Abstract
“Hiding in Plain Sight” (HIPS) strategies for Personal Health Information (PHI) replace PHI with surrogate values to hinder re-identification attempts. We evaluate three different HIPS strategies for PHI replacement, a standard Consistent replacement strategy, a Random replacement strategy, and a novel Markov model strategy. We evaluate the privacy-preserving benefits and relative utility for information extraction of these strategies on both a simulated PHI distribution and real clinical corpora from two different institutions using a range of false negative error rates (FNER). The Markov strategy consistently outperformed the Consistent and Random substitution strategies on both real data and in statistical simulations. Using FNER ranging from 0.1% to 5%, PHI leakage at the document level could be reduced from 27.1% to 0.1% and from 94.2% to 57.7% with the Markov strategy versus the standard Consistent substitution strategy, at 0.1% and 0.5% FNER, respectively. Additionally, we assessed the generated corpora containing synthetic PHI for reuse using a variety of information extraction methods. Results indicate that modern deep learning methods have similar performance on all strategies, but older machine learning techniques can suffer from the change in context. Overall, a Markov surrogate generation strategy substantially reduces the chance of inadvertent PHI release.
1. Introduction
The removal of Personal Health Information (PHI) from medical records to prevent patient re-identification is driven by privacy considerations, legal mandates or other ethical imperatives. Synthetic text may be useful in some circumstances [1,2,3], but real text typically serves as the ground truth for evaluation even when synthetic text is used for training. The removal of PHI in the healthcare context is termed “de-identification”. In the United States, the governing legal framework for privacy in healthcare is the Health Insurance Portability and Accountability Act (HIPAA), which under section 164.512 of the Privacy Rule provides both the standard and implementation guidelines for two methods of de-identification. The “safe harbor” de-identification method specifies which data elements to remove. Although these elements may be targeted by de-identification software [4], this software may not replace identified PHI with realistic surrogate text. For instance, redacted text may be replaced by the category label of the PHI removed and not resemble the original text, potentially making it more difficult to train machine learning algorithms where training and test data should be similar. The challenge of creating high quality de-identified synthetic text for use with machine learning may lead even large, multi-national corporations to skip the process entirely, leading to PHI data leaks [5]. Most patient re-identification occurs when data is not de-identified according to existing standards [6].
1.1. Replacement Text in Existing Software
Software has been developed to de-identify clinical text; however, cost, availability, and licensing restrictions have limited public evaluations to only a subset of software [7,8]. Software evaluated has included Amazon’s Comprehend Medical [9], CliniDeID [10,11], National Library of Medicine’s Scrubber [12], NeuroNER [4] and MITRE Identification Scrubber Toolkit [13,14], Massachusetts Institute of Technology de-identification software [15] and Emory Health Information DE-identification (HIDE) software [16]. However, most de-identification software does not provide the option to output realistic replacement text, which is unfortunate because PHI identification at detection time could suggest a more appropriate replacement. Some exceptions to this include CliniDeID, Stanford’s HIPS [17] and nference De-Id [18]. CliniDeID uses entity-specific randomly selected values that are applied consistently across the record as surrogates, HIPS [17] uses curated lists to generate realistic replacements and nference De-Id [18] uses a more complex strategy including applying “entity-specific rules and heuristics to improve the fidelity of the surrogate” including pronoun and ethnicity adjustments. A summary of this software is shown in Table 1.

Table 1.
Comparison of de-identification tools by surrogate substitution strategy and open-source availability. Simple indicates placeholder-style substitution (e.g., [NAME]), Label indicates structured identifiers (e.g., [PATIENT_ID_001]), Consistent indicates that a given PHI entity is always replaced with the same realistic surrogate. Markov indicates the realistic surrogate replacement using a Markov-chain strategy described here in the Section 2. A ✓ indicates the presence of that feature and a ✗ indicates its absence.
1.2. Single Occurrence Re-Identification Vulnerability in Existing HIPS Replacement Methods
A recent systematic review [22] of different de-identification methods indicates that contemporary machine learning and hybrid approaches can yield binary token-level F1-scores of over 98%. Although this number may appear high at first glance, the high occurrence of PHI in a sufficiently realistic and large corpus will still generate false negatives for HIPAA-covered entities, thereby leaking PHI. Realistic surrogate text generation strategies can address the “residual PHI problem” [23] by limiting the ability of human readers to identify approximately 70% of PHI in clinical text [24]. This “hiding-in-plain sight” (HIPS) [13] strategy is based on similarity between the original text and the synthetic replacement, making the leaked PHI inconspicuous. Thus, a HIPS approach is preferable from a re-identification perspective versus simply replacing PHI with the detected entity type, a common default output of PHI Named Entity Recognition (NER) software and used in the widely distributed MIMIC corpus. However, existing HIPS implementations consistently replace the PHI so that the patient or provider name is always replaced by the same surrogate value. The advantage of this consistent replacement strategy is the provisioning of more realistic text, the disadvantage is that the presence of a single critical missing personal identifier (such as a patient name) gives an attacker the ability to identify the patient, even without complex statistical analysis. Thus, a HIPS corpus utilizing a consistent strategy of surrogate replacement has the same “Single Occurrence Re-identification Vulnerability” (SORV) as PHI replaced by entity names. This vulnerability could expose two-thirds of the leaked PHI in an attack utilizing machine learning; therefore, further improvements to ensure privacy protection are warranted [24] given the increasing prevalence of machine learning. Another machine learning approach that HIPS is vulnerable to are “parrot attacks”, where an attacker employs a similar annotation and de-identification method on the target text as the de-identification corpus provider. The attacker then examines any false negatives to infer that PHI has been missed [25]. A HIPS approach that removed the SORV would be preferable.
1.3. Need for Improved HIPS Strategies
The increasing power of Large Language Models (LLMs) can render traditional HIPS strategies (“Consistent” substitution) less effective for PHI leakage. Using a similar strategy to ours, Patsakis and Lykousas [26] used the Faker library to perform a synthetic substitution of PHI to use surrogates for 11 identifiers for short snippets of text describing 20 famous celebrities. Each celebrity had 46 to 61 descriptions, with a mean count of 54 descriptions. Initially, GPT-3 [27] could identify 72.6% of celebrity descriptions, but none when celebrities were de-identified using a “Consistent” strategy. However, simple prompt engineering allowed for the identification of 69.44% of the descriptions, indicating that a “Consistent” HIPs strategy is not sufficient for the protection of PII/PHI when using LLMs. Recent advances in LLM are likely to exacerbate this situation.
1.4. Contribution: Evaluation of Novel HIPS Strategies
In this paper, we implement and evaluate novel HIPS surrogate replacement strategies in which PHI mentions are replaced by surrogate text without the “traditional” constraint that such a replacement must be consistent with previous replacements of the same type. For example, each mention of a patient name in a document could be replaced by a randomly selected name in our “Random” strategy or by a name selected from a Markov chain in our “Markov” strategy. We implement these two strategies and statistically quantify their privacy preserving benefits relative to the traditional “Consistent” surrogate replacement strategy. To our knowledge, no statistical evaluation has been done on these PHI substitution strategies using either synthetic or realistic clinical data sets and we provide the first such implementation. Finally, we quantify the impact of these substitution strategies on selected information extraction tasks on three different data sets, to determine the extent to which these strategies may impair the utility of the text for information extraction tasks since. This is because although the effect of surrogates is believed to be minimal “where identifiers are not crucial”, it is otherwise believed to have “a great impact on data utility” [28], although this has not been explored.
2. Materials and Methods
2.1. Software Implementation
We make our tool (version 0.4) and source code publicly available at https://github.com/uabnlp/BRATsynthetic (accessed on 6 September 2025). BRATsynthetic uses the BRAT [29] annotation format and the associated text file (if available) to create surrogate PHI independent of the software originally used to identify that PHI. BRATsynthetic generates realistic text for 27 categories of PHI as described in the Section 2.2.1 below. Dates and ages are offset by a random number. PHI categories are stored as entities (“T” labels) in BRAT files; associated, non-PHI annotations (attributes “A” and events “E”) may also be stored in these files, and BRATsynthetic properly maintains mapping of non-PHI annotations to PHI categories after synthetic replacements. Prior to replacement, we normalize line endings, require paired .txt/.ann files (example Figure 1 and Figure 2), restrict substitutions to the PHI tag set, and update spans in reverse-offset order to preserve alignment. Synthetic values are produced with Faker 13.7 using a defined seed from the configuration, with format and case-preserving rules for IDs, names, and codes. Each PHI category uses a custom Maker class with unique rules for handling regular expression patterns, edge cases, and Faker function calls. Within-document mention repetition follows a two-state first-order Markov policy (reuse vs. resample) as shown in Figure 3. Automated and manual verification includes counting the number of entities, events, and attributes before and after synthetic replacement, and ensuring spans correctly align for non-PHI annotations with their corresponding synthetically replaced PHI categories. The header text, “PERSONAL IDENTIFYING INFORMATION (PHI) IN THIS DOCUMENT HAS BEEN REPLACED WITH SYNTHETIC TEXT. ANY CORRESPONDENCE TO THE PHI OF ANY REAL PERSON IS UNINTENTIONAL” is prefixed to the start of the file to reduce the risk of potential legal issues associated with the unintentional match to real persons that can occur in all HIPS strategies.
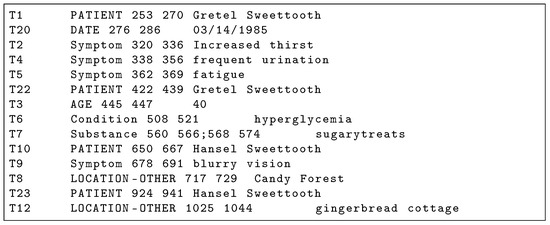
Figure 1.
BRAT Annotation .ann file corresponding to Figure 2 clinical note. PHI annotations are capitalized.

Figure 2.
Example synthetic clinical note with identified PHI highlighted in yellow. PHI leakage results from false negatives in the de-identification software, an example of which is the MRN highlighted in the black box. Hansel and Gretel fairytale inspired synthetic clinical note was generated using Microsoft Copilot.
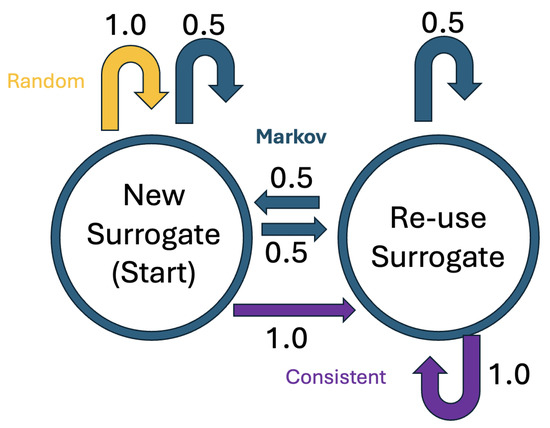
Figure 3.
State transition diagram for HIPS replacement strategies.
2.2. Evaluation Corpora
To assess the impact of surrogate substitution strategy on privacy protection, we evaluated BRATsynthetic on an in-house corpus of de-identified clinical text from 165 patients collected under IRB Protocol #300002331 (NIH Grant R01AG060993-03S1, Automating Delirium Identification and Risk Prediction in Electronic Health Records). Those notes were de-identified under IRB Protocol #300002212 “U-BRITE Deidentified Translational Data Repository for Research and Education” at the University of Alabama at Birmingham that we previously used [30] to generate .ann files as input for BRATsynthetic. The corpus includes all available EHR notes for those patients, specifically 3617 documents including 1,489,362 critical entities from both inpatient and outpatient encounters from 2014 to 2021. The UAB Discharge corpus is a subset of the main UAB Corpus containing only discharge summaries. The MIMIC Corpus is a set of MIMIC discharge summaries that was obtained as part of participation in SemEval 2014 Task 7 “Analysis of Clinical Text” [31] and are sourced from outside UAB [32]. The Opioid Use Disorder (OUD) Corpus [33] is a UAB-derived data set consisting of 3295 clinical notes from 59 patients (23 controls) from physician case referrals between 2016 and 2021. These data sets contain human-annotated BRAT files and are used to evaluate changes in the information extraction performance of the different HIPS strategies, specifically if a particular strategy uniquely affects document structure and downstream language model evaluation tasks.
2.2.1. Resynthesis Elements
BRATsynthetic generates a superset of i2b2 2014 personal health information entity types [34], including standard HIPAA Safe Harbor Categories, as well as more specific category types under the BRATsynthetic “Replacement Type” column in Table 2. There is a many-to-many correspondence, where some data elements map to multiple BRATsynthetic types (ex. NAMES), others are a one-to-one mapping (ex. EMAIL), and others condense different Safe Harbor Elements into a single BRATsynthetic type (ex. IDNUM). Table 3 summarizes this and other label differences between Safe Harbor Elements and BRATsynthetic categories. BRATsynthetic does provide additional resynthesis capacity by the inclusion of the PROFESSION category (shared with I2B2) and the TIME category (not required either under HIPAA Safe Harbor or used by the I2B2), which we include due to the increasing availability of second and sub-second data collected from personal devices not anticipated in the original guidelines. BRATsynthetic currently lacks UNIQUE text resynthesis ability, where UNIQUE corresponds to a section of text that could potentially reveal personal identity such as “governor’s wife” or details of unusual accidents.

Table 2.
BRATsynthetic safe harbor element replacement.

Table 3.
Differences between safe harbor elements and BRATsynthetic.
2.2.2. Surrogate Substitution Strategies
BRATsynthetic implements three HIPS substitution strategies: Consistent, Random, and Markov as well as a baseline “Simple” non-HIPS strategy, where identified PHI entities are replaced by the entity name. An example substitution is shown in Table 4. All three HIPS strategies are implemented as a Markov chain with two states, one that creates a new surrogate value and another that repeats the previous surrogate value. The traditional Consistent strategy can be considered a simple, degenerate Markov chain with self-transition probability of 1.0. The initial state always creates a new surrogate value, but the subsequent transitions vary between strategies, as shown in Figure 3. The Consistent strategy always maintains the previous surrogate value, the Markov strategy selects a new surrogate half the time, and the Random strategy always selects a new surrogate. The surrogate values are selected at random from the faker [35] library for each occurrence of each type of entity.

Table 4.
Substitution strategies in BRATsynthetic. All are HIPS strategies except “Simple”. MSRS refers to the “Maximum Surrogate Repeat Size” and NA means Not Applicable.
2.2.3. Maximum Surrogate Repeat Size (MSRS)
The “Single Occurrence Re-identification Vulnerability” (SORV) occurs with the Consistent strategy of surrogate generation because the use of the same replacement value means that a single occurrence of a false negative (FN) will be identifiable as PHI. However, should an FN occur once in the de-identified document for that entity type (leaving in the patient’s real name for example), it may not be immediately obvious with the Random strategy that the real name was not just another surrogate value. This is true when the number of FN errors of that critical type is lower than the maximum number of times the same surrogate value is repeated when more than one surrogate value is used. We refer to this latter value as the “Maximum Surrogate Repeat Size” (MSRS). The Markov substitution strategy operates in the same way as the Random strategy, but previously used surrogate values are much more likely to occur due to the 0.5 state transition probability. This generates a larger maximum number of times a surrogate value is repeated (larger MSRS) for each critical entity so it can better “mask” FN errors. A sample run of these strategies is shown on an entity note with six names in Table 4. BRATsynthetic applies substitutions at the document level, a patient level replacement strategy operating over multiple documents is also desirable, but not currently implemented.
2.3. PHI Leakage Evaluation on Real Corpora
We evaluate the protective ability of surrogate addition on the UAB Corpus, UAB Discharge Corpus, and the MIMIC Corpus by focusing on “critical” PHI as specified in Table 2 under the “Critical PHI” column. Table 5 provides corpus statistics on the critical entity distributions of these corpora. We define an entity as “critical” if a single FN for that entity (MRN is one such example) is sufficient to identify a patient and require immediate legal disclosure of the breach. An FN by de-identification software or humans in this critical category of PHI may be commonly identified even in a haphazard fashion by a casual reader of the document when the document employs no surrogate substitution (like MIMIC) or uses substituted text as shown in Table 4 under the Consistent strategy. However, a single FN would not be obvious to a reader if a Random or Markov strategy is used, since the real name would not constitute the largest minority surrogate class. Thus, the ability to detect the real name as shown in Table 4 is dependent on the overall FNER, the distribution of critical entities and FN errors within those entities and the substitution strategy used. Using our UAB and MIMIC Corpora, we simulate and assess three different resynthesis strategies for surrogate information: “Consistent”, “Random”, and “Markov” under four different FNER (0.1%, 0.5%, 1%, and 5%). Leakage rates are computed at the document level. A leak is considered to occur if the PHI signal (measured by the critical PHI FN count) exceeds the injected noise as measured by MSRS. In essence, MSRS represents the maximum frequency with which a given surrogate can repeat within a document; when the number of false negatives does not exceed this number, any leaked PHI is statistically indistinguishable from synthetic surrogates. In practice, this means that for the Consistent strategy where the same surrogate is repeated, any FN in a critical entity constitutes a document leak, whereas for the Random, and Markov strategies this only occurs when FN errors exceed the MSRS. A leak at the patient level as one or more document leaks. Since results can vary depending on the distribution of FN errors in a corpus, we average our results over 1000 FN error generation simulations using different random seeds. Our leakage calculations and simulations are available at https://github.com/uabnlp/BRATsynthetic (accessed on 6 September 2025) [21] and were performed using Python version 3.9.

Table 5.
Critical entity distribution by corpus.
2.4. PHI Leakage Evaluation on Simulated Corpora
We calculated the expected leakage probability of critical information over a range of corpus sizes, ranging from 10 to 10,000 documents. For this calculation, we considered a leak to occur if the expected number of unremoved (false negative) PHI entities in the corpus was greater than a set threshold for each of the substitution methods. For Consistent substitution, the threshold was set at 0, since even a single instance of a real patient entity can be readily distinguished from the single fake entity. For Random and Markov substitution, the threshold was set as the expected number of fake entities [36] based on a pool of 1000 fake entities from which to randomly select using the transition probabilities described above. The threshold for a Random substitution was calculated as 1.015 and for a Markov substitution it was 2.028. We allowed the number of entities per document to assume the values of 15, 150, and 1500, and allowed the FNER to assume the values of 1% and 5%. We then calculated the expected number of real patient entities in the corpus as the product of the FNER times the number of entities per document times the number of documents. We used the binomial distribution to determine the p-value of whether the expected number of real patient entities exceeded the threshold. Calculations were performed using R v4.1.2.
2.5. Assessment of HIPS Strategy on Information Extraction Efficacy
As shown in Table 4, the Random and Markov strategies generate text that differs from the original text or a Consistent HIPS strategy. This can potentially cause problems for Natural Language Processing (NLP) algorithms seeking to extract information from corpora generated using these substitution strategies. To assess the impact of this, we compare the relative performance of the different strategies on a variety of information extraction NLP tasks using different generations of NLP software. This includes the popular MedSpacy [37] software, a third-party evaluation by TriNetx that utilized a Support Vector Machine (SVM) [38] based genomic marker pipeline from Averbis and a BioBERT [39]-based encoder models for Named Entity Recognition (NER) and clinical modifiers from our prior work [40]. While it is not feasible to test all NLP tasks, the clinical modifier pipeline was selected as likely to be particularly susceptible to PHI context, especially for the “Subject” modifier, which evaluates whether an extracted Named Entity is associated with the patient or someone else, a task made more challenging when the surrogate name for the patient may change in the same document. For the MedSpacy [37] evaluation, we used the “en_core_sci_sm” model on the OUD corpus with default parameters for all tasks but “Spans”, which utilizes MedSpacy custom “TargetRules” to identify OUD entities with regular expressions. Sentences were segmented with PyRuSH, a python implementation of RuSH [41]. We computed Jenson–Shannon divergence [42] to compare distributions. The Averbis genomic marker pipeline utilized by TriNetX, first tokenizes the input text, then performs dictionary lookup for mention finding, disambiguates with a linear Support Vector Machine (SVM) that considers the context, applies rules for specific patterns, and finally performs the final classification with a linear SVM. Evaluation on the genomic marker pipeline was done on a set of 144 documents, and the results are reported and errors analyzed only for the five documents where results differed. For the BERT-based evaluation, we replicated our previous work [40], but replaced PHI using Simple replacement or one of the three HIPS strategies. We used Shapiro–Wilks tests to examine the normality of the mean F1 scores, with p > 0.05 indicating that the scores for Consistent, Markov, Random, and Simple are from the same normal distribution and are not significantly different from results for the original text.
2.6. BRATsynthetic Runtime Experiment
Runtime performance was evaluated on a superset of the UAB corpus consisting of 28,547 documents that have been de-identified using a BERT [43] based de-identification tool [44], but have not been manually reviewed. Corpus statistics and experimental parameters are in Table 6.

Table 6.
BRATsynthetic Runtime Statistics.
3. Results
3.1. Surrogate Replacement Strategy: PHI Leakage Assessment
Figure 4 shows the Markov strategy shows a lower document leakage rate on the UAB corpus. At a lower FNER (1% or less) its impact is more pronounced, with leakage eliminated at the lowest error rate. Even at a 5% error rate the strategy still performs substantially better than the two other strategies, which leak PHI close to 100% of the time. Figure 5 compares PHI leakage between the UAB and MIMIC corpus at the document level for discharge summaries only and indicates a similar substitution strategy performance between the two corpora, but dramatically lowers PHI leakage rates for the MIMIC corpus which contains significantly fewer critical PHI entities per discharge summary relative to UAB. The Markov substitution also has the highest performance on the synthetic corpora in Figure 6 and Figure 7.
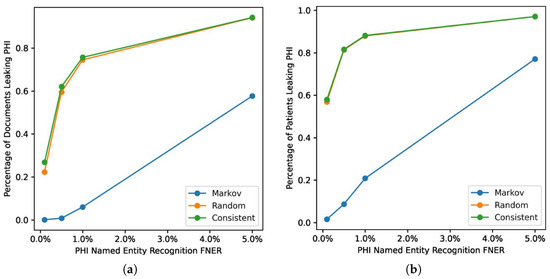
Figure 4.
Impact of surrogate substitution strategies on PHI leakage on UAB corpus. This shows the computed leakage rate for (a) document level and (b) patient level.
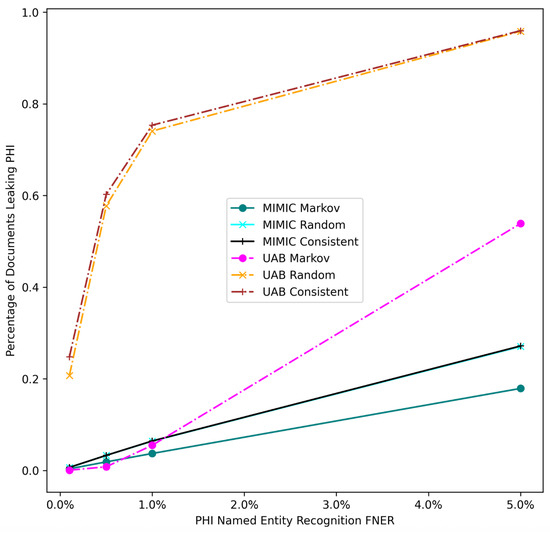
Figure 5.
UAB and MIMIC corpus PHI document leakage. FNER is the false negative error rate.
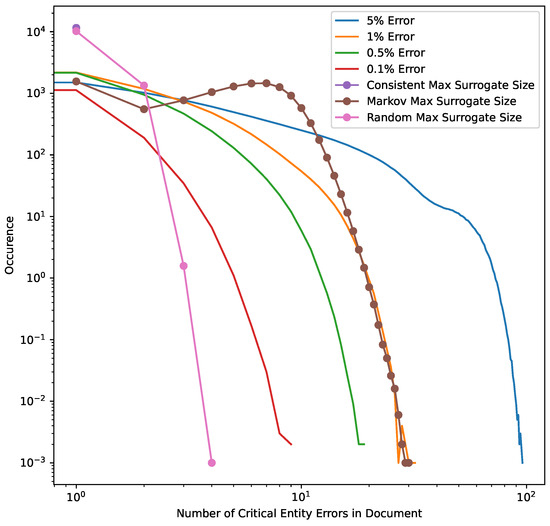
Figure 6.
Model error masking simulation on UAB corpus. The distribution of errors across 1000 simulations, superimposed over the distribution of the MSRS for all three HIPS substitution strategies. Consistent substitution always reapplies the same surrogate replacement value for each class of critical PHI, so any false negatives are detectable. The Random substitution strategy allows for same surrogate replacement, but since the critical entity surrogate pool size is set at 1000 almost all entities use a unique surrogate. Thus, for Random the MSRS is usually 1, but it some cases it ranges from 2 to 4. The Markov strategy utilizes a state transition probability of 0.5 and so will generate a higher MSRS and generally “cover” error rates of 1% or less in our data set.
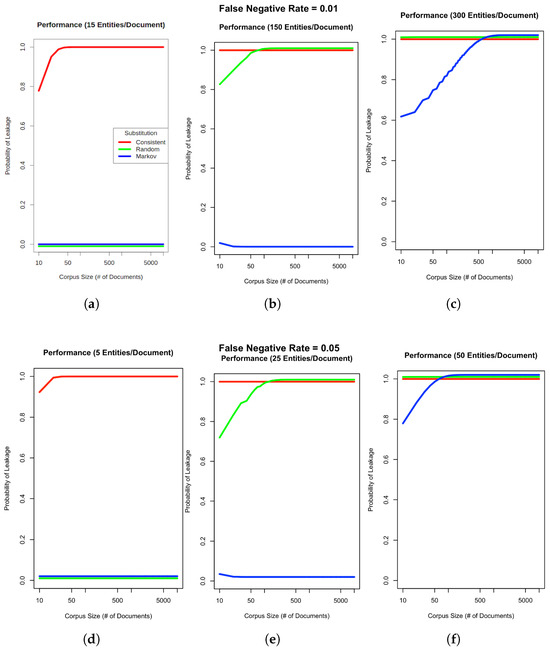
Figure 7.
PHI leakage on synthetic corpora. Charts (a–c) show the probability of PHI leaking given a 1% PHI FNER and varying input corpus sizes with either 15, 150, or 300 entities (critical or otherwise) per document. Charts (d–f) assume a higher FNER of 5% with 5, 25, or 50 entities per document. Although leakage results are reported at the document level, patient level calculations would be the almost same but would use a pooled entity count given that multiple documents from the same patient would be aggregated. Red, green and blue lines indicate Consistent, Random and Markov substitution respectively.
3.2. Surrogate Replacement Strategy: Effect on Information Extractions Tasks
Our results on the OUD dataset (Table 7) indicate that for context sensitive information extraction tasks, an encoder model fine-tuned on the target data set is not impacted by the surrogate substitution strategy. This is also true for most other information extraction tasks (Table 8), including entity extraction, even when the entity extraction model is not fine-tuned for the target dataset as is the case for MedSpacy. However, MedSpacy rule-based context prediction was impacted (1st row of Table 8) as was the SVM-based Averbis genomic marker pipeline as shown in Table 9. However, all MedSpacy divergences were bits, confirming negligible distributional change. A sample of errors for MedSpacy POS-tagging and dependency parsing are shown in Table 10 and Table 11 respectively. In general, the differences between the output of different surrogate substitution strategies did not correspond to our human intuition of the expected error types, as conceptually insignificant changes in numbers for ages and dates could result in unanticipated changes in the POS tag or the dependency class.

Table 7.
Clinical modifier extraction macro F1 score on the OUD dataset with a fine-tuned BioBERT pipeline. NER results are reported based on partial matches. None indicates identified text.

Table 8.
HIPS strategy Jensen–Shannon divergence [bits] on the OUD dataset using standard MedSpacy. We provided the Entity identification framework with a custom set of OUD-specific rules.

Table 9.
Averbis genomic marker pipeline genomic marker counts and error analysis for five OUD documents where results differed from the original data. A red text color indicates any hypothesized surrogate-induced error we could identify. Note that the MRN 1324616 shown in the Consult Note is synthetic and not a real patient MRN. Abbreviations for HIPS substitution strategies are Consistent (Cons), Random (Rand), and Markov (Mark).

Table 10.
Universal Part of Speech tagset [45] comparison of two examples mentioned, bold text indicates for that portion of the mention text indicates the dependencies for that text are shared by all substitution methods. POS tags are color-coded: PROPN, NOUN, NUM.

Table 11.
Comparison between incoming dependencies for 2 example mentions, bold text indicates for that portion of the mention text indicates the dependencies for that text are shared by all substitution methods. Dependencies are color-coded: CONJ, APPOS, NUMMOD, Compound, PUNCT, ADVMOD, NPADVMOD, ROOT.
4. Discussion
4.1. PHI Leakage Replacement Strategy Evaluation
Our results indicate that a Markov replacement strategy can significantly reduce the chance of PHI leakage relative to a corpus released without substitution or using a traditional Consistent substitution strategy. This effect is seen in simulated PHI distributions (Figure 7) as well as real data from the UAB Discharge and MIMIC corpora (Figure 5), with the effect much more pronounced in the MIMIC corpora, which has almost no leakage at lower error rates. This discrepancy is due to the lower number of critical PHI entities contained in MIMIC corpus versus UAB Discharge. Since the release of our BRATsynthetic tool [46] and preprint [47], Simancek and Vydiswaran [48] have shown the effectiveness of a Markov chain-based surrogate replacement strategy using a Longformer BERT model for clinical text de-identification on a set of 400 discharge summaries. In this real-world data set, the average name leakage rate decreased from 13.1% with random replacement to 3.8% with Markov replacement of surrogates. In both real-world and simulated data the Markov substitution strategy is effective because it adds a variable amount of noise to a document data set (see Figure 6), at levels exceeding any original PHI signal that may not have been removed by the de-identification process. This shape of the Markov substitution strategy curve in Figure 5 is controlled by the state transition rate, which determines how often a surrogate is repeated. This could be adjusted in future work to reflect the expected number of FN errors in any entity to better conceal PHI leakage.
4.2. Larger Corpus Size
Our work suggests that the Markov substitution strategy allows for a larger release corpus size with the same or lower PHI leakage risk relative to Consistent or Random substitution strategies. This is true for a range of PHI entity counts and FNERs, as shown in Figure 7. Our results in Figure 4, Figure 5 and Figure 6 focus on critical PHI, but the protective effect shown in Figure 7 applies to any PHI entity. We evaluated critical PHI because of the difficulty in assessing the impact or liability of inadvertent non-critical PHI release, similar to other work that focused on names only [48].
4.3. HIPS Strategy vs NER for PHI Protection
Our results in Figure 4 for real-world data and Figure 7 for simulated data show that the implementation of an effective surrogate replacement strategy using Markov models can have a greater impact on patient privacy protection than marginal gains in PHI NER performance provided by additional training data. For example, Dernoncourt et al. [4] show increased performance for artificial neural network and conditional random field de-identification algorithms with additional training data, but the total improvement as measured by F1 score shows that the maximum improvement difference was only about 2% between using 5% of available training data versus using all available training data for the MIMIC dataset. Our results indicate that the surrogate replacement strategy can have a greater impact in protecting PHI compared to even a 4% improvement in recall (from 5% to a 1%) in PHI NER software, depending on the total amount and distribution of PHI.
4.4. Impact on Information Extraction Tasks
Our results in Section 3.2 show that common information extraction tasks are not significantly impaired by the use of non-traditional PHI substitution strategies such as Markov or Random substitutions. The only clear exception to this is the MedSpacy “Context” task that includes the determination of the subject of a medical condition, something which Random or Markov replacement could interfere with. However, this was not replicated with the fine-tuned BioBERT used to generate results for Table 7, suggesting that this result may be more a limitation of the MedSpacy model.
4.5. Comparison to LLM-Based De-Identification Methods
LLM-based de-identification methods can produce contextually natural text, often exceeding template-driven systems in generation quality. However, they are computationally intensive, more expensive to operate and raise concerns about auditability and potential privacy leakage if safeguards are not rigorously applied. BRATsynthetic, on the contrary, is an inexpensive, efficient, and transparent deterministic surrogate substitution without specialized hardware, although at the cost of a more constrained narrative variety. In practice, these approaches should be viewed as complementary, with LLMs excelling in realism and adaptability, while BRATsynthetic offers practical scalability and predictable control, and its relative weakness in creating smooth transitions is less problematic in clinical documentation, where narratives are typically fragmented.
4.6. Limitations
BRATsynthetic uncouples surrogate generation from PHI NER and can thus leverage any independent improvements in PHI NER. The tradeoff is that PHI NER software is best positioned to recognize an appropriate substitution (such as one that maintains non-standard date formatting or even typos) whereas BRATsynthetic relies on PHI categories or its own regular expression-based interpretation of the text (if available) to generate the surrogate.
In assessing the impact of non-traditional replacement strategies for re-use, we sample only a limited set of task and datasets. However, we selected a clinical modifier task particularly well suited for the evaluation of surrogate changes, since it incorporates modifiers for Subject and DocRelTime whose mentions would be modified heavily in a Random or Markov corpus.
Both the faker library and BRATsynthetic are open source code, which could potentially assist an attacker since some synthetic surrogates are generated from lists including patient and location names. This vulnerability will be mitigated by private, user-provided pools of PHI for use as replacement values in an updated version of BRATsynthetic. Other forms of surrogate values, such as dates and randomly generated identifiers consisting of alphanumeric text, do not have this vulnerability. Finally, we have not performed a complexity or runtime analysis on the faker library, but scaling on large datasets is feasible, as shown in Table 6.
5. Conclusions
We show that a Markov substitution strategy can reduce PHI leakage on two real world corpora and on synthetic PHI distributions using a range of realistic false negative PHI NER rates and entity distributions and make our software freely available. Although our work here shows the theoretical and statistical utility of the Markov-based surrogate substitution method, further evaluation of the software implementation on a wider range of benchmarks including human validation would be useful, as would an assessment against more complex re-identification attempts including parrot [25] and LLM-based attacks [26].
Author Contributions
J.D.O. wrote the first draft of the paper, M.D.C. converted the paper to LATEX from Word and all authors assisted with critical revisions and writing of the manuscript for important intellectual content. BRATsynthetic was updated with contributions from T.O., M.D.C., J.D.O., A.T., A.N., A.M. and C.C. Surrogate analysis was performed by A.T., A.I.A., L.M.-G., R.E.K. and J.D.O., S.M.A., A.T. and R.E.K. performed statistical analysis. Funding provided by J.R.C., J.D.O. and R.E.K. All authors have read and agreed to the published version of the manuscript.
Funding
We acknowledge the support of 5P30AR072583 “Building and InnovatinG: Digital heAlth Technology and Analytics (BIGDATA)” (software development), and 5R01AG060993-03S1 “Automating Delirium Identification and Risk Prediction in Electronic Health Records (Supplement)” and the Alabama Department of Mental Health OUD Center of Excellence for the generation of the de-identified corpora.
Institutional Review Board Statement
This study was approved by the Institutional Review Board (IRB) at the University of Alabama at Birmingham (UAB). Clinical notes from UAB patients were collected under IRB Protocol #300002331 and #121114001 Using Text Mining to Extract Information from Text Documents in the Electronic Health Record. Those notes were de-identified under IRB Protocol #300002212 “U-BRITE Deidentified Translational Data Repository for Research and Education”.
Informed Consent Statement
Patient consent was waived due to the minimal risk to patients since all de-identification work took place within secure servers designated by the UAB Health System as HIPAA compliant.
Data Availability Statement
The deidentified Delirium and Opiate Use Disorder (OUD) dataset will be made available to interested parties who wish to replicate results with a Data Use Agreement (DUA) prohibiting distribution and re-identification attempts.
Acknowledgments
We would like to thank Ahana Chatterjee and Mojisola Fasokun for de-identification of the delirium corpus, the FAKER project which BRATsynthetic leverages for surrogate generation and Maio Danila for assistance with editing the paper. Finally, we would like to thank TriNetX, including Garo Stone-DerHagopian and his team for running the Averbis genomic pipeline.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Li, J.; Zhou, Y.; Jiang, X.; Natarajan, K.; Pakhomov, S.V.; Liu, H.; Xu, H. Are synthetic clinical notes useful for real natural language processing tasks: A case study on clinical entity recognition. J. Am. Med. Inform. Assoc. 2021, 28, 2193–2201. [Google Scholar] [CrossRef]
- Kumichev, G.; Blinov, P.; Kuzkina, Y.; Goncharov, V.; Zubkova, G.; Zenovkin, N.; Goncharov, A.; Savchenko, A. Medsyn: Llm-based synthetic medical text generation framework. In Proceedings of the Joint European Conference on Machine Learning and Knowledge Discovery in Databases; Springer: Cham, Switzerland, 2024; pp. 215–230. [Google Scholar]
- Hiebel, N.; Ferret, O.; Fort, K.; Névéol, A. Clinical Text Generation: Are We There Yet? Annu. Rev. Biomed. Data Sci. 2025, 8, 173–198. [Google Scholar] [CrossRef] [PubMed]
- Dernoncourt, F.; Lee, J.Y.; Uzuner, O.; Szolovits, P. De-identification of patient notes with recurrent neural networks. J. Am. Med. Inform. Assoc. 2016, 24, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Schneble, C.O.; Elger, B.S.; Shaw, D.M. Google’s Project Nightingale highlights the necessity of data science ethics review. EMBO Mol. Med. 2020, 12, e12053. [Google Scholar] [CrossRef]
- Emam, K.E.; Jonker, E.; Arbuckle, L.; Malin, B. A Systematic Review of Re-Identification Attacks on Health Data. PLoS ONE 2011, 6, e28071. [Google Scholar] [CrossRef] [PubMed]
- Heider, P.M.; Obeid, J.S.; Meystre, S.M. A Comparative Analysis of Speed and Accuracy for Three Off-the-Shelf De-Identification Tools. AMIA Summits Transl. Sci. Proc. 2020, 2020, 241–250. [Google Scholar]
- Steinkamp, J.M.; Pomeranz, T.; Adleberg, J.; Kahn, C.E.; Cook, T.S. Evaluation of Automated Public De-Identification Tools on a Corpus of Radiology Reports. Radiol. Artif. Intell. 2020, 2, e190137. [Google Scholar] [CrossRef]
- Guzman, B.; Metzger, I.; Aphinyanaphongs, Y.; Grover, H. Assessment of Amazon Comprehend Medical: Medication Information Extraction. arXiv 2020, arXiv:2002.00481. [Google Scholar] [CrossRef]
- CliniDeID. 2022. Available online: https://github.com/Clinacuity/CliniDeID (accessed on 1 January 2025).
- Meystre, S.M.; Underwood, G.; Heider, P. CliniDeID: An Open Source Solution for Accurate Clinical Text De-Identification. Technical Report, ResearchGate. 2023. Available online: https://www.researchgate.net/profile/Stephane-Meystre/publication/371303503_CliniDeID_an_Open_Source_Solution_for_Accurate_Clinical_Text_De-Identification/links/647def6279a722376513593b/CliniDeID-an-Open-Source-Solution-for-Accurate-Clinical-Text-De-Identification.pdf (accessed on 1 June 2025).
- Kayallp, M.; Browne, A.C.; Dodd, Z.A.; Sagan, P.; McDonald, C.J. An Easy-to-Use Clinical Text De-identification Tool for Clinical Scientists: NLM Scrubber. 2015. Available online: https://www.researchgate.net/publication/319914511_An_Easy-to-Use_Clinical_Text_De-identification_Tool_for_Clinical_Scientists_NLM_Scrubber?channel=doi&linkId=59c193170f7e9b21a8265f57&showFulltext=true (accessed on 1 June 2025).
- Aberdeen, J.; Bayer, S.; Yeniterzi, R.; Wellner, B.; Clark, C.; Hanauer, D.; Malin, B.; Hirschman, L. The MITRE Identification Scrubber Toolkit: Design, training, and assessment. Int. J. Med. Inform. 2010, 79, 849–859. [Google Scholar] [CrossRef]
- MIST v2.04. Updated Version of MITRE MIST Tool. 2019. Available online: https://mist-deid.sourceforge.net/ (accessed on 1 June 2025).
- Douglass, M.; Clifford, G.; Reisner, A.; Moody, G.; RG, M. Computer-assisted de-identification of free text in the MIMIC II database. In Proceedings of the Computers in Cardiology, Chicago, IL, USA, 19–22 September 2004; pp. 341–344. [Google Scholar] [CrossRef]
- Gardner, J.; Xiong, L. HIDE: An Integrated System for Health Information DE-identification. In Proceedings of the 2008 21st IEEE International Symposium on Computer-Based Medical Systems, Jyvaskyla, Finland, 17–19 June 2008; pp. 254–259, ISSN 1063-7125. [Google Scholar] [CrossRef]
- Chambon, P.J.; Wu, C.; Steinkamp, J.M.; Adleberg, J.; Cook, T.S.; Langlotz, C.P. Automated deidentification of radiology reports combining transformer and “hide in plain sight” rule-based methods. J. Am. Med. Inform. Assoc. 2022, 30, 318–328. [Google Scholar] [CrossRef]
- Murugadoss, K.; Rajasekharan, A.; Malin, B.; Agarwal, V.; Bade, S.; Anderson, J.R.; Ross, J.L.; Faubion, W.A.; Halamka, J.D.; Soundararajan, V.; et al. Building a best-in-class automated de-identification tool for electronic health records through ensemble learning. Patterns 2021, 2, 100255. [Google Scholar] [CrossRef]
- Microsoft Presidio. Presidio: Data Protection and De-Identification SDK. Available online: https://microsoft.github.io/presidio/ (accessed on 1 June 2025).
- Neamatullah, I.; Douglass, M.; Lehman, L.H.; Goldberger, A. De-Identification Software Package v1.1 PhysioNet. 2007. Available online: https://doi.org/10.13026/C20M3F (accessed on 1 June 2025).
- Trotter, A.; O’Leary, T.; Cochran, M.D.; Coffee, C.; Nadimpalli, A.; Osborne, J.D. BRATsynthetic. 2025. Available online: https://github.com/uabnlp/BRATsynthetic (accessed on 20 September 2025).
- Kovačević, A.; Bašaragin, B.; Milošević, N.; Nenadić, G. De-identification of clinical free text using natural language processing: A systematic review of current approaches. Artif. Intell. Med. 2024, 102845. [Google Scholar] [CrossRef]
- Carrell, D.; Malin, B.; Aberdeen, J.; Bayer, S.; Clark, C.; Wellner, B.; Hirschman, L. Hiding in plain sight: Use of realistic surrogates to reduce exposure of protected health information in clinical text. J. Am. Med. Inform. Assoc. 2013, 20, 342–348. [Google Scholar] [CrossRef]
- Carrell, D.S.; Malin, B.A.; Cronkite, D.J.; Aberdeen, J.S.; Clark, C.; Li, M.R.; Bastakoty, D.; Nyemba, S.; Hirschman, L. Resilience of clinical text de-identified with “hiding in plain sight” to hostile reidentification attacks by human readers. J. Am. Med. Inform. Assoc. 2020, 27, 1374–1382. [Google Scholar] [CrossRef]
- Carrell, D.S.; Cronkite, D.J.; Li, M.R.; Nyemba, S.; Malin, B.A.; Aberdeen, J.S.; Hirschman, L. The machine giveth and the machine taketh away: A parrot attack on clinical text deidentified with hiding in plain sight. J. Am. Med. Inform. Assoc. 2019, 26, 1536–1544. [Google Scholar] [CrossRef]
- Patsakis, C.; Lykousas, N. Man vs the machine in the struggle for effective text anonymisation in the age of large language models. Sci. Rep. 2023, 13, 16026. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Jonnagaddala, J.; Wong, Z.S.Y. Privacy preserving strategies for electronic health records in the era of large language models. npj Digit. Med. 2025, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Stenetorp, P.; Pyysalo, S.; Topić, G.; Ohta, T.; Ananiadou, S.; Tsujii, J. brat: A Web-based Tool for NLP-Assisted Text Annotation. In Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics; Segond, F., Ed.; Association for Computational Linguistics: Avignon, France, 2012; pp. 102–107. Available online: https://aclanthology.org/E12-2021/ (accessed on 1 September 2025).
- Osborne, J.D.; Booth, J.S.; O’Leary, T.; Mudano, A.; Rosas, G.; Foster, P.J.; Saag, K.G.; Danila, M.I. Identification of Gout Flares in Chief Complaint Text Using Natural Language Processing. AMIA Annu. Symp. Proc. 2020, 2020, 973–982. [Google Scholar]
- Pradhan, S.; Elhadad, N.; Chapman, W.; Manandhar, S.; Savova, G. SemEval-2014 Task 7: Analysis of Clinical Text. In Proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014), Dublin, Ireland, 23–24 August 2014; pp. 54–62. [Google Scholar] [CrossRef]
- Johnson, A.; Bulgarelli, L.; Pollard, T.; Horng, S.; Celi, L.A.; Mark, R. *MIMIC-IV* (version 0.4). PhysioNet, 2020. RRID:SCR_007345. Available online: https://physionet.org/content/mimiciv/0.4/ (accessed on 1 September 2025).
- Almudaifer, A.I.; Feldman, S.S.; O’Leary, T.; Covington, W.L.; Hairston, J.; Deitch, Z.; Crisan, E.; Riggs, K.; Walters, L.; Osborne, J.D. Annotation of Opioid Use Disorder Entity Modifiers in Clinical Text. In MEDINFO 2023—The Future Is Accessible; IOS Press: Amsterdam, The Netherlands, 2024; pp. 1458–1459. [Google Scholar]
- Stubbs, A.; Uzuner, O. Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/UTHealth corpus. J. Biomed. Inform. 2015, 58, S20–S29. [Google Scholar] [CrossRef]
- Curella, F.; Faraglia, D. Faker Project. Available online: https://faker.readthedocs.io/en/master/ (accessed on 1 June 2025).
- Sigman, K. Expected number of Visits of a Finite State Markov Chain to a Transient State. 2016. Available online: http://www.columbia.edu/~ks20/4106-18-Fall/Notes-Transient.pdf (accessed on 1 January 2022).
- Eyre, H.; Chapman, A.B.; Peterson, K.S.; Shi, J.; Alba, P.R.; Jones, M.M.; Box, T.L.; DuVall, S.L.; Patterson, O.V. Launching into clinical space with medspaCy: A new clinical text processing toolkit in Python. AMIA Annu. Symp. Proc. 2022, 2021, 438. [Google Scholar]
- Hearst, M.A.; Dumais, S.T.; Osuna, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Almudaifer, A.I.; Covington, W.; Hairston, J.; Deitch, Z.; Anand, A.; Carroll, C.M.; Crisan, E.; Bradford, W.; Walter, L.A.; Eaton, E.F.; et al. Multi-task transfer learning for the prediction of entity modifiers in clinical text: Application to opioid use disorder case detection. J. Biomed. Semant. 2024, 15, 11. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Mowery, D.L.; Doing-Harris, K.M.; Hurdle, J.F. RuSH: A Rule-based Segmentation Tool Using Hashing for Extremely Accurate Sentence Segmentation of Clinical Text. In Proceedings of the AMIA Annual Symposium; American Medical Informatics Association: Bethesda, MD, USA, 2016; pp. 1587–1596. Available online: https://api.semanticscholar.org/CorpusID:35505279 (accessed on 1 July 2025).
- Endres, D.M.; Schindelin, J.E. A new metric for probability distributions. IEEE Trans. Inf. Theory 2003, 49, 1858–1860. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers); Association for Computational Linguistics: Minneapolis, MN, USA, 2019; pp. 4171–4186. [Google Scholar]
- Osborne, J.D.; O’Leary, T.; Mudano, A.; Booth, J.; Rosas, G.; Peramsetty, G.S.; Knighton, A.; Foster, J.; Saag, K.; Danila, M.I. *Gout Emergency Department Chief Complaint Corpora* (Version 1.0). PhysioNet, 2020. RRID:SCR_007345. Available online: https://physionet.org/content/emer-complaint-gout/1.0/ (accessed on 1 September 2025).
- Petrov, S.; Das, D.; McDonald, R. A universal part-of-speech tagset. arXiv 2011, arXiv:1104.2086. [Google Scholar]
- O’Leary, T.; Osborne, J.D. *uabnlp/BRATsynthetic: Version 0.3*. Zenodo. 2022. Available online: https://zenodo.org/records/7250621 (accessed on 1 January 2024).
- Osborne, J.D.; O’Leary, T.; Nadimpalli, A.; Kennedy, R.E. Bratsynthetic: Text de-identification using a markov chain replacement strategy for surrogate personal identifying information. arXiv 2022, arXiv:2210.16125. [Google Scholar] [CrossRef]
- Simancek, D.; Vydiswaran, V.V. Handling Name Errors of a BERT-Based De-Identification System: Insights from Stratified Sampling and Markov-based Pseudonymization. In Proceedings of the Workshop on Computational Approaches to Language Data Pseudonymization (CALD-pseudo 2024), St. Julian’s, Malta, 21 March 2024; pp. 1–7. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).