RAG (TrustRAG) | AES-256 encrypted passages, provenance tagging, audit logs. | Privacy and compliance emphasis. | Meets HIPAA/GDPR; builds clinician trust with provenance notes. |
| Agentic RAG | LangChain agent with tools (EntityExtraction, TrialSearch, GuidelineLookup). | Decomposes queries into multi-step reasoning. | Mirrors how clinicians break down complex diagnostic questions. |
| Real-Time Knowledge Integration (CRAG) | Supplements PubMed index with bioRxiv/arXiv/NIH preprints via async retrieval. | Keeps knowledge base up to date. | Ensures coverage of emerging biomedical evidence. |
| Haystack Pipeline | DPR dual-encoder + BM25 → cross-encoder re-ranking. | End-to-end optimized retrieval. | Achieves state-of-the-art accuracy; strong overall baseline. |
| Method | P@5 | MRR | nDCG@10 | Hallucination Rate (%) | Latency (ms) |
|---|---|---|---|---|---|
| Dense Retrieval | 0.62 | 0.58 | 0.59 | 18.4 | 150 |
| Sparse Retrieval | 0.55 | 0.52 | 0.53 | 20.1 | 120 |
| Hybrid (RRF) | 0.68 | 0.62 | 0.67 | 11.2 | 180 |
| GraphRAG | 0.64 | 0.59 | 0.63 | 15.0 | 300 |
| Long RAG | 0.63 | 0.57 | 0.62 | 14.0 | 165 |
| Adaptive Retrieval | 0.66 | 0.60 | 0.66 | 12.5 | 170 |
| Multimodal RAG | 0.65 | 0.61 | 0.63 | 13.0 | 180 |
| SELF-RAG | 0.65 | 0.60 | 0.66 | 5.8 | 220 |
| CRAG | 0.63 | 0.58 | 0.63 | 14.3 | 200 |
| TrustRAG | 0.64 | 0.59 | 0.64 | 13.8 | 180 |
| Agentic RAG | 0.67 | 0.61 | 0.66 | 12.0 | 350 |
| Haystack (DPR + BM25 + Cross-Encoder) | 0.69 | 0.64 | 0.69 | 10.5 | 240 |
| RAG Variant | Accuracy | Hallucination Rate | Latency | Interpretability | Domain Adaptability | Resource Cost |
|---|---|---|---|---|---|---|
| Dense (FAISS, embeddings) | High (large corpora) | Medium | Low | Low (black-box embeddings) | Medium | High (GPU-heavy) |
| Sparse (BM25/elastic) | Medium | Medium–High | Very low | Medium–High (keywords visible) | Low (weak semantic reasoning) | Low |
| Hybrid (Dense + Sparse, RRF) | High | Medium–Low | Medium | Medium | High (works across domains) | High |
| Graph (GraphRAG, UMLS ontologies) | High (ontology-rich tasks) | Low | Medium | Very High (transparent reasoning) | High (needs ontologies) | Medium–High |
| Self-reflective | High (with citations) | Very Low | Medium–high (extra refinement steps) | High | Medium (needs high-quality citations) | Medium |
| Multimodal (text + imaging) | High | Low | High (multi-source fusion) | High | High (radiology, genomics, etc.) | Very High |
| Long-context (sliding window) | Medium–High | Medium | High (long input sequences) | Medium | High (good for large clinical docs) | High |
| Adaptive (two-stage/noise reduction) | High | Low | Medium | Medium | Medium–high (dynamic retrieval pipelines) | Medium |
| Real time (CRAG, live updates) | High (up-to-date retrieval) | Medium–Low | Medium | Medium | High (handles evolving corpora) | Medium–High |
| Security-focused (privacy-preserving) | Medium | Medium | High (encryption overhead) | Medium | High (healthcare, compliance) | High |
| Agentic (LLM-driven pipelines) | High | Low | High (multi-agent orchestration) | High | High (flexible tasks) | Very High |
| Haystack optimized pipelines | Medium–High | Medium | Medium | Medium | High (production-ready, modular) | Medium |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wołk, K. Evaluating Retrieval-Augmented Generation Variants for Clinical Decision Support: Hallucination Mitigation and Secure On-Premises Deployment. Electronics 2025, 14, 4227. https://doi.org/10.3390/electronics14214227
Wołk K. Evaluating Retrieval-Augmented Generation Variants for Clinical Decision Support: Hallucination Mitigation and Secure On-Premises Deployment. Electronics. 2025; 14(21):4227. https://doi.org/10.3390/electronics14214227
Chicago/Turabian StyleWołk, Krzysztof. 2025. "Evaluating Retrieval-Augmented Generation Variants for Clinical Decision Support: Hallucination Mitigation and Secure On-Premises Deployment" Electronics 14, no. 21: 4227. https://doi.org/10.3390/electronics14214227
APA StyleWołk, K. (2025). Evaluating Retrieval-Augmented Generation Variants for Clinical Decision Support: Hallucination Mitigation and Secure On-Premises Deployment. Electronics, 14(21), 4227. https://doi.org/10.3390/electronics14214227







