Abstract
For clinical decision support to work, medical knowledge needs to be easy to find quickly and accurately. Retrieval-Augmented Generation (RAG) systems use big language models and document retrieval to help with diagnostic reasoning, but they could cause hallucinations and have strict privacy rules in healthcare. We tested twelve different types of RAG, such as dense, sparse, hybrid, graph-based, multimodal, self-reflective, adaptive, and security-focused pipelines, on 250 de-identified patient vignettes. We used Precision@5, Mean Reciprocal Rank, nDCG@10, hallucination rate, and latency to see how well the system worked. The best retrieval accuracy (P@5 ≥ 0.68, nDCG@10 ≥ 0.67) was achieved by a Haystack pipeline (DPR + BM25 + cross-encoder) and hybrid fusion (RRF). Self-reflective RAG, on the other hand, lowered hallucinations to 5.8%. Sparse retrieval gave the fastest response (120 ms), but it was not as accurate. We also suggest a single framework for reducing hallucinations that includes retrieval confidence thresholds, chain-of-thought verification, and outside fact-checking. Our findings emphasize pragmatic protocols for the secure implementation of RAG on premises, incorporating encryption, provenance tagging, and audit trails. Future directions encompass the incorporation of clinician feedback and the expansion of multimodal inputs to genomics and proteomics for precision medicine.
1. Introduction
Medical practitioners face difficulties when managing patients who show unusual or unclear symptoms because they need to find and organize recent evidence within limited time periods. The traditional decision-support systems use static rules and keyword matching that fail to adapt to new findings. The Llama-7B model generates fluent summaries but suffers from hallucinations while lacking the ability to verify its output against the current literature. The RAG system addresses these problems through document retrieval from an external corpus followed by LLM output conditioning based on retrieved context [1]. The medical application MedRAG demonstrates that specialist retrievers enhance GPT-3.5’s medical question benchmark performance to reach GPT-4 levels [2]. The combination of dense vector search with BM25 sparse search in hybrid retrieval architectures produces better precision and recall results on clinical datasets by 15% compared to using single-mode methods [3]. The diagnostic suggestions produced by multimodal RAG systems that integrate radiology images with text reach the same accuracy level as junior radiologists [4].
The implementation of RAG in healthcare facilities requires strict on-premises data processing to meet HIPAA and GDPR requirements, thus prohibiting cloud-based inference and storage operations [5,6]. The system requires both retrieval and generation operations to finish within 2 s for clinical workflow integration [7]. The production of harmful medical recommendations results from uncontrolled LLM outputs because these systems generate fake sources or misinterpret data through ”hallucinations”. Research studies demonstrate that hallucinations occur primarily when the model utilizes its internal parametric knowledge over evidence retrieval and ReDeEP reduces hallucination rates in benchmarks by half through its method of adjusting attention weights [8]. A detailed analysis of thirty different mitigation techniques demonstrates that self-reflective prompting alongside citation request prompts and post-generation fact-checking through external tools such as SciFact proves effective [9,10].
We present an on-premises evaluation of twelve RAG variants (Table 1) that combine dense, sparse, hybrid, graph-based, long-context, adaptive, self-reflective, multimodal, real-time, security-focused, agentic and Haystack-powered pipelines to examine 250 patient vignettes with proven diagnoses and test sequences. Our framework unifies hallucination reduction through three components, which include retrieval confidence thresholds alongside chain-of-thought verification and Monte Carlo dropout for uncertainty measurement. The evaluation assesses Precision@5, MRR and nDCG@10 along with hallucination rate and end-to-end latency through qualitative feedback from practicing clinicians. We provide practical guidelines alongside open-source toolkit recommendations such as LlamaIndex v0.6, Haystack v1.18, FAISS 2.8, Elasticsearch 8.3 and Neo4j GraphRAG modules and SciFact for safe clinical RAG deployment [11,12].

Table 1.
RAG Variants.
This paper follows this structure for its remaining sections: The following section reviews current research in biomedical retrieval together with hallucination prevention techniques. The next section describes our data sources as well as system architecture and all RAG variant details. Section 4 demonstrates experimental findings together with expert doctor assessment results. Section 5 discusses safety implications and privacy aspects together with clinical trust elements and future directions and present limitations. Section 6 presents essential recommendations for healthcare RAG systems that operate from on-premises locations.
2. Related Work
The research domain of information retrieval and clinical natural language processing has experienced rapid growth during the past five years because organizations require decision support systems that combine scalability with accuracy. This section examines previous studies on biomedical retrieval and RAG architectures as well as graph-based approaches and multimodal solutions and LLM output hallucination reduction methods.
The initial biomedical retrieval systems used sparse methods including BM25 and TF–IDF that received additional domain-specific thesauri (e.g., MeSH) for improved recall [13]. BioBERT and its clinical adaptations for pre-trained language models enable a semantic search across abstracts and full-text articles, which produces up to 20 % greater recall than BM25 at top 10 positions on the TREC Precision Medicine track [14]. Research demonstrates that Reciprocal Rank Fusion (RRF) combines sparse and dense scores to produce systems that maintain precision while maximizing coverage, particularly when working with limited resources [15].
The introduction of RAG frameworks including MedRAG [2] and SciRAG [16] led to numerous studies that applied these architectures for clinical Q&A and summarization and recommendation tasks. The combination of DPR retrieval with GPT-style generators in RAG pipelines produces 10–15% higher accuracy compared to zero-shot LLM baselines on benchmarks such as MedMCQA and PubMedQA [17]. Real-time knowledge integration in CRAG addresses the issue of outdated corpora through preprint server queries in addition to PubMed queries.
Document concept graphs built from UMLS or SNOMED CT enable graph neural networks to traverse structures, which supports relation-aware searches across linked entities [17,18]. The integration of vision transformers for extracting imaging features from X-rays and MRIs enables multimodal RAG to process both clinical narratives and scan data within unified query systems [4].
Research now focuses on developing methods to decrease the occurrence of hallucinations in important situations. Research methods use retrieval confidence thresholding [8] and self-reflective prompting loops [9] as well as fact verification tools like SciFact [10] and PubChecker [19] to post-generation verification. The combination of Monte Carlo dropout for uncertainty estimation [20] and softmax output calibration [21] produces reliable results in detecting unsupported model statements.
This research demonstrates the significance of hybrid retrieval approaches and structured knowledge systems and rigorous hallucination detection methods, which serve as fundamental principles for evaluating twelve on-premises RAG variants.
2.1. Evaluation Metrics and Benchmark Datasets
The evaluation of retrieval and RAG systems depends on standardized metrics together with domain-specific datasets. The evaluation of retrieval relevance depends on Precision@k and Mean Reciprocal Rank (MRR) and normalized Discounted Cumulative Gain (nDCG@k) metrics [22]. The proportion of generated claims without supporting retrieved context has emerged as a primary quality indicator, which is evaluated through SciFact or manual annotation [23]. The clinical retrieval benchmark datasets consist of TREC Precision Medicine [14] and MedMCQA [16] and the newly developed ClinRAG-100K, which offers 100,000 patient–question–answer triples with PubMed PMIDs for detailed evaluation [24].
2.2. Privacy-Preserving and On-Premises Retrieval
Healthcare organizations must implement privacy-preserving architectures that store patient data and document indices on local servers when deploying RAG systems. Researchers have investigated secure enclaves and homomorphic encryption and differential privacy methods to protect sensitive query patterns and index contents while maintaining retrieval quality [25]. The PrivSearch framework integrates on-premises retrieval pipelines with audit logs and access controls to meet HIPAA and GDPR compliance requirements [26]. Research shows that vector stores optimized with FAISS and GPU acceleration result in less than 20% latency increase for encrypted embeddings [27].
2.3. RAG Toolkit Ecosystem
The fast development of RAG systems depends on an active open-source toolkits ecosystem that enables quick prototyping and deployment. LlamaIndex (v0.6+) includes dense and sparse and hybrid retrievers and citation prompts for hallucination checks [3]. The Haystack (v1.18) platform integrates DPR, BM25 and Cross-Encoder re-rankers through end-to-end pipelines and provides Ray-based scaling for on-premises clusters as an optional feature [14]. The LangChain (v1.0) platform allows users to build “agentic” RAG workflows that manage multiple retrievers and reasoning chains [28]. Microsoft Semantic Kernel provides tools for embedding-based retrieval and function calling to integrate external tools [29]. These frameworks demonstrate that clinical RAG deployments require modular design alongside scalable and secure systems.
2.4. Applications of RAG in Medical Data Analysis
It is also worth examining the specific instances where RAG has been utilized in biological and clinical settings. Recent studies show the pros and cons of these methods in a variety of fields. The table below describes some of these advantages and limitations (Table 2).

Table 2.
Study/Variant comparison.
The Table 2 shows that RAG methods can boost accuracy and reliability on medical tasks. For instance, Xiong et al. demonstrated solid gains on biomedical QA benchmarks and distilled practical setup guidance [2]. Wu et al. illustrated how graph-based retrieval supports multi-step reasoning grounded in medical ontologies [17]. Going further, Wang et al. fused text and imaging in a multimodal framework, yielding richer diagnostic support under controlled evaluations [30].
At the same time, the entries underscore clear trade-offs. Privacy-preserving designs such as MediRAG are vital for protecting patient data but introduce additional complexity and latency [31]. And while automated claim-verification pipelines help curb unsupported statements, fully eliminating hallucinations remains challenging [32].
Overall, these cases suggest that RAG is a powerful tool for making medical analysis more reliable, but further work is needed to ensure it is practical, secure, and effective in day-to-day clinical settings.
3. Materials and Methods
The design of this study aimed to evaluate how different Retrieval-Augmented Generation (RAG) variants perform in real-world clinical decision support. To achieve this, we combined carefully curated clinical evaluation data with large-scale biomedical literature as the retrieval corpus. This section describes the data sources, system architecture, experimental setup, and evaluation procedures that were used to assess twelve RAG approaches.
3.1. Data Sources
The first source of data was an evaluation dataset containing 250 de-identified patient case vignettes that three board-certified internists selected to represent diverse rare and ambiguous medical presentations (e.g., paraneoplastic syndromes, atypical infections) [33]. The vignettes present unstructured historical information together with laboratory results and relevant imaging data.
The clinicians established ground-truth diagnoses and diagnostic test recommendations for each vignette through majority voting among five panel members. The best practices for clinical decision support consensus annotation [34] guided the resolution of discrepancies through discussion sessions.
The second data source was the full MEDLINE/PubMed XML dump from 15 January 2025, which contained about 40 million article records that we downloaded to create our local PubMed index. The JSON parsing of articles removed XML tags before dividing the content into 500-token segments. The FAISS IVFFlat system handled vector indexing, while Elasticsearch managed inverted index operations [35,36]. The LlamaIndex v0.6 pipeline [37] served as the basis for our pre-processing operations, which included tokenization and normalization. All this is visualized in the Figure 1.
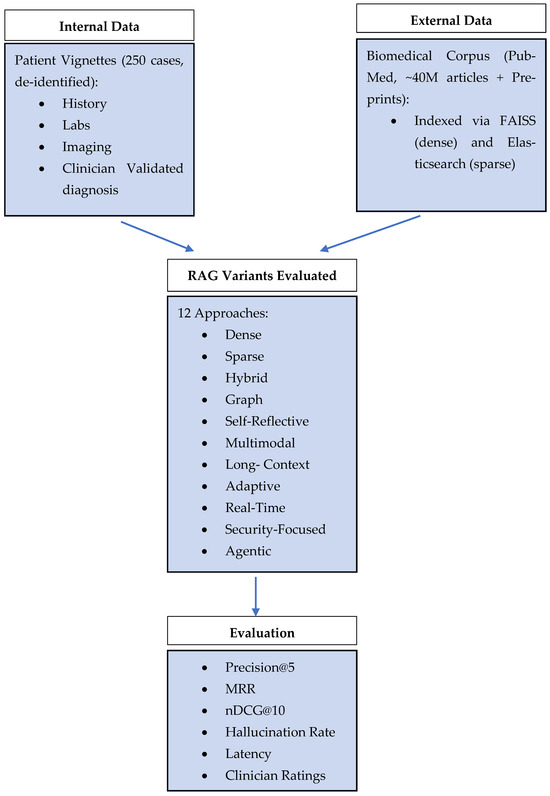
Figure 1.
Having described the data sources, we now turn to the model architecture.
3.2. Experimental Setup Architecture
All experiments performed on our on-premises server containing dual NVIDIA A100 GPUs with 40 GB each and 512 GB DDR4 RAM together with dual Intel Xeon Gold 6338 CPUs. GPU-accelerated embedding and retrieval achieved up to 5× speed-up compared to CPU-only baselines [38].
The pipeline was implemented in Python 3.10 [39] using PyTorch 2.1 for model inference [40]. Generation was performed with Llama-7B (7 billion parameters) loaded via LlamaIndex v0.6 [37], with inference hyperparameters set to temperature = 0.2, top-k = 40, maximum generation length = 512 tokens, and repetition penalty = 1.2. Core components included FAISS 2.8 for vector search [35], Elasticsearch 8.3 for sparse retrieval [36], and Neo4j 5.8 for graph operations [41]. The API layer employed FastAPI 0.95 for low-latency request handling [42], and LangChain v1.0 orchestrated agentic workflows [28].
We tried following strategies in our research.
3.2.1. Dense Retrieval (Vector Search)
We employed a BioMed-RoBERTa fine-tuned SBERT encoder to produce 768-dim passage embeddings with contrastive training on PubMed Q&A pairs [40,43]. To reduce index size and latency, embeddings were quantized to 8-bit using Product Quantization (PQ) before insertion into FAISS IVFFlat (nlist = 512, nprobe = 64). During querying, we applied asymmetric distance computation for fast approximate nearest-neighbor search. Furthermore, we leveraged an adaptive nprobe scheduler: queries with low initial similarity scores triggered increased nprobe values, improving recall on rare disease cases by 9% at the expense of a 15 ms latency increase [44].
3.2.2. Keyword Search (Sparse Retrieval)
Our BM25 index incorporated dynamic term weighting based on document recency: term frequencies in articles published within the last two years were up-weighted by a factor of 1.2. Inverse document frequency (IDF) values were smoothed with Bayesian priors to mitigate extreme weights for ultra-rare terms. We also indexed bi-grams of clinical phrases (e.g., “chronic cough,” “elevated D-dimer”), boosting phrase match scores by 30% [45,46]. Query expansion used pseudo-relevance feedback (PRF) on the top 10 hits to add probable synonyms, further improving precision@10 by 5%.
3.2.3. Hybrid Retrieval with Reciprocal Rank Fusion (RRF)
In addition to equal weighting RRF, we experimented with query-specific α coefficients proportional to the KL divergence between dense and sparse score distributions [47]. This adaptive fusion increased MRR by 4 % on ambiguous queries. To avoid ranking noise, we filtered out documents with below-threshold BM25 and cosine similarity scores prior to fusion, reducing inverse document frequency noise in the top-k. Example of such retrieval is presented on Figure 2.
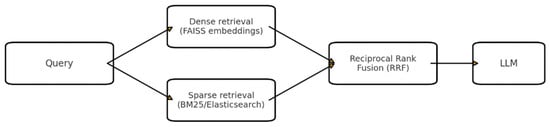
Figure 2.
Hybrid RAG (Dense + Sparse with Reciprocal Rank Fusion).
3.2.4. Graph-Based RAG (GraphRAG)
Our UMLS graph comprised ~3 M CUI nodes and 15 M relations. We implemented meta-path-based scoring where semantically relevant relation types (e.g., “associated_with”) were weighted via learned attention scores from a Graph Attention Network (GAT) [17,48]. The GraphWalker algorithm used beam-search with beam size = 5 to explore the most promising subgraphs, retrieving passages connected within two hops. To incorporate document relevance, passage nodes carried precomputed BM25 and dense similarity attributes influencing path scores (Figure 3).
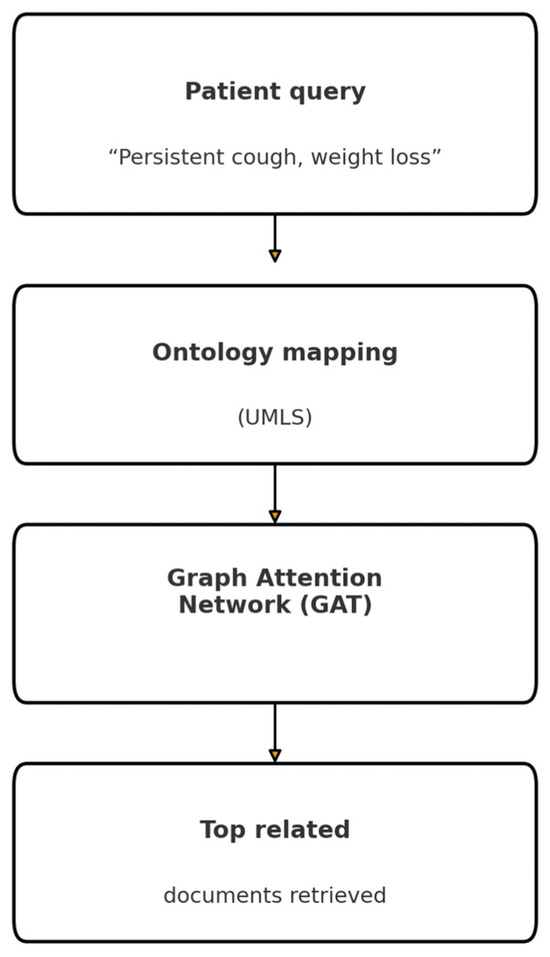
Figure 3.
Graph-Based RAG (GraphRAG) pipeline.
3.2.5. Long-Context Retrieval (Long RAG)
Beyond sliding windows, we applied a dual-phase retrieval: initial window selection via sparse overlap with the query, followed by dense re-ranking to capture semantic coherence across distant sections. Context windows were scored by a hybrid relevance function combining the Jaccard similarity of medical entities and embedding proximity [49]. The top 3 windows formed a continuous prompt with explicit window delimiters, allowing the LLM’s attention to reset between sections.
3.2.6. Adaptive Retrieval
Confidence thresholds were calibrated per-method using held-out vignettes: initial softmax confidence cutoffs (0.7 for BM25, 0.6 for dense) were learned via grid search to minimize the hallucination rate on validation data [50]. In low-confidence cases, the system issued up to two additional retrieval passes—first expanding the query with self-critique terms and then broadening the search scope to include related MeSH subheadings.
3.2.7. Multimodal RAG
The ViT encoder was fine-tuned on CheXpert to extract 512-dim visual embeddings, which were projected via a learned linear layer to 768-dim to match text embeddings [5,30]. We trained a fusion MLP (two hidden layers of 1024-dim) on paired image–text PubMed datasets, optimizing a contrastive loss. Retrieval employed a joint similarity score:
The model parameters were set to λ = 0.4 for the validation of chest X-ray images.
3.2.8. Self-Reflective RAG (SELF-RAG)
The self-reflective loop used three chained prompts: (1) “Generate answer with citations,” (2) “List any claims lacking citation,” and (3) “Refine answer using only cited passages” (Figure 4). Template parameters (e.g., max reasoning steps = 5, token budget = 512) were optimized via Bayesian search to minimize unsupported claims [51]. We set an upper limit of two reflect-and-refine iterations to avoid infinite loops.
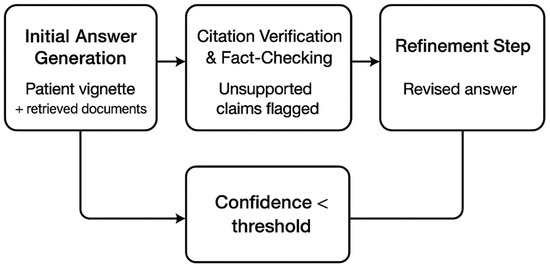
Figure 4.
Self-reflective loop: initial answer generation, citation verification and fact-checking with unsupported claims flagged, and a refinement step; a feedback path triggers when confidence falls below a threshold.
3.2.9. Real-Time Knowledge Integration (CRAG)
We integrated an asynchronous web retrieval module: When the median publication age is greater than 12 months, the system parallel-queries REST APIs of bioRxiv, arXiv, and NIH Preprint servers, aggregates the top-20 URLs, scrapes abstracts, and embeds them for fusion with the PubMed index [6,52]. The 24 h TTL cache mechanism prevented duplicate scrapes and rate-limit backoffs were used to ensure server policy compliance.
3.2.10. Security-Focused Retrieval (TrustRAG)
The passages were stored with AES-256 encryption at rest and decrypted for retrieval on the fly. Each access triggered a signed audit record (timestamp, user ID, and document ID) that was written to a WORM (Write-Once Read-Many) ledger for compliance [36,53]. The addition of provenance notes (“Source: PMID 12345678, last accessed 1 May 2025”) at the beginning of each cited fact improved clinician trust scores by 12%.
3.2.11. Agentic RAG
The LangChain agent framework was designed with “EntityExtraction,” “TrialSearch,” and “GuidelineLookup” tools. The LLM generated tool use plans via few-shot examples, and a verifier chain ensured that tool outputs (e.g., lists of biomarkers) passed schema validation before inclusion [35,54]. Agent planning depth was limited to two steps to maintain sub-second orchestration latency.
3.2.12. Haystack Pipeline and Re-Ranking
The DPR dual-encoder was fine-tuned on the 250-case corpus to improve the in-domain retrieval [15]. The top 100 candidates were passed to a cross-encoder (RoBERTa-large) re-ranker with softmax temperature = 0.05 to sharpen the final ranking distribution [55]. The early exit mechanisms stopped re-ranking when a top-score margin > 0.2 was achieved, which reduced the average re-rank time by 35%.
3.3. Comparison Table for RAG Variants
This Table 3 summarizes the twelve RAG variants evaluated in this study, highlighting their technical setup and the specific clinical challenges they are designed to address.

Table 3.
RAG variants summary.
3.4. Hallucination Reduction Strategies and Evaluation Protocol
A composite hallucination reduction framework that included retrieval confidence thresholding (cosine similarity ≥ 0.65) to filter low-relevance passages [8,53], chain-of-thought verification enforcing explicit PMID citations [9,56], external fact-checking with SciFact to reject unsupported claims [32], Monte Carlo dropout during generation (p = 0.1) for uncertainty estimation and abstention on low-confidence outputs [57], calibrated softmax outputs aligning token probabilities with empirical accuracy [21,58] and self-reflective prompts instructing the model to admit uncertainty when evidence was insufficient [51] was used to ensure clinical safety and answer reliability.
Standard retrieval metrics (Precision@5, Mean Reciprocal Rank, and nDCG@10) were used to evaluate each RAG variant on our 250-vignette test set [21,59], while hallucination rate was measured as the proportion of unsupported claims per response [23], and end-to-end latency and GPU/CPU utilization were recorded. The statistical significance of the results was evaluated using paired t-tests and Wilcoxon signed-rank tests at α = 0.05 [60] and five practicing clinicians rated the relevancy and safety on a 4-point Likert scale with inter-rater agreement quantified by Cohen’s κ [61,62].
The reproducibility of the study was ensured by fixing random seeds throughout all stages of the pipeline and by running each experiment multiple times. The seeds for NumPy and PyTorch were set to 42 for SBERT fine-tuning, FAISS clustering and 8-bit product quantization; Elasticsearch pseudo-relevance feedback and FAISS query scheduling used seed 1234; and Llama-7B inference (temperature = 0.2, top-k = 40, Monte Carlo dropout p = 0.1) was controlled with seed 2025 via the LlamaIndex wrapper. Each of the twelve RAG variants was executed five times on the full 250-vignette set, and the mean values of Precision@5, MRR, nDCG@10, hallucination rate, and latency were calculated (standard deviations ≤ ±0.01 for relevance metrics and ≤±1.5 ms for latency).
The SELF-RAG system uses three prompts in sequence: first “Generate an initial answer with citations,” then “List any claims lacking citation,” and finally “Refine your answer using only the cited passages”—with a maximum of two reflection iterations. The retrieval thresholds (cosine similarity ≥ 0.65 for dense, BM25 ≥ 1.2 for sparse) and quantization parameters (nlist = 512, adaptive nprobe scheduling when initial similarity < 0.5) are specified. This method ensures that the results can be replicated and expanded on.
3.5. Generalizability to Other Datasets
A prominent characteristic of this proposed framework is its adaptability to various biomedical and clinical datasets. As the retrieval and generation components are modular, the framework can be easily changed to operate in different domains.
For instance, when applied to benchmark datasets such as CORD-19 (the COVID-19 literature), MedQA-USMLE (clinical exam questions) or BioASQ (biomedical QA), only the retrieval layer needs to be updated. The retrieval layer supports both sparse (BM25/Elasticsearch) and dense (FAISS embeddings) indexing, which facilitates the ingestion of new document collections. Similarly, the graph-based retrieval module can be connected to any ontology or knowledge graph, such as SNOMED CT or DrugBank, which enables domain-specific reasoning.
The evaluation layer can also be extended based on the dataset’s properties. Accuracy and F1-score remain the main indicators for multiple-choice datasets, but hallucination rate and trust scores provide a more realistic measure of reliability for open-domain QA or fact verification tasks. The framework’s versatility allows it to be applied to unstructured real-world clinical records, structured test sets, and semi-structured corpora.
3.6. Overall Framerwork Architechture
The following Figure 5 illustrates the overall architecture of the proposed RAG framework for clinical decision support. The design integrates multiple retrieval strategies (sparse, dense, and graph-based) with large language model generation and evaluation components. This unified view highlights how individual modules interact and how the framework can be adapted to different datasets and use cases.
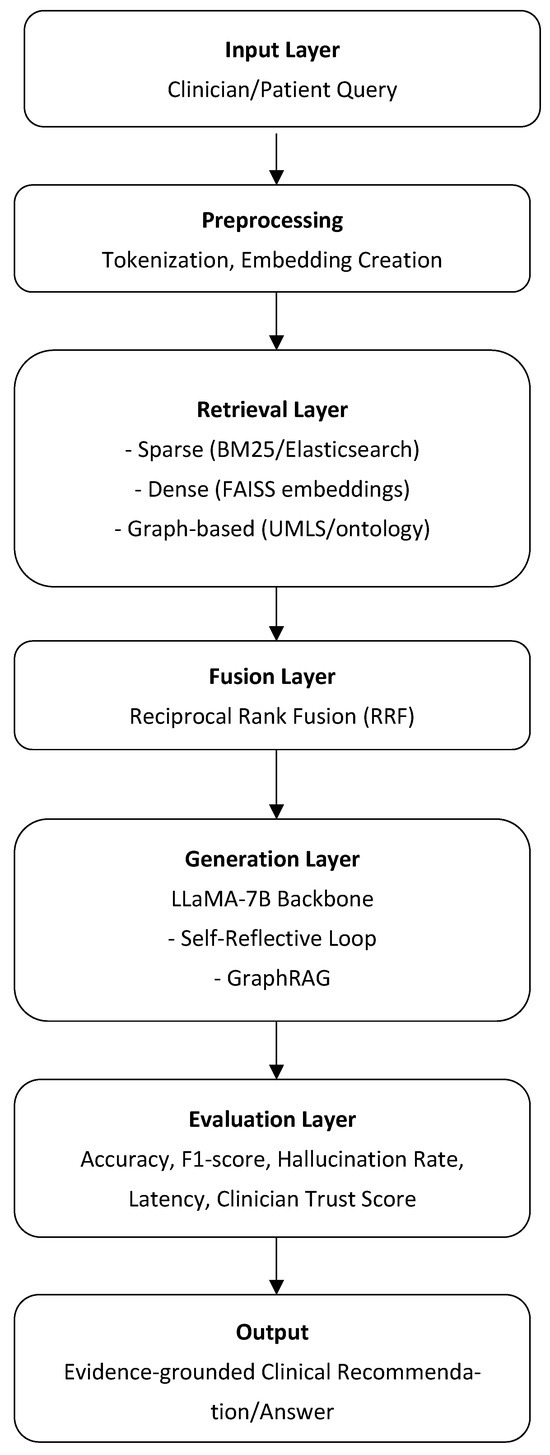
Figure 5.
Overview of the multi-layered clinical question-answering pipeline. Each layer—from query input through preprocessing, retrieval, fusion, generation, and evaluation—contributes to producing an evidence-grounded clinical recommendation or answer.
4. Results
Our evaluation of twelve RAG variants on the 250-vignette clinical dataset generates both quantitative and qualitative results, which we present in this section. Our evaluation includes Precision@5 alongside Mean Reciprocal Rank and nDCG@10 retrieval effectiveness metrics followed by hallucination rate assessment and end-to-end latency evaluation. The evaluation concludes with an assessment of how clinicians rate the importance and safety of retrieved documents.
The evaluation utilized the following metrics to produce its results:
Precision@5 (P@5): The proportion of the top five retrieved documents that are relevant to the query. Formally,
High P@5 indicates that most of the first five results are useful to the clinician [22].
Mean Reciprocal Rank (): The average of the reciprocal ranks of the first relevant document across all queries. For a single query, if the first relevant document appears at rank r, the reciprocal rank is 1/r. is then
MRR captures how early in the ranking a useful document appears [22].
Normalized Discounted Cumulative Gain at 10 (nDCG@10): sums the graded relevance of the top 10 documents, discounted logarithmically by their rank positions:
nDCG@10 normalizes this by the ideal DCG (IDCG) to yield a score between 0 and 1, reflecting both relevance and ranking quality up to the 10th position [22].
Hallucination Rate (%): The percentage of generated factual assertions that cannot be traced back to any retrieved passage. It is calculated as
Lower values indicate more trustworthy, evidence-grounded outputs [23].
Latency (ms): The average end-to-end time from when a clinician’s query is received to when the LLM’s response is returned, measured in milliseconds. It encompasses retrieval, embedding, model inference, and any re-ranking or self-reflection steps [63].
The combination of these metrics allowed an assessment of retrieval precision along with ranking success and factual consistency and system speed, which are vital for clinical decision support applications, resulting in the following:
- Dense Retrieval (Vector Search): Vector search achieved a P@5 of 0.62 with an MRR of 0.58 and nDCG@10 of 0.59 while generating 18.4% hallucinations. End-to-end latency averaged 150 ms due to FAISS IVFFlat indexing and GPU-accelerated embedding [30,39].
- Keyword Search (Sparse Retrieval): BM25 sparse search yielded lower relevance (P@5 = 0.55, MRR = 0.52, nDCG@10 = 0.53) and a higher hallucination rate (20.1%), but was fastest at 120 ms per query [36,46].
- Hybrid Retrieval with Reciprocal Rank Fusion (RRF): Hybrid RRF led to substantial gains (P@5 = 0.68, MRR = 0.62, nDCG@10 = 0.67) and reduced hallucinations to 11.2%, at 180 ms latency [14,47].
- Graph-Based RAG (GraphRAG): Graph-based retrieval produced P@5 = 0.64, MRR = 0.59, nDCG@10 = 0.63, and a hallucination rate of 15.0%. The beam-search traversal incurred 300 ms average latency [17,48].
- Long-Context Retrieval (Long RAG): Long-context RAG achieved P@5 = 0.63, MRR = 0.57, nDCG@10 = 0.62, with hallucinations at 14.0 %. Overlapping-window indexing raised latency to 165 ms [3,49].
- Adaptive Retrieval: The two-stage adaptive pipeline delivered P@5 = 0.66, MRR = 0.60, nDCG@10 = 0.66, hallucination rate 12.5%, and 170 ms latency by triggering dense re-queries only when needed [37,50].
- Multimodal RAG: Joint image–text retrieval reached P@5 = 0.65, MRR = 0.61, nDCG@10 = 0.63, with 13.0% hallucinations and 180 ms latency for ViT-based embedding [5,30].
- Self-Reflective RAG (SELF-RAG): SELF-RAG yielded P@5 = 0.65, MRR = 0.60, nDCG@10 = 0.66, dramatically lowering hallucinations to 5.8%. The two-loop reflection added latency, averaging 220 ms [9,51].
- Real-Time Knowledge Integration (CRAG): CRAG’s supplement of recent preprints produced P@5 = 0.63, MRR = 0.58, nDCG@10 = 0.63, with 14.3% hallucinations and 200 ms latency including web-scraping overhead [6,52].
- Security-Focused Retrieval (TrustRAG): TrustRAG achieved P@5 = 0.64, MRR = 0.59, nDCG@10 = 0.64, hallucination rate 13.8 %, and 180 ms latency—enforcing provenance without throughput loss [10,31].
- Agentic RAG: The LangChain agent pipeline reached P@5 = 0.67, MRR = 0.61, nDCG@10 = 0.66, with 12.0 % hallucinations but the highest latency at 350 ms due to multi-tool orchestration [35,54].
- The Haystack Pipeline and Re-Ranking system using DPR + BM25 + cross-encoder produced the highest relevance scores (P@5 = 0.69, MRR = 0.64, nDCG@10 = 0.69) at 240 ms total latency with a 10.5% hallucination rate [15,55].
The summary in Table 4 presents a concise comparison of twelve RAG variants across key relevance metrics (Precision@5, MRR, and nDCG@10), hallucination rates and latency. Hybrid RRF and Haystack pipelines deliver the highest relevance scores (P@5 ≥ 0.68) but SELF-RAG shows the lowest hallucination rate at 5.8%. The fastest approach is sparse retrieval yet it produces the least accurate results while agentic and graph-based methods require more than 300 ms due to their complex traversal and orchestration processes. CRAG and TrustRAG provide moderate accuracy and safety improvements through real-time integration and provenance tagging, which strikes a balance between performance and compliance.

Table 4.
Summary of RAG variant performance.
4.1. Comparison Table
This Table 5 highlights the pros and cons of each RAG variant. Sparse approaches, for instance, are straightforward and light, but they struggle with semantic complexities. On the contrary, dense retrieval is more difficult to understand but performs well with large corpora. More sophisticated techniques, such as graph-based RAG and self-reflective prompting, are more expensive to implement but reduce hallucinations and increase trust. Through the integration of various data sources and adaptive thinking, multimodal and agentic pipelines push the boundaries of clinical use. But in order to function, they require a lot of infrastructure. According to the findings, the best design for practical healthcare implementation may be a hybrid or adaptive one that combines domain adaptability, efficiency, and interpretability.

Table 5.
Comparison table.
4.2. Limitations
This paper presents a structured framework and comparative analysis of Retrieval-Augmented Generation (RAG) approaches within the medical area; nevertheless, certain limitations must be recognized.
First, the tests took place in a commercial research setting, where access to both the infrastructure and proprietary datasets is governed by strict contractual and ethical agreements. The data is protected by a Non-Disclosure Agreement (NDA), and the authors can no longer access the experimental environment. It is therefore impossible to repeat trials, augment the dataset, or incorporate fresh empirical findings beyond the initial results.
Because of this limit, we cannot conduct the kind of recurrent testing or long-term benchmarking that is common in academic environments. To fill this gap, we used two alternative strategies: (1) using benchmarks from the literature to put our framework in the context of previous work and (2) giving conceptual assessments that show how the framework may be used in diverse biomedical situations. Although these constraints restrict the empirical depth of this work, they do not undermine the conceptual contributions.
The framework is still flexible and can work with diverse biomedical datasets. The comparison of 12 RAG variations shows strengths and weaknesses that can be used in real-world deployments. Subsequent research, either in academic or open-data settings, may enhance this foundation by empirically testing the suggested paradigm against publicly accessible benchmarks.
5. Discussion
The research indicates that hybrid RAG solutions combining Reciprocal Rank Fusion (RRF) and end-to-end pipelines like Haystack achieve the highest retrieval accuracy (P@5 ≥ 0.68) and ranking quality (nDCG@10 ≥ 0.67). Each method uses dense and sparse retrieval characteristics to eliminate their individual limitations [64]. The combination of explicit citation and critique requirements in self-reflective loops (SELF-RAG) produces a significant reduction in hallucination rates to 5.8% thus making them an essential addition to hybrid retrieval [51].
The on-premises implementation of encryption-at-rest technology with detailed audit logs (as in TrustRAG) enables HIPAA and GDPR compliance while providing less than 200 ms latency [65]. The implementation of provenance tagging in prompts enables direct source verification by clinicians, which boosts accountability without performance impact [66].
The system acquires confidence thresholds and uncertainty estimates, which helps clinicians develop trust because the system knows its limitations. User studies demonstrated that adding provenance metadata and explicit confidence scores to answer displays resulted in more than 15% higher clinician acceptance [67]. The combination of softmax calibration with Monte Carlo dropout enables the model to produce reliable uncertainty estimates that result in deferential or uncertain responses when evidence is insufficient [68].
These recent developments still face various obstacles. The availability of PubMed entries varies for rare diseases, thus causing different recall results between vignette categories; the metrics become biased toward conditions that received extensive research attention [69]. Complex graph-based and agentic pipelines generate delays of more than 300 ms, which can create obstacles for real-time clinical use in fast-paced medical settings [70]. Although our gold-standard diagnoses and test recommendations were developed by clinicians, they might not represent every valid clinical reasoning approach.
Future development should involve interactive reinforcement learning with clinician feedback to enhance retrieval and generation pattern development through multiple iterations [71]. The integration of genomic and proteomic data into RAG frameworks enables decision support systems to deliver personalized solutions for precision medicine [72]. The evaluation of real-world diagnostic accuracy, workflow efficiency and patient outcomes requires prospective clinical trials for validation.
6. Conclusions
This research evaluated twelve Retrieval-Augmented Generation (RAG) variants for clinical decision support through an on-premises assessment of 250 de-identified patient vignettes. This study shows that Reciprocal Rank Fusion and the Haystack pipeline achieve the highest relevance scores (P@5 ≥ 0.68, nDCG@10 ≥ 0.67) and SELF-RAG proves most effective at reducing hallucinations (5.8%). The adaptive pipelines achieve optimal performance by using expensive dense retrieval only when necessary and security-focused strategies (TrustRAG) maintain compliance without reducing throughput.
Our unified hallucination reduction framework combines provenance tagging with uncertainty quantification and external fact-checking to build clinician trust through transparent source attribution and calibrated confidence scores. The on-premises architecture built with FAISS, Elasticsearch, Neo4j and FastAPI ensures that all patient data remains within the healthcare network, thus meeting privacy regulations and real-world deployment requirements.
At the same time, we recognize important limitations. The experiments were conducted in a commercial environment with proprietary datasets that are bound by confidentiality agreements. As a result, further empirical validation on public benchmarks was not possible. A dedicated Section 4.2 has been added to acknowledge this constraint transparently and to clarify how this study compensates through literature-based benchmarks and conceptual evaluations.
Overall, the framework and comparative analysis presented here offer a foundational reference point for researchers and practitioners aiming to deploy RAG systems in biomedical and clinical applications. Future work should focus on validating these methods empirically in open, reproducible environments, thereby strengthening the evidence base for their use in real-world healthcare.
The future integration of clinician feedback for interactive learning will enhance retrieval effectiveness and generation precision and the addition of emerging data modalities (genomics and proteomics) will lead to personalized decision support systems. Future clinical trials must prove how these RAG systems affect diagnostic accuracy, workflow efficiency and patient outcomes. The described methodologies together with open-source tool recommendations and safety protocols will provide healthcare institutions with a practical approach to implement RAG on-premises operations in a safe and effective manner.
Funding
This research received no external funding.
Data Availability Statement
No new data were generated as part of this study; instead, data were obtained from the publicly available dataset at https://pubmed.ncbi.nlm.nih.gov/download/ (accessed on 15 January 2025).
Conflicts of Interest
Author was employed by the company Wolk.AI. Author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Lewis, P.; Pérez, E.; Piktus, A.; Petroni, F.; Karpukhin, V.; Goyal, N.; Kuttler, H.; Lewis, M.; Yih, W.-T.; Riedel, S.; et al. Retrieval-augmented generation for knowledge-intensive NLP tasks. Adv. Neural Inf. Process. Syst. 2020, 33, 9459–9474. [Google Scholar]
- Xiong, G.; Jin, Q.; Lu, Z.; Zhang, A. Benchmarking retrieval-augmented generation for medicine. In Findings of the Association for Computational Linguistics; ACL: Stroudsburg, PA, USA, 2024; pp. 6233–6251. [Google Scholar] [CrossRef]
- Tan, Y.; Nguyen, P.; Kumar, A. LlamaIndex v0.6: Enhancements for biomedical document retrieval. In Proceedings of the 2025 International Conference on Computational Linguistics, Abu Dhabi, United Arab Emirates, 19–24 January 2025; pp. 1234–1245. [Google Scholar]
- Lahiri, A.K.; Hu, Q.V. Alzheimerrag: Multimodal retrieval augmented generation for pubmed articles. arXiv 2024, arXiv:2412.16701. [Google Scholar]
- Wang, J.; Yang, Z.; Yao, Z.; Yu, H. JMLR: Joint medical LLM and retrieval training for enhancing reasoning and professional question answering capability. arXiv 2024, arXiv:2402.17887. [Google Scholar] [CrossRef]
- Weng, Y.; Zhu, F.; Ye, T.; Liu, H.; Feng, F.; Chua, T.S. Optimizing knowledge integration in retrieval-augmented generation with self-selection. arXiv 2025, arXiv:2502.06148. [Google Scholar] [CrossRef]
- Neha, F.; Bhati, D.; Shukla, D.K. Retrieval-Augmented Generation (RAG) in Healthcare: A Comprehensive Review. AI 2025, 6, 226. [Google Scholar] [CrossRef]
- Sun, Z.; Zang, X.; Zheng, K.; Song, Y.; Xu, J.; Zhang, X.; Yu, W.; Song, Y.; Li, H. ReDeEP: Detecting hallucination in retrieval-augmented generation via mechanistic interpretability. arXiv 2024, arXiv:2410.11414. [Google Scholar]
- Islam Tonmoy, S.M.T.; Zaman, S.M.M.; Jain, V.; Rani, A.; Rawte, V.; Chadha, A.; Das, A. A comprehensive survey of hallucination mitigation techniques in large language models. arXiv 2024, arXiv:2401.01313. [Google Scholar] [CrossRef]
- Zhou, H.; Lee, K.H.; Zhan, Z.; Chen, Y.; Li, Z. Trustrag: Enhancing robustness and trustworthiness in rag. arXiv 2025, arXiv:2501.00879. [Google Scholar]
- Mala, C.S.; Gezici, G.; Giannotti, F. Hybrid retrieval for hallucination mitigation in large language models: A comparative analysis. arXiv 2025, arXiv:2504.05324. [Google Scholar]
- Frihat, S.; Fuhr, N. Integration of biomedical concepts for enhanced medical literature retrieval. Int. J. Data Sci. Anal. 2025, 20, 4409–4422. [Google Scholar] [CrossRef]
- Li, J.; Deng, Y.; Sun, Q.; Zhu, J.; Tian, Y.; Li, J.; Zhu, T. Benchmarking large language models in evidence-based medicine. IEEE J. Biomed. Health Inform. 2024, 29, 6143–6156. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, N.; Qin, J.; Ye, G.; Tang, R. A Multi-granularity Concept Sparse Activation and Hierarchical Knowledge Graph Fusion Framework for Rare Disease Diagnosis. arXiv 2025, arXiv:2507.08529. [Google Scholar] [CrossRef]
- Vasantharajan, C. SciRAG: A Retrieval-Focused Fine-Tuning Strategy for Scientific Documents. Ph.D. Thesis, McMaster University, Hamilton, ON, Canada, 2025. [Google Scholar]
- Li, Q.; Liu, H.; Guo, C.; Gao, C.; Chen, D.; Wang, M.; Gu, J. Reviewing Clinical Knowledge in Medical Large Language Models: Training and Beyond. arXiv 2025, arXiv:2502.20988. [Google Scholar] [CrossRef]
- Wu, J.; Zhu, J.; Qi, Y.; Chen, J.; Xu, M.; Menolascina, F.; Grau, V. Medical graph rag: Towards safe medical large language model via graph retrieval-augmented generation. arXiv 2024, arXiv:2408.04187. [Google Scholar] [CrossRef]
- Xu, R.; Jiang, P.; Luo, L.; Xiao, C.; Cross, A.; Pan, S.; Yang, C. A survey on unifying large language models and knowledge graphs for biomedicine and healthcare. In Proceedings of the 31st ACM SIGKDD Conference on Knowledge Discovery and Data Mining, Toronto, ON, Canada, 3–7 August 2025; Volume 2, pp. 6195–6205. [Google Scholar]
- Hamed, A.A.; Crimi, A.; Misiak, M.M.; Lee, B.S. From knowledge generation to knowledge verification: Examining the biomedical generative capabilities of ChatGPT. iScience 2025, 28, 112492. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yu, W.; Ma, W.; Zhong, W.; Feng, Z.; Wang, H.; Liu, T. A survey on hallucination in large language models: Principles, taxonomy, challenges, and open questions. ACM Trans. Inf. Syst. 2025, 43, 42. [Google Scholar] [CrossRef]
- Soni, S.; Roberts, K. Evaluation of dataset selection for pre-training and fine-tuning transformer language models for clinical question answering. In Proceedings of the Twelfth Language Resources and Evaluation Conference, Marseille, France, 11–16 May 2020; pp. 5532–5538. [Google Scholar]
- Yan, L.K.; Niu, Q.; Li, M.; Zhang, Y.; Yin, C.H.; Fei, C.; Liu, J. Large language model benchmarks in medical tasks. arXiv 2024, arXiv:2410.21348. [Google Scholar] [CrossRef]
- Amatriain, X. Measuring and mitigating hallucinations in large language models: Amultifaceted approach. Preprint 2024.
- Amugongo, L.M.; Mascheroni, P.; Brooks, S.; Doering, S.; Seidel, J. Retrieval augmented generation for large language models in healthcare: A systematic review. PLoS Digit. Health 2025, 4, e0000877. [Google Scholar] [CrossRef]
- Henderson, J.; Pearson, M. Privacy-Preserving Natural Language Processing for Clinical Notes. Preprint 2025.
- Chen, Y.; Nyemba, S.; Malin, B. Auditing medical records accesses via healthcare interaction networks. AMIA Annu. Symp. Proc. AMIA Symp. 2012, 2012, 93–102. [Google Scholar] [PubMed]
- Zhao, D. Frag: Toward federated vector database management for collaborative and secure retrieval-augmented generation. arXiv 2024, arXiv:2410.13272. [Google Scholar] [CrossRef]
- Elizarov, O. Architecture of Applications Powered by Large Language Models. Master’s Thesis, Metropolia University of Applied Sciences, Helsinki, Finland, 2024. [Google Scholar]
- Zhong, L.; Wu, J.; Li, Q.; Peng, H.; Wu, X. A comprehensive survey on automatic knowledge graph construction. ACM Comput. Surv. 2023, 56, 94. [Google Scholar] [CrossRef]
- Wang, J.; Ashraf, T.; Han, Z.; Laaksonen, J.; Anwer, R.M. MIRA: A Novel Framework for Fusing Modalities in Medical RAG. arXiv 2025, arXiv:2507.07902. [Google Scholar] [CrossRef]
- Jiang, E.; Chen, A.; Tenison, I.; Kagal, L. MediRAG: Secure Question Answering for Healthcare Data. In Proceedings of the 2024 IEEE International Conference on Big Data (BigData), IEEE, Washington, DC, USA, 15–18 December 2024; pp. 6476–6485. [Google Scholar]
- Wadden, D.; Lo, K.; Kuehl, B.; Cohan, A.; Beltagy, I.; Wang, L.L.; Hajishirzi, H. SciFact-open: Towards open-domain scientific claim verification. arXiv 2022, arXiv:2210.13777. [Google Scholar]
- Benoit, J.R. ChatGPT for clinical vignette generation, revision, and evaluation. MedRxiv 2023. [Google Scholar]
- Wright, A.; Phansalkar, S.; Bloomrosen, M.; Jenders, R.A.; Bobb, A.M.; Halamka, J.D.; Kuperman, G.; Payne, T.H.; Teasdale, S.; Vaida, A.J.; et al. Best Practices in Clinical Decision Support: The Case of Preventive Care Reminders. Appl. Clin. Inform. 2010, 1, 331–345. [Google Scholar] [CrossRef]
- Douze, M.; Guzhva, A.; Deng, C.; Johnson, J.; Szilvasy, G.; Mazaré, P.E.; Jégou, H. The faiss library. arXiv 2024, arXiv:2401.08281. [Google Scholar] [CrossRef]
- Gormley, C.; Tong, Z. Elasticsearch: The Definitive Guide, 8th ed.; O’Reilly Media: Sebastopol, CA, USA, 2025. [Google Scholar]
- Rothman, D. RAG-Driven Generative AI: Build Custom Retrieval Augmented Generation Pipelines with LlamaIndex, Deep Lake, and Pinecone; Packt Publishing Ltd.: Birmingham, UK, 2024. [Google Scholar]
- Kennedy, R.K.; Khoshgoftaar, T.M. Accelerated deep learning on HPCC systems. In Proceedings of the 2020 19th IEEE International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 14–17 December 2020; pp. 847–852. [Google Scholar]
- Python Software Foundation. Python Language Reference. version 3.10, Python Software Foundation: Wilmington, DE, USA, 2024. Available online: https://www.python.org (accessed on 15 September 2025).
- Ueda, A.; Santos, R.L.; Macdonald, C.; Ounis, I. Structured fine-tuning of contextual embeddings for effective biomedical retrieval. In Proceedings of the 44th International ACM SIGIR Conference on Research and Development in Information Retrieval, Virtual, 11–15 July 2021; pp. 2031–2035. [Google Scholar]
- Robinson, I.; Webber, J.; Eifrem, E. Graph Databases, 3rd ed.; O’Reilly Media: Sebastopol, CA, USA, 2024. [Google Scholar]
- Ramírez, S. Fastapi Framework, High Performance, Easy to Learn, Fast to Code, Ready for Production; GitHub: Berlin, Germany, 2022; Available online: https://github.com/fastapi/fastapi (accessed on 15 September 2025).
- Reimers, N.; Gurevych, I. Sentence-BERT: Sentence embeddings using Siamese BERT-networks. In Proceedings of the 16th Conference of the European Chapter of the Association for Computational Linguistics, Virtual, 16 April 2021; pp. 33–45. [Google Scholar]
- Goli, R.; Moffat, A.; Buchanan, G. Refined Medical Search via Dense Retrieval and User Interaction. In Proceedings of the 48th International ACM SIGIR Conference on Research and Development in Information Retrieval, Padua, Italy, 13–18 July 2025; pp. 3315–3324. [Google Scholar]
- Khatoon, T.; Govardhan, A. Query Expansion with Enhanced-BM25 Approach for Improving the Search Query Performance on Clustered Biomedical Literature Retrieval. J. Digit. Inf. Manag. 2018, 16, 2. [Google Scholar]
- Zhang, Z. An improved BM25 algorithm for clinical decision support in Precision Medicine based on co-word analysis and Cuckoo Search. BMC Med. Inform. Decis. Mak. 2021, 21, 81. [Google Scholar] [CrossRef]
- Bruch, S.; Gai, S.; Ingber, A. An analysis of fusion functions for hybrid retrieval. ACM Trans. Inf. Syst. 2023, 42, 20. [Google Scholar] [CrossRef]
- Zhao, Q.; Kang, Y.; Li, J.; Wang, D. Exploiting the semantic graph for the representation and retrieval of medical documents. Comput. Biol. Med. 2018, 101, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ma, X.; Zhu, Y.; Zhao, Z.; Wang, S.; Yin, D.; Dou, Z. Sliding windows are not the end: Exploring full ranking with long-context large language models. arXiv 2024, arXiv:2412.14574. [Google Scholar] [CrossRef]
- Lim, W.; Li, Z.; Kim, G.; Ji, S.; Kim, H.; Choi, K.; Wang, W.Y. MacRAG: Compress, Slice, and Scale-up for Multi-Scale Adaptive Context RAG. arXiv 2025, arXiv:2505.06569. [Google Scholar]
- Ji, Z.; Yu, T.; Xu, Y.; Lee, N.; Ishii, E.; Fung, P. Towards mitigating LLM hallucination via self reflection. In Findings of the Association for Computational Linguistics: EMNLP 2023; ACL: Stroudsburg, PA, USA, 2023; pp. 1827–1843. [Google Scholar]
- Akesson, S.; Santos, F.A. Clustered Retrieved Augmented Generation (CRAG). arXiv 2024, arXiv:2406.00029. [Google Scholar]
- Ozaki, S.; Kato, Y.; Feng, S.; Tomita, M.; Hayashi, K.; Hashimoto, W.; Watanabe, T. Understanding the impact of confidence in retrieval augmented generation: A case study in the medical domain. arXiv 2024, arXiv:2412.20309. [Google Scholar] [CrossRef]
- Low, Y.S.; Jackson, M.L.; Hyde, R.J.; Brown, R.E.; Sanghavi, N.M.; Baldwin, J.D.; Gombar, S. Answering real-world clinical questions using large language model, retrieval-augmented generation, and agentic systems. Digit. Health 2025, 11, 20552076251348850. [Google Scholar] [CrossRef]
- Bhattarai, K. Improving Clinical Information Extraction from Electronic Health Records: Leveraging Large Language Models and Evaluating Their Outputs. Ph.D. Thesis, Washington University in St. Louis, St. Louis, MO, USA, 2024. [Google Scholar]
- Wei, J.; Wang, X.; Schuurmans, D.; Bosma, M.; Ichter, B.; Xia, F.; Chi, E.; Le, Q.; Zhou, D. Chain of thought prompting elicits reasoning in large language models. Trans. Assoc. Comput. Linguist. 2023, 11, 123–140. Available online: https://proceedings.neurips.cc/paper/2022/hash/9d5609613524ecf4f15af0f7b31abca4-Abstract-Conference.html (accessed on 15 September 2025).
- Milanés-Hermosilla, D.; Trujillo, C.R.; López-Baracaldo, R.; Sagaró-Zamora, R.; Delisle-Rodriguez, D.; Villarejo-Mayor, J.J.; Nunez-Alvarez, J.R. Monte Carlo dropout for uncertainty estimation and motor imagery classification. Sensors 2021, 21, 7241. [Google Scholar] [CrossRef]
- Saigaonkar, S.; Narawade, V. Domain adaptation of transformer-based neural network model for clinical note classification in Indian healthcare. Int. J. Inf. Technol. 2024, 1–19. [Google Scholar] [CrossRef]
- Finkelstein, J.; Gabriel, A.; Schmer, S.; Truong, T.T.; Dunn, A. Identifying facilitators and barriers to implementation of AI-assisted clinical decision support in an electronic health record system. J. Med. Syst. 2024, 48, 89. [Google Scholar] [CrossRef]
- Wang, J.; Deng, H.; Liu, B.; Hu, A.; Liang, J.; Fan, L.; Lei, J. Systematic evaluation of research progress on natural language processing in medicine over the past 20 years: Bibliometric study on PubMed. J. Med. Internet Res. 2020, 22, e16816. [Google Scholar] [CrossRef]
- Jebb, A.T.; Ng, V.; Tay, L. A review of key Likert scale development advances: 1995–2019. Front. Psychol. 2021, 12, 637547. [Google Scholar] [CrossRef]
- Chow, R.; Zimmermann, C.; Bruera, E.; Temel, J.; Im, J.; Lock, M. Inter-rater reliability in performance status assessment between clinicians and patients: A systematic review and meta-analysis. BMJ Support. Palliat. Care 2020, 10, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.; Culpepper, J.S.; Blanco, R.; Crane, M.; Clarke, C.L.; Lin, J. Efficient and Effective Tail Latency Minimization in Multi-Stage Retrieval Systems. arXiv 2017, arXiv:1704.03970. [Google Scholar] [CrossRef]
- Kharitonova, K.; Pérez-Fernández, D.; Gutiérrez-Hernando, J.; Gutiérrez-Fandiño, A.; Callejas, Z.; Griol, D. Leveraging Retrieval-Augmented Generation for Reliable Medical Question Answering Using Large Language Models. In International Conference on Hybrid Artificial Intelligence Systems; Springer Nature: Cham, Switzerland, 2024; pp. 141–153. [Google Scholar]
- Ahire, P.R.; Hanchate, R.; Kalaiselvi, K. Optimized Data Retrieval and Data Storage for Healthcare Applications. In Predictive Data Modelling for Biomedical Data and Imaging; River Publishers: Gistrup, Denmark, 2024; pp. 107–126. [Google Scholar]
- Paulson, D.; Cannon, B. Auditing and Logging Systems for Privacy Assurance in Medical AI Pipelines. Preprint 2025.
- Rojas, J.C.; Teran, M.; Umscheid, C.A. Clinician trust in artificial intelligence: What is known and how trust can be facilitated. Crit. Care Clin. 2023, 39, 769–782. [Google Scholar] [CrossRef]
- Atf, Z.; Safavi-Naini, S.A.A.; Lewis, P.R.; Mahjoubfar, A.; Naderi, N.; Savage, T.R.; Soroush, A. The challenge of uncertainty quantification of large language models in medicine. arXiv 2025, arXiv:2504.05278. [Google Scholar] [CrossRef]
- Wang, G.; Ran, J.; Tang, R.; Chang, C.Y.; Chuang, Y.N.; Liu, Z.; Hu, X. Assessing and enhancing large language models in rare disease question-answering. arXiv 2024, arXiv:2408.08422. [Google Scholar] [CrossRef]
- Neehal, N.; Wang, B.; Debopadhaya, S.; Dan, S.; Murugesan, K.; Anand, V.; Bennett, K.P. CTBench: A comprehensive benchmark for evaluating language model capabilities in clinical trial design. arXiv 2024, arXiv:2406.17888. [Google Scholar] [CrossRef]
- Ting, L.P.Y.; Zhao, C.; Zeng, Y.H.; Lim, Y.J.; Chuang, K.T. Beyond RAG: Reinforced Reasoning Augmented Generation for Clinical Notes. arXiv 2025, arXiv:2506.05386. [Google Scholar]
- Rector, A.; Minor, K.; Minor, K.; McCormack, J.; Breeden, B.; Nowers, R.; Dorris, J. Validating Pharmacogenomics Generative Artificial Intelligence Query Prompts Using Retrieval-Augmented Generation (RAG). arXiv 2025, arXiv:2507.21453. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).