1. Introduction
Cells rely on the interaction of multiple proteins for life activities. A protein complex, formed through interactions, consists of molecules with similar functions. Detecting protein complexes in protein–protein interaction (PPI) networks facilitates the exploration of the relationships between network structures and function modules. Moreover, it plays a crucial role in annotating the proteins with unknown functions and gaining insights into the organization and functionality of cells [
1].
Researchers have proposed many experimental methods to identify the interactions between proteins, including yeast two-hybrid (Y2H) [
2,
3] and tandem affinity purification (TAP) [
4]. These methods have generated a vast amount of protein–protein interaction (PPI) data, which serve as valuable support for the application of data mining techniques in protein complex detection.
A PPI dataset is usually abstracted as an undirected network, wherein proteins are nodes and the interactions between proteins are edges. A PPI network contains different protein function modules [
5]. Generally, a protein complex is a biological functional module [
6] comprising two or more proteins that share the same function. Proteins in the same protein complex exhibit strong connections, whereas the proteins belonging to different complexes have weaker connections. Detecting protein complexes from PPI networks aims to discover sets of proteins with dense connections. This process can be viewed as a network clustering task, wherein clusters are determined based on topological features, where the connection strength within a cluster is greater than that between clusters [
7,
8]. This process yields disjoint or overlapping clusters as its outcome [
9].
Various network clustering algorithms for identifying protein complexes have been developed. In general, these algorithms include graph partition algorithms, density-based local search algorithms, and algorithms based on graph embedding [
10,
11,
12].
The clustering algorithm based on graph partition divides nodes into clusters according to an objective function, aiming to identify an optimal partitioned network. It maximizes the similarity between nodes within each cluster while minimizing the similarity between different clusters. One well-known algorithm in this category is the Markov algorithm (MCL) [
13,
14]. MCL begins by constructing the initial flow matrix based on a PPI network and then simulates random flow through the network using the concept of random walk to partition the entire network into sub-graphs with high connectivity probability. The collection of nodes within each sub-graph represents a protein complex. However, MCL does not handle overlapping clusters. To address this limitation, the soft regularized MCL (SR-MCL) algorithm was developed, which enables the identification of overlapping clusters.
The density-based local search clustering algorithm focuses on identifying dense sub-graphs based on the characteristic of connection density. Among the various network clustering methods, one approach aims to find
k-closely connected sub-network modules, such as the Closely Connected Percolation Method (CPM) [
15]. CPM initially identifies closely connected subnets within the network and subsequently identifies
k-closely connected subnet modules based on these initial subnets. A few approaches are also known as the seed expansion method. They select a node as a seed and expand around the seed to a cluster according to certain rules. One example of the seed expansion method is the density peak clustering (DPClus) algorithm [
16]. DPClus introduces the concept of “cluster periphery” in protein interaction networks. It assigns edge weights based on common neighbor counts between interacting proteins, while node weights are determined by the sum of their adjacent edges’ weights. The peripheral value of a node within a cluster is determined as the ratio of its adjacent nodes to the total number of nodes in the cluster. The algorithm starts by selecting the highest-weighted node as the seed for the initial cluster. Edge weights are influenced by common neighbor counts, and node weights reflect the density of immediate neighbors. If nodes satisfy both the custom threshold for local density and the threshold for cluster peripheral value, DPClus iteratively adds the nodes to obtain the final cluster. To account for the minimum diameter and average node distance characteristics of protein complexes, the improved DPClus algorithm (IPCA) [
17] enhances DPClus through the integration of sub-graph diameters and interaction probabilities, which provide insights into the density of the network. Other methods in this category include SEGC [
18], Core [
19], etc.
Network clustering algorithms based on graphs embed map network nodes onto a lower-dimensional vector space by encoding their properties [
20]. This mapping preserves the topological characteristics of the nodes as much as possible. Subsequently, a network clustering was performed in this transformed vector space [
21,
22]. One example of such an algorithm is the ensemble learning framework for density peak clustering (ELF-DPC) [
23]. ELF-DPC first maps the PPI network to the vector space and constructs a weighted network to identify core edges. By integrating structural modularity and trained voting regression models, the algorithm creates an ensemble learning model. ELF-DPC then expands the core edges into clusters based on this learning model.
The PPI network, as a type of complex network, exhibits intricate network topology characteristics [
24,
25,
26]. The fundamental features used to describe the network topology are primarily derived into three levels. Firstly, micro-topological structure metrics focus on individual nodes or edges, including measures such as node degree and centrality [
27,
28]. Secondly, meso-topological metrics analyze groups of nodes, such as community structure [
29], modules, and motifs. Lastly, macro-topological metrics consider the entire network, encompassing aspects such as degree distribution and community size distribution. Developing a network clustering algorithm that incorporates these network features can enhance the accuracy of community detection [
30]. At present, seed expansion methods can effectively utilize network features. However, existing algorithms mainly consider local micro-topological structure features [
31] and ignore the potential distribution characteristics of community size at a macro-global level. The distribution of community sizes in the PPI network exhibits a certain correlation with power-law distribution [
32].
In this paper, we present a novel network clustering approach that incorporates the characteristics of power-law distribution to identify protein complexes. Our proposed algorithm, named GCAPL, encompasses two main stages: cluster generation and cluster determination. During the cluster generation stage, the GCAPL algorithm incorporates node degree and clustering coefficient to assign weights to nodes. The unclustered nodes with the highest weight were selected as seeds. Following that, a cluster generation model leveraging the scale-free power-law distribution was given to discovery clusters with dense centers and sparse peripheries. Through an iterative process, candidate nodes were added to the seeds to form candidate clusters using the cluster generation model. In the cluster determination stage, we constructed a power-law distribution function about the distribution of cluster sizes and the cluster total number. The function acts as a criterion to regulate the presence of clusters of various sizes. By applying the power-law distribution function, we can assess whether a candidate cluster qualifies as a final cluster.
This paper makes several significant contributions: (1) Integrating multiple available basic micro-topological structural information into the k-order neighborhood of a node for seed selection; (2) Constructing a cluster generation model considering scale-free power-law distribution to obtain inherent organization information of functional modules; (3) Giving a cluster determination model based on macro-topological structure characteristic of the number distribution of clusters of different sizes to constrain final clusters; (4) Verifying the proposed network clustering algorithm fused with topological structural information could effectively mine functional modules by the experiment results on the real datasets.
The other sections of our paper are as follows.
Section 2 introduces preliminary concepts and symbols.
Section 3 presents a network clustering algorithm fused with power-law distribution characteristics.
Section 4 reports the relevant experiments to verify the effectiveness of the network clustering algorithm.
Section 5 provides conclusions.
3. Methods
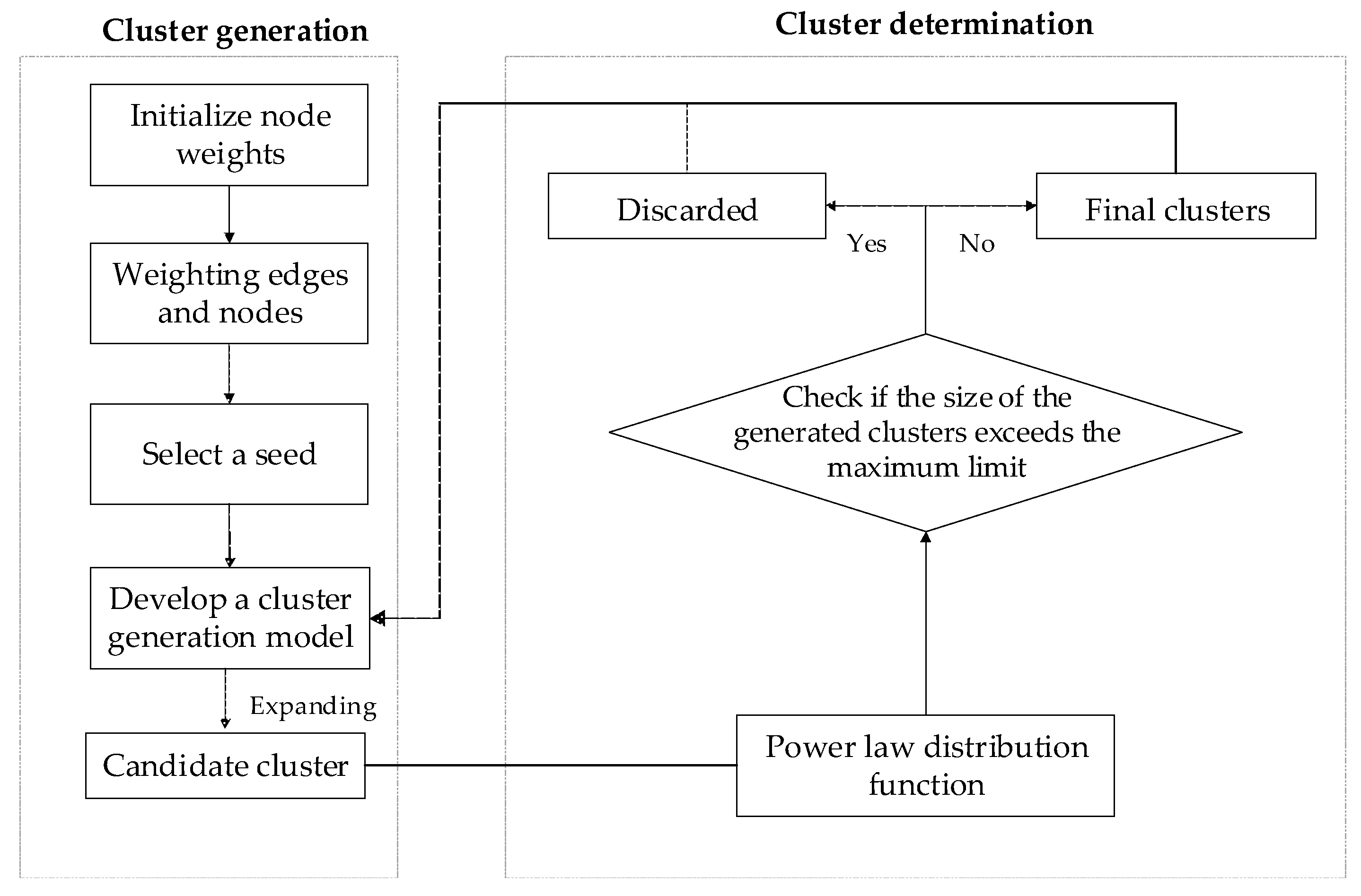
GCAPL algorithm consists of two stages: cluster generation and cluster determination. In the first stage, the algorithm calculates the weights of nodes and edges by incorporating micro-topological structure metrics. A seed is the node that has the highest weight among the unclustered nodes. The seed is expanded by a cluster generation model considering a scale-free power-law distribution to a candidate cluster. In the second stage, we established the cluster determination model with a power-law distribution of the cluster numbers with different cluster sizes. This cluster decision model was utilized to determine the final clusters.
Figure 1 shows the algorithm flow chat.
3.1. Cluster Generation
In the cluster generation stage, the GCAPL algorithm initially selects seeds based on node weights and subsequently expands these seeds using the cluster generation model to obtain candidate clusters.
To identify a suitable seed node, a node with a higher weighted degree may be a good seed node in network community mining. A node with a higher weighted degree may serve as a useful seed node in network community mining. The weighted degree of a node
is calculated based on its directly adjacent edges and the weights associated with these edges, and was defined as:
For an edge
, the endpoints of the edge and the common adjacent nodes between these endpoints are tightly surrounding this edge. We can obtain the edge weight of
according to the importance of these nodes in topological characteristics. The micro-topological structure metrics, such as clustering coefficient and node degree, are employed to capture the topological characteristics and assign weights to nodes. For a dense submodule in a network, nodes with high clustering coefficients and low node degrees may serve as important central nodes. The topological characteristics of a node
is expressed by the ratio of its clustering coefficient
to its node degree
, i.e.,
. More comprehensively, the global information of a network is introduced. A network
G’s clustering coefficient
FC(G) is defined as the average value of the clustering coefficients of all nodes in the node set
V, i.e.,
. Similarly, the
G’s node degree
FD(G) is the average of all node degrees in the network, i.e.,
. In the network G, the connection strength of a node
is related to
. Therefore, the weight of the edge
can be defined as follows:
Furthermore, Equation (4) from the previous section only considers the information of the node’s direct neighbors. To highlight the importance of an edge within a large network module, the edge weight in its
t neighborhood can be defined as follows:
Here,
t is a predefined parameter that determines the extent of the neighborhood. After the
t-th iteration, the node weight can be defined as:
Initially, the node weights are set to for all nodes, indicating that the initial importance of all nodes is the same.
Once the node weight calculation is completed, the next step is to select a seed node v from the node set V whose node weight is highest. Following that, the seed node is used to establish the cluster generation model, which allows for the expansion of the seed into a candidate cluster.
The cluster generation model aims to expand seed nodes into candidate clusters based on connection strength. The obtained seed node
v serves as the initial cluster
CS(v), and candidate nodes from the neighborhood
NE(C(v)) are considered for addition based on the compactness of
CS(v) and the connection strength between
CS(v) and a candidate node
u to expand the initial cluster
CS(v). The compactness
of the cluster
CS(v) quantifies the connection density within the cluster and is defined as
, where
represents a set of nodes that make up
C(v), and
denotes the node
u’s direct neighbor nodes. The connection strength
of a candidate node
u reflects the peripheral edges of the cluster and is defined as
. The cluster generation model requires a variable function to combine the compactness of the cluster and the peripheral edges of the cluster, so that as the cluster size increases, the contribution of the cluster’s compactness to the cluster generation gradually decreases while the contribution of the cluster’s peripheral connections to the cluster generation gradually increases. A suitable choice for this function is the scale-free power-law distribution function, which is a monotonic function. It serves as a foundation for constructing the variable function that effectively fuses the above two kinds of connection information. A power-law distribution function is
and let
,
,
, then we can define the variable function as:
where
is a parameter to control the change of
. Then, define the cluster generation model as:
When
is set to 1,
CT tends to prioritize the formation of dense clusters. On the other hand, when
is set to 0, nodes with lower degrees are more likely to be added to
CS(v). The
enables the cluster generation model to find both dense clusters and clusters with dense cores and sparse peripheries, providing flexibility in capturing different types of cluster structures. For each candidate node
u and threshold
, if
and
(
is a user-defined threshold), then the node
u is added to the cluster
CS(v). This process is repeated for each node in
NE(CS(v)), resulting in the initial formation of a candidate cluster
CS(v).
3.2. Cluster Determination
In complex networks, the distribution of community size exhibits heterogeneity. Smaller communities tend to be more abundant in number, while larger communities are relatively scarce. This inverse relationship between size and number also holds in PPI networks, where the sizes and numbers of protein complexes are inversely proportional. It is assumed that the number of complexes follows a power-law distribution that is defined as follows:
where
x and
y represent positive random variables.
Let the size of a protein complex be
. The corresponding number of the complexes under this size is given by a cluster determination model:
where
c and
k are positive parameters.
The cluster determination model aims to effectively regulate the number of clusters, considering their varying sizes, from a global perspective. To accomplish this, we defined two sequences: is a predefined sequence with uniform values representing the cluster sizes, and is a sequence obtained through the cluster generation model representing the corresponding cluster numbers.
Let the cluster CS(v)’s size be denoted as |V(CS(v))| and be a parameter that refers to the allowable difference or deviation in the size of a cluster. Following that, we can find a value with in , assuming that clusters of size have been generated at the current stage. We calculated the maximum number of clusters corresponding to a given cluster size according to the power-law distribution function. If , the candidate cluster CS(v) is considered a final cluster. Otherwise, CS(v) is discarded.
The two stages of cluster generation and cluster determination are repeated alternately until all nodes have been clustered.
3.3. Complexity Analysis
The GCAPL algorithm utilizes linked lists to construct a graph. First, it calculates the weights of all nodes using Formula (6). Following that, it selects the node with the highest weight as the seed and treats it as the initial cluster. Subsequently, following the cluster generation model, neighbor nodes of the initial cluster are incrementally added to create candidate clusters. Finally, the algorithm determines the final clusters by the cluster determination model. The specific process of the GCAPL algorithm is shown in Algorithm 1.
| Algorithm 1: GCAPL Algorithm. |
| Input: Network , Parameters iter, , for cluster generation, Parameters c, k, for Cluster determination |
| output: Set of final clusters PC |
| 1: Initialize , and the unclustered nodes set, ; |
| 2: Compute edge and node weights by utilizing information within the t-neighborhood; |
3: Determine the cluster size set ;
4: Calculate the upper limit of the number of clusters , corresponding to the cluster size using Equation (9);
5: while , do
6: Select a node v with the largest weight in UV as a seed, and the initial cluster is CS(v);
7: Iteratively select the node set AN among the neighbor nodes of CS(v), such that each node u in AN satisfies and ;
8: ;
9: Compute the cluster CS(v)’s size as , and compute the number of generated clusters with size as ;
10: Find in , and
11: Compute the number of generated clusters of size as
12: if then
13: ,
14: return PC |
The time cost of the GCAPL algorithm lies in two parts: cluster generation and cluster determination.
Assuming a network G has n nodes and m edges. In the cluster generation stage, the node weighting process revealed a time cost of . The time cost of seed selection based on node weights is . The expansion of seeds into clusters also has a time cost of . Therefore, is the total time complexity of the cluster generation phase.
In the cluster determination phase, the worst-case scenario is when each candidate cluster size needs to be compared with each element in the sequence . As a result, this phase revealed a time cost of . Therefore, algorithm GCAPL’s overall time complexity is , considering both the cluster generation and cluster determination phases.
4. Experiments and Results
4.1. Datasets
The protein interaction networks used in the experiments are presented in
Table 2. These datasets were processed to remove self-intersections and duplicate interactions.
The gold standard complex datasets CYC2008 [
38] and MIPS [
39] were utilized for parameter analysis and evaluation of the clustering results.
4.2. Evaluation Metrics
The evaluation of the effectiveness of the GCAPL algorithm was performed using the F-measure and Accuracy metrics as evaluation criteria.
The F-measure [
40] provides a balanced measure of precision and recall. It serves as a quantitative metric of the agreement between a predicted complex set and a benchmark complex set, capturing the level of similarity between them. Precision measures the agreement between the generated clusters and known complexes, while recall quantifies the agreement between the known complexes and the generated clusters.
Given the generated cluster as
and the gold standard complex as
, the affinity score within the neighborhood
is employed for quantifying the similarity between the generated cluster
and the standard complex
, and
,
. A higher
value indicates a stronger resemblance between
and
. Assuming a threshold of
[
40,
41], if
,
and
can be considered as matched. Let
represent the set of correct predictions, where each generated cluster exhibits some correspondence with at least one known protein complex in the set
TC, and
. Additionally, let
be the set of known complexes, where each complex matches at least one complex in the generated cluster set
PC, and
.
Precision is quantitatively calculated as the ratio of the number of correctly predicted instances to the total number of predicted instances, i.e.,
. Recall is defined as
. F-measure is quantitatively calculated as
Accuracy, as another evaluation metric, is computed as the geometric mean of the positive predictive value (PPV) and sensitivity (Sn). PPV represents the proportion of correctly identified positive instances among the predicted instances, while Sn measures the proportion of correctly identified positive instances among all actual positive instances.
Suppose
T is a
matrix, in which the
i-th row of
T represents the
i-th prediction cluster
and the
j-th column represents the
j-th annotation complex
.
denotes the count of shared proteins between the predicted complex
and the known complex
and quantifies the degree of overlap or similarity between these two complexes.
PPV is characterized by
Accuracy [
39] is then calculated as
4.3. Parametric Analysis
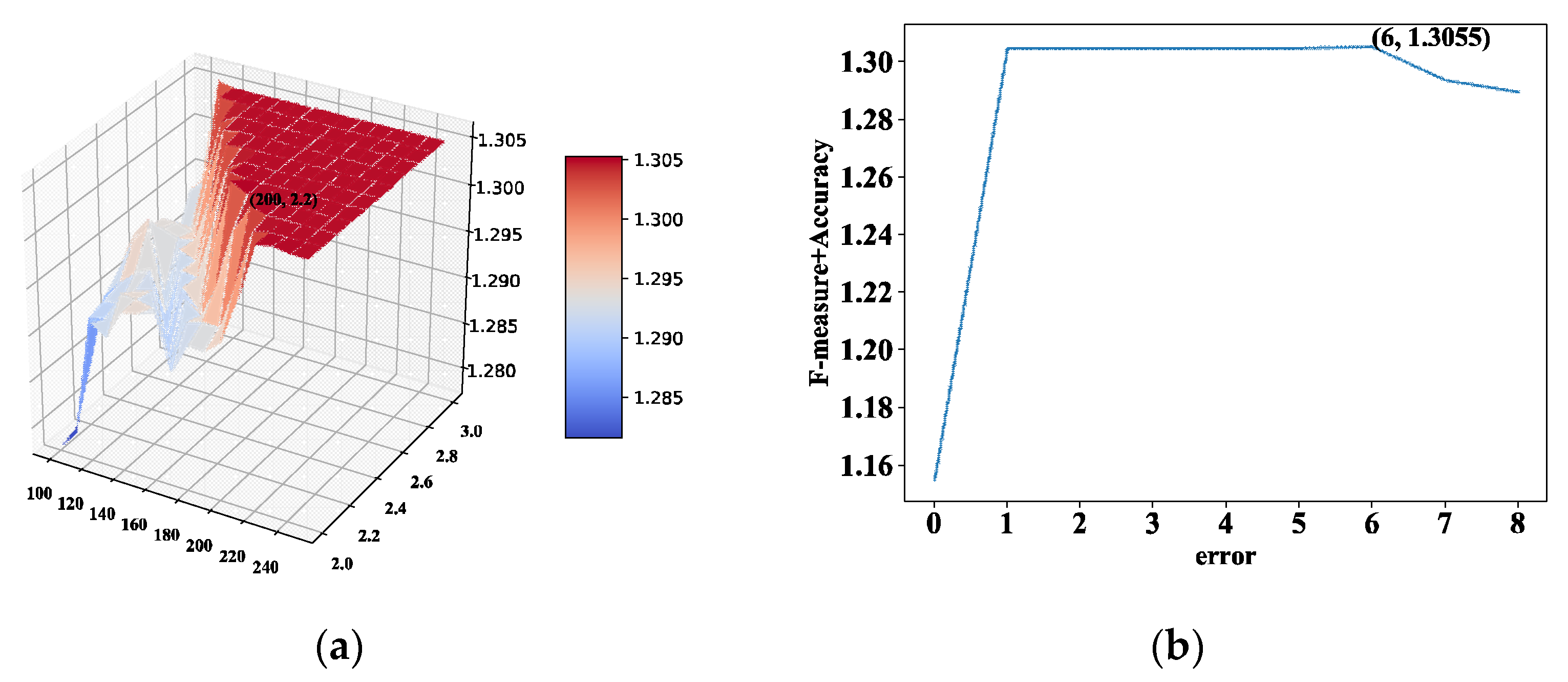
GCAPL encompasses several predefined parameters, including c, , , iter, , and . The coefficients c and k correspond to the coefficients and exponents of the power-law distribution function, respectively. The is a cluster size error. The iter refers to the count of repetitive steps. The stands for an adaptive parameter. The is defined as the compactness threshold. The BioGrid dataset serves as a standard protein interaction network dataset, wherein all interactions are derived from reliable and precise low-throughput theoretical interactions. Consequently, on this dataset, the parameter optimization aims to maximize the value of , prompting a thorough parameter analysis to identify the optimal parameter value.
The analysis of parameters c, k, and was performed to investigate the impact of these parameters on the algorithm. The coefficient c and the exponent k were utilized to generate the sequences and based on the power-law distribution function. Meanwhile, the parameter was employed to regulate the error tolerance in cluster size. Initially, the analysis focuses on varying c and k while keeping the parameter constant. Subsequently, the investigation shifts to studying the influence of the parameter while maintaining c and k at constant values.
We first fixed
, and experiments were conducted on the BioGrid PPI network to investigate the impact of the parameters
c and
k. The values of
c ranged from 100 to 250, while
k varied from 2.0 to 3.0. These experiments aimed to assess how the changes in
c and
k influenced the results and outcomes of the study. When the values of
and
are set, the
metric attains a higher value. Next, we first fixed
,
and
is maximized at
. We set
,
,
. In
Figure 2a, the impact of parameters
c and
k on
is illustrated, with
. The relationship between the parameter
and
are depicted in
Figure 2b, with
and
.
Next, we kept the values of , , and fixed, and analyzed the real-valued discrete parameters: the number of iterations iter, the adjustment parameter of the change rate, and the tightness threshold . Considering the interdependence among these parameters, an orthogonal matrix was employed to identify the optimal parameter combination with a high likelihood. During the experimental design phase, each parameter variable was treated as an independent factor. Feasible values corresponding to these factors are assigned as distinct levels. The complete set of parameter combinations represents the experimental space. An orthogonal array L36 (63 × 37) is employed, which comprises 36 parameter combinations. There are parameters, , , and , that we exclusively consider the initial three columns of the orthogonal array to facilitate the analysis. Among the 36 parameter combinations, the one with the highest is selected as the optimal configuration. Through the experiments, the parameters are set to , , and .
4.4. Power-Law Distribution Analysis
This subsection examines the power-law distribution of network clustering results, taking the BioGRID dataset as an example. The clustering result of this dataset was utilized to explore the relationship between the cluster size and number.
Assume that the cluster size is represented by
x and the corresponding number of clusters is denoted by
y. According to Equation (9), we have
. By performing logarithm operations on both sides of the equation, it represents that
It was observed that and exhibit a linear relationship. Thus, the analysis of the power-law distribution of x and y was transformed into a linear relationship analysis of and .
In the clustering result of the BioGRID dataset, we took the logarithm of the cluster size
x and the corresponding cluster number
y, resulting in transformed variables
and
. To explore whether there is a linear relationship between
and
, a linear fitting method was performed on
and
. The results of the linear fitting analysis conducted on
and
is shown in
Figure 3, providing valuable insights into the nature of their relationship.
Table 3 presents the calculated
p-value and
for the linear fitting analysis conducted on
and
. A small
p-value indicates a strong fit of the clustering result, demonstrating good fitting effectiveness. Similarly, a large value of
suggests a favorable fit. In
Table 3, the obtained
p-value is
, and the value of
is 0.5. Thus, the sizes of clusters generated by the proposed algorithm in the PPI network follow a power-law distribution, along with the corresponding numbers of these clusters.
4.5. Comparative Experiment
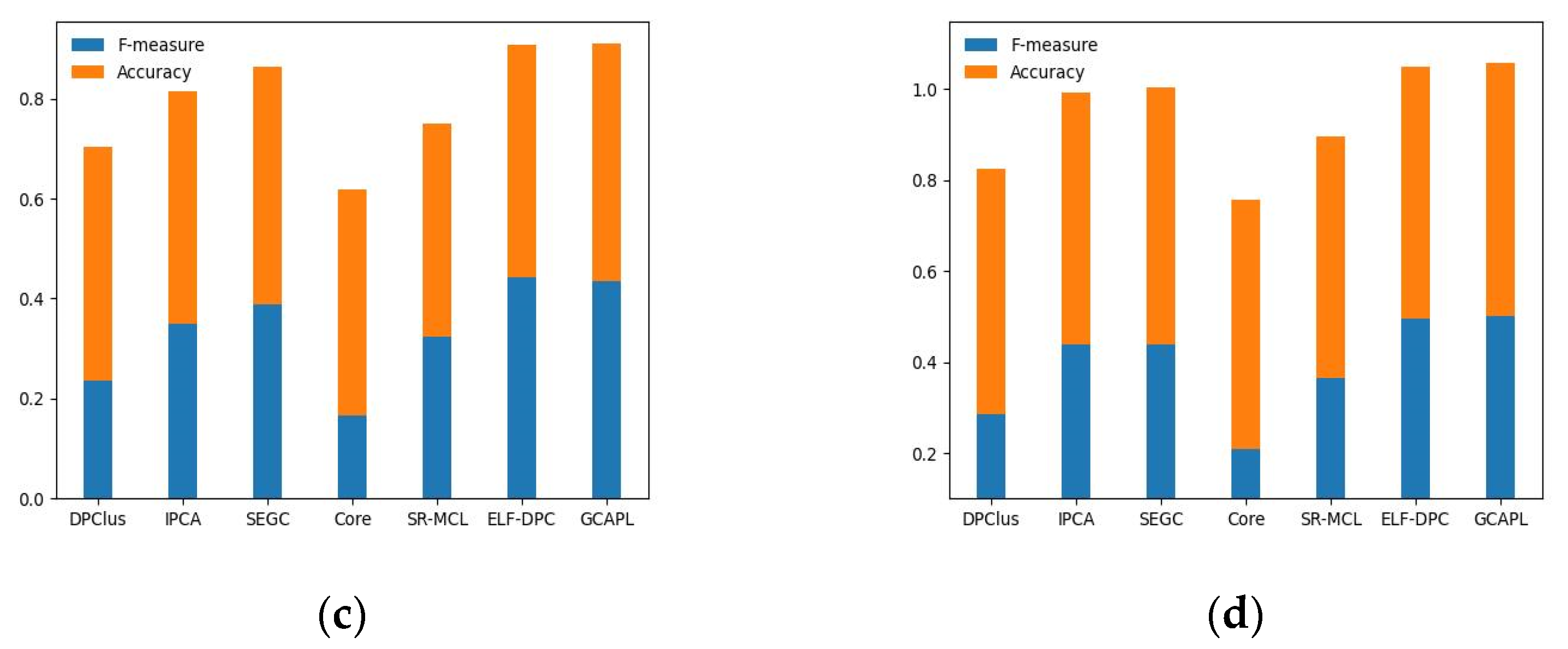
To assess the algorithm’s performance, we compared the GCAPL algorithm with several other algorithms, namely DPClus, IPCA, SEGC, Core, SR-MCL, and ELF-DPC.
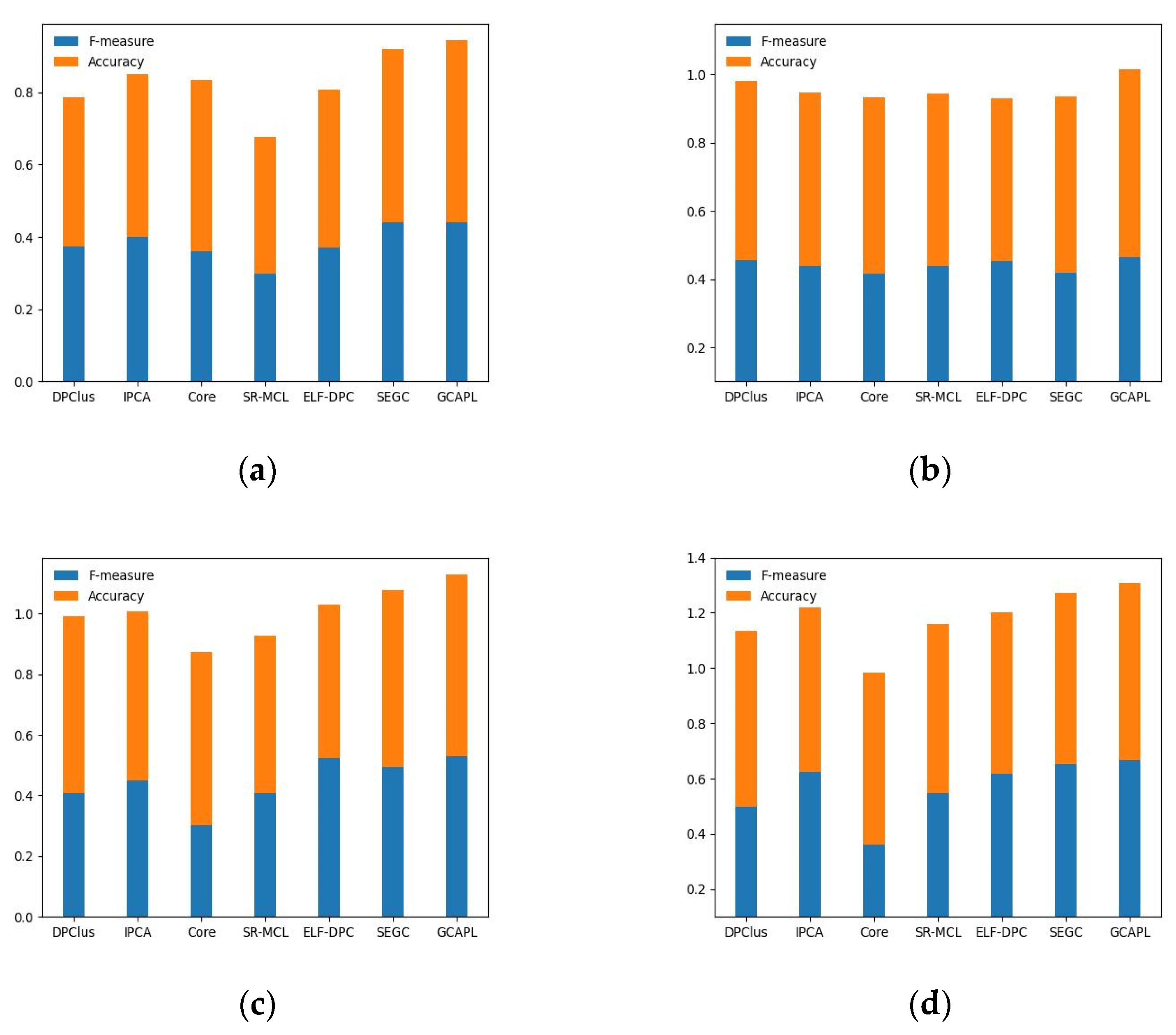
Figure 4a–d present the experimental results on the Gavin02, Gavin06, K-extend, and BioGRID datasets, using CYC2008 as the standard set. The results demonstrate that, compared to other algorithms, the GCAPL algorithm achieves comparable or higher F-measure and Accuracy values. The GCAPL algorithm performs well in terms of
. Compared with other algorithms, the
F-measure + Accuracy of GCAPL exhibits an average improvement of 13.12%, 6.97%, 14.43%, and 14.39% on Gavin02, Gavin06, K-extend, and BioGRID. In addition, the SEGC algorithm demonstrates lower F-measure and Accuracy performance compared to GCAPL on the Gavin02, K-extend, and BioGRID datasets. On the Gavin06 dataset, the DPClus algorithm performs better than other algorithms, except for the GCAPL algorithm. The GCAPL algorithm has a similar framework to the two algorithms mentioned above, and incorporating macro-topological information contributes to improving complex detection performance. By considering both the micro-topological structure of a network and the macro-topological structure feature of the power-law distribution, the GCAPL algorithm effectively detects protein complexes.
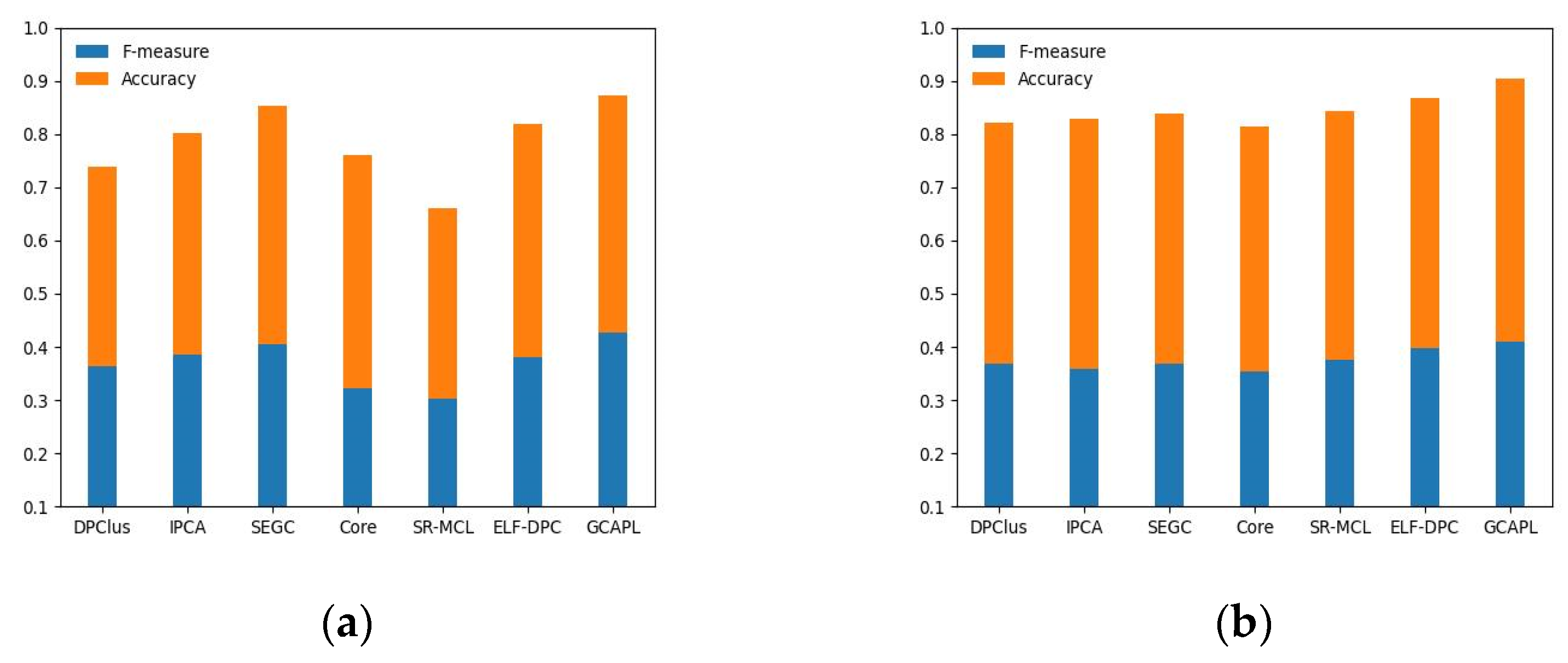
Figure 5a–d illustrate the evaluation results of the DPClus, IPCA, SEGC, Core, SR-MCL, ELF-DPC, and GCAPL algorithms on the Gavin02, Gavin06, K-extend, and BioGRID datasets, respectively, using MIPS as the standard set. The GCAPL algorithm consistently exhibits superior values of F-measure and Accuracy across the four different PPI datasets compared to compared algorithms. Compared with other algorithms, the
F-measure + Accuracy of GCAPL exhibits an average increase of 9.90%, 7.01%, 14.34%, and 13.63% on Gavin02, Gavin06, K-extend, and BioGRID. This indicates that the GCAPL algorithm performs well in terms of its ability to detect protein complexes.
In summary, the GCAPL algorithm has good performance in detecting protein complexes. The GCAPL algorithm uses not only micro-topological structure metrics but also the macro-topological structure characteristic of the power-law distribution about clusters, and it can obtain better results in complex detection. The GCAPL algorithm further explores the relationship between network topological characteristics and functional modules in PPI networks, which is of great significance for improving the accuracy of protein complex detection.
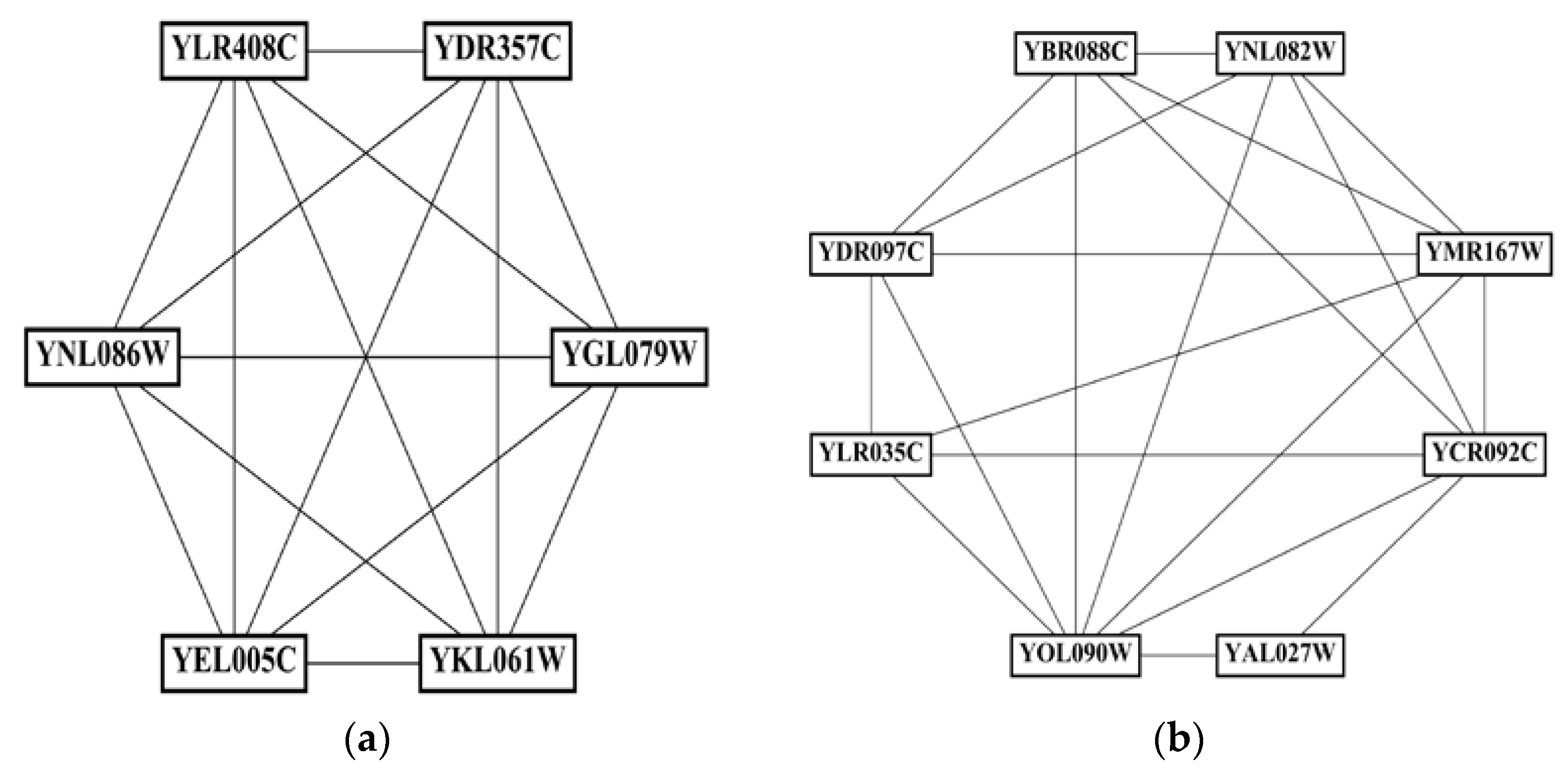
4.6. Examples of Predicted Complexes
In this subsection, four predicted protein complexes with different sizes detected by the GCAPL algorithm are exhibited, and their corresponding network topology structures are shown in
Figure 6. The predicted complex in
Figure 6a is a fully interconnected network.
Figure 6b shows a cluster that has a dense sub-graph with a relatively sparse periphery.
Figure 6c,d show two clusters that are dense sub-graphs.
Table 4 presents the Gene Ontology annotations of these predicted protein complexes in three aspects of biological processes, molecular functions, and cell components with corresponding significance
p-values. The obtained
p-values are notably small, indicating that these clusters have significant biological significance. The effectiveness of the GCAPL algorithm is demonstrated in its ability to identify protein complexes with multiple network structures.
5. Conclusions
Detecting protein complexes is of great significance for understanding biological mechanisms. This paper proposes a network clustering algorithm fused with power-law distribution for protein complex detection. The algorithm begins by calculating node weights, taking into account micro-topological structure metrics. Subsequently, the algorithm selects the non-clustered nodes with the higher weights as seeds and forms initial clusters around the seeds. Next, the algorithm greedily adds candidate nodes into the initial clusters based on the characteristics of scale-free power-law distribution to generate candidate clusters. A power-law distribution function, based on the macro-topological structure feature of power-law distribution about cluster size and number, is established to guide the cluster generation process. The power-law distribution function is employed to determine whether a candidate cluster qualifies as a final cluster. Compared with other algorithms, the F-measure + Accuracy of GCAPL improves by an average of 12.23% and 10.97% on the CYC2008 and MIPS benchmarks, respectively. The experimental analysis reveals that the proposed algorithm exhibits distinct advantages over other approaches.
The GCAPL algorithm mainly considers the biological network whose community size conforms to the power-law distribution characteristics. The algorithm does not take into account other distribution characteristics of the community size and fully considers the preferential attachment. The above information may further improve the performance of our algorithm to detect protein complexes. In addition, in real PPI networks, the connections between nodes are subject to constant changes, leading to variations in network topological structures. To mine functional modules in dynamic PPI networks, our future work will also focus on constructing dynamic networks and developing dynamic protein complex identification methods.