Abstract
Automated diagnosis for the quality of bovine in vitro-derived embryos based on imaging data is an important research problem in developmental biology. By predicting the quality of embryos correctly, embryologists can (1) avoid the time-consuming and tedious work of subjective visual examination to assess the quality of embryos; (2) automatically perform real-time evaluation of embryos, which accelerates the examination process; and (3) possibly avoid the economic, social, and medical implications caused by poor-quality embryos. While generated embryo images provide an opportunity for analyzing such images, there is a lack of consistent noninvasive methods utilizing deep learning to assess the quality of embryos. Hence, designing high-performance deep learning algorithms is crucial for data analysts who work with embryologists. A key goal of this study is to provide advanced deep learning tools to embryologists, who would, in turn, use them as prediction calculators to evaluate the quality of embryos. The proposed deep learning approaches utilize a modified convolutional neural network, with or without boosting techniques, to improve the prediction performance. Experimental results on image data pertaining to in vitro bovine embryos show that our proposed deep learning approaches perform better than existing baseline approaches in terms of prediction performance and statistical significance.
1. Introduction
Each month, during human female ovulation, the left or right ovary produces and releases an egg for fertilization. Early development of an embryo starts with the fertilization of an egg by a sperm at the fallopian tube. The fertilized egg (embryo), where the egg and the sperm unite, develops and goes through several stages, reaching the blastocyst stage before implanting in the wall of the uterus. The implantation of the embryo is a critical point during pregnancy for embryologists, who aim to accurately identify the quality of an embryo at this point [1]. Due to the lack of consistent noninvasive methods to determine the quality of embryos, the process of identifying the quality of an embryo is a challenging task [2]. As bovine and human females share various characteristics in terms of folliculogenesis, the cow has been used as a model to investigate folliculogenesis events in humans [3,4]. Bó et al. [5] described the principles of evaluating bovine embryos, which are important for deciding whether the embryo is worth transferring or frozen for a given recipient and whether the embryo is eligible to be exported for human use. The conventional evaluation method of the quality of bovine embryos is subjective visual evaluation of embryo images by embryologists using stereomicroscopy, followed by a scoring system issued by the International Embryo Technology Society (IETS). The scores produced determine whether the quality of the bovine embryo is excellent, fair, or good. Figure 1 shows the process of bovine embryo production through in vitro fertilization [6,7,8].
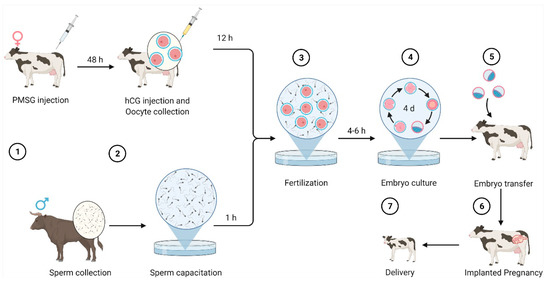
Figure 1.
In vitro fertilization in cows and delivery, shown as steps (1) to (7). Figure created with Biorender.
Deep Learning (DL) techniques have been successfully applied to solve many real biological problems, including various medical imaging problems [9,10,11,12,13,14,15]. Specifically, DL techniques aim to improve the prediction performance in a given task via learning complex concepts from simpler ones [16]. DL methods provide promising solutions in artificial intelligence when applied to medical imaging and could improve the performance of several medical imaging problems, including predicting the quality of embryos, in the coming years.
Predicting the quality of embryos helps embryologists to (1) improve the embryo transfer process, which leads to better genetics and viable embryos [17]; (2) reduce the time spent searching for viable embryos, which is important for clients in the cattle industry who need to freeze or transfer the embryos to recipients [18,19]; and (3) avoid abortion at later stages of pregnancy caused by poor quality of embryos [20]. Due to the importance of predicting the quality of embryos, researchers from different domains, including developmental biology and machine learning, are involved in this work. For example, Rocha et al. [21] proposed a machine learning approach to predict the quality of bovine embryos that works as follows. A preprocessing step aims to construct feature vectors via extracting 36 features from images of in vitro bovine images. Then, the constructed features are provided as input to a neural network, to build a model, which is then used to classify unseen images. Manna et al. [22] proposed an ensemble approach utilizing features extracted from the foreground image of the embryo to construct feature vectors, which are then provided to an ensemble of neural networks to train and predict the quality of unseen embryos. Filho et al. [23] proposed a machine learning approach to predict the grading of human embryos using extracted image features, provided as input to support vector machines (SVM), to yield a model. The resulting model is then used to evaluate the model using images of unseen human embryos obtained via two independent experts. Other machine learning and deep learning approaches have been developed for other prediction tasks related to embryos [24,25]. For example, Miyagi et al. [26] developed a deep learning approach utilizing a convolutional neural network consisting of two convolutional layers for feature extraction, two pooling layers for dimensionality reduction, and fully connected layers for classification. The developed approach is used to predict the number of live births from images of blastocysts. Experimental results showed that the deep learning approach performs significantly better than the baseline method.
The work described here differs from [5,21,22,23,26] in three ways. (1) We present three deep learning approaches, where the first deep learning approach (DL) exploits a convolutional neural network consisting of a convolutional layer for feature extraction, an average pooling layer for dimensionality reduction and fully connected, and output layers for the classification. The second deep learning approach (B1DL) extends the DL approach and is coupled with a boosting technique to improve the prediction performance by selecting an ensemble with the highest area under the curve during the training, which is then used to perform predictions on unseen embryo images. The third deep learning approach (B2DL) is different from the B1DL approach in terms of utilizing a different boosting technique. (2) The proposed deep learning approaches are compared against other deep learning approaches under a supervised learning setting, using only images of in vitro bovine embryos. (3) We conduct statistical tests and evaluate the performance of all approaches using several performance measures.
Conventional approaches have relied on the experience and expertise of humans, following evaluation principles to determine the quality of bovine embryos [5]. To automate the intelligence of the expert, ML approaches require extracting image features to construct a dataset, which are then provided to a ML algorithm (artificial neural networks) to create a model that automates human intelligence for predicting the quality of unseen bovine embryos [21]. However, these approaches are (1) not accurate enough as they rely on manually hand crafted features; (2) based on the subjective evaluation of experts who spend much time examining embryo images and can mislabel some due to oversight or fatigue; and (3) based on a set of subjective features that could affect the generalizability of models [25]. In contrast to previous approaches that rely on a set of specified features, sophisticated deep learning approaches can be used to improve prediction performance without specifying a set of hand-crafted features [27,28]. However, these deep learning approaches require extensive computational resources and the adopting of concepts that improve the learning process [29,30].
Contributions. Our contributions in this paper are summarized as follows.
- -
- We present new deep learning approaches for developmental biology, enabling deep learning algorithms to achieve high-performance results from images of in vitro bovine embryos.
- -
- The proposed approaches adopt modified versions of deep convolutional neural networks and boosting techniques, and are the first, to the best of our knowledge, to be applied in the developmental biology discipline pertaining to predicting the quality of embryos.
- -
- In contrast to previous works, we assess our approaches by employing several classification performance measures against baselines, including existing deep learning algorithms such as LeNet and AlexNet, based only on in vitro data.
- -
- We perform an experimental study to report the predictive performance of all deep learning approaches. The results demonstrate that our approaches significantly outperformed the baseline approaches.
Organization. The rest of this paper is organized as follows. We review existing research related to this paper in Section 2. Section 3 and Section 4 present our proposed deep learning approaches and report performance results of the proposed approaches against the baselines. We provide a discussion of the results in Section 5. Lastly, we conclude the paper and point out directions for future work in Section 6.
2. Related Work
There are three main bodies of research related to our work: AdaBoost [31,32], LeNet [33,34], and AlexNet [35,36].
2.1. AdaBoost
AdaBoost aims to generate a highly accurate classifier via taking the weighted majority vote of several classifiers. Algorithm 1 outlines the details of AdaBoost, which works as follows. For a given training set S consisting of m examples (line 1), all m examples are assigned the same weight (see lines 2–4). The for loop in lines 5–13 aims to produce an ensemble of T classifiers as follows. In line 6, the training set S and corresponding weights Dt are provided as inputs to a machine learning algorithm to induce a model ht at the tth iteration. The error of the ht classifier is calculated based on misclassified examples and corresponding weights in Dt (lines 7–8). The weight αt of ht is calculated (Line 9). Lines 10–12 perform a weight updating mechanism that aims to increase the weight of the ith training example xi during the next iteration (i.e., t + 1) to focus on if the classifier ht incorrectly predicts xi. Otherwise, its weight is decreased. Zt is a normalization factor that makes Dt+1 normally distributed. This process (i.e., lines 5–13) is then performed for an additional t − 1 times. For a given test example , a prediction is performed based on the sign of weighted T classifiers obtained from lines 5–13.
| Algorithm 1: AdaBoost. |
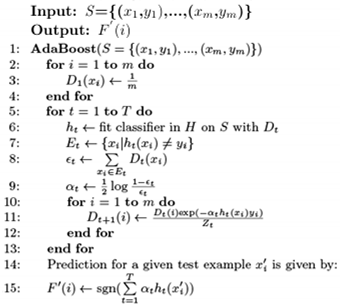 |
2.2. LeNet
Figure 2 shows the architecture of the convolutional neural network (CNN) LeNet, which has a 32 × 32 (INPUT)–28 × 28 × 6 (CONV)–14 × 14 × 6 (AVGPOOL)–10 × 10 × 16 (CONV)–5 × 5 × 16 (AVGPOOL)–120 (FC)–84 (FC)–10 (OUTPUT) structure. The LeNet consists of six layers, excluding the input layer shown in the first. The input is a grayscale image with 32 × 32 pixels. The first layer is a convolutional layer (CONV) that filters the input with six kernels each of size 5 × 5 with a stride of 1 pixel. The convolutional layer produces six feature maps each of size 28 × 28, followed by an average pooling layer that reduces the dimensionality of each feature map by a factor of 2. The third layer is a convolutional layer that filters the output of the average pooling layer with 16 kernels each of size 5 × 5 with a stride of 2 pixels. The third layer produces 16 feature maps each of size 10 × 10, followed by an average pooling layer (i.e., L4) that reduces the dimensionality of each feature map by a factor of 2. The average pooling layer is then flattened and immediately followed by two fully connected layers and an output layer, which has a number of units corresponding to the class labels. The convolutional and fully connected layers are composed of rectified linear units (ReLUs) as ReLUs lead to faster training [37].
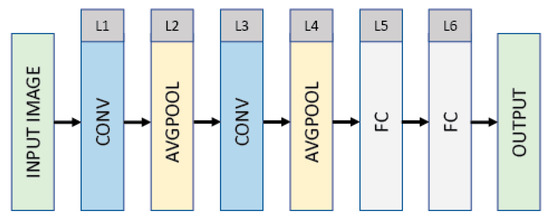
Figure 2.
Architecture of the convolutional neural network (CNN) LeNet, consisting of six layers, excluding the input and output layers.
2.3. AlexNet
Figure 3 shows the architecture of AlexNet, which has a 224 × 224 × 3 (INPUT)–54 × 54 × 96 (CONV)–27 × 27 × 96 (MAXPOOL)–23 × 23 × 256 (CONV)–11 × 11 × 256 (MAXPOOL)–9 × 9 × 384 (CONV)–7 × 7 × 384 (CONV)–5 × 5 × 256 (CONV)–2 × 2 × 256 (MAXPOOL)–4096 (FC)–4096 (FC)–1000 (OUTPUT) structure. The AlexNet consists of five convolutional layers (interspersed with normalization layers), three max pooling layers, two fully connected layers (interspersed with dropout with a ratio of 0.4), and an output layer. The input is an RGB image of 224 × 224 × 3 pixels. The first layer is a convolutional layer (CONV) that filters the input with 96 kernels each of size 11 × 11 with a stride of 4 pixels. The convolutional layer produces 96 feature maps each of size 54 × 54, followed by a max pooling layer that reduces the dimensionality of each feature map by a factor of 2. The third layer is a convolutional layer that filters the output of the max pooling layer with 256 kernels each of size 5 × 5 with a stride of 1 pixel. The third layer produces 256 feature maps each of size 23 × 23, followed by a max pooling layer (i.e., L4) that reduces the dimensionality of each feature map by a factor of 2. The fifth layer produces 384 feature maps each of size 9 × 9, followed by two convolutional layers that produce 384 and 256 feature maps of sizes 7 × 7 and 5 × 5, both with a stride of 1 pixel. The output of layer 7 is provided to a max pooling layer, which reduces the dimensionality of each feature map by a factor of 2. The output of the max pooling layer (i.e., L8) is then flattened and immediately followed by two fully connected layers and an output layer, which is a classification layer that has a number of units corresponding to the 1000 categorical labels. The convolutional and fully connected layers (excluding the output using Softmax) are composed of rectified linear units (ReLUs) as ReLUs lead to faster training.
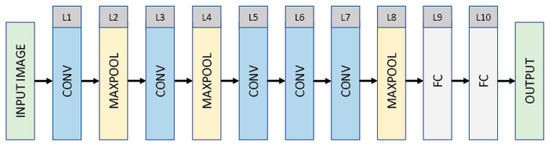
Figure 3.
Architecture of convolutional neural network AlexNet consisting of 10 layers, excluding the input and output layers.
3. Our Proposed Approaches
3.1. The Deep Learning Approach (DL)
The CNN variant (see Figure 4) takes an RGB input image. The details of the proposed architecture (CNN variant) employed via DL are shown in Figure 4, which has a 20 × 20 × 3 (INPUT)–20 × 20 × 120 (CONV)–10 × 10 × 120 (AVGPOOL)–512 (FC)–512 (FC)–2 (OUTPUT) structure. The CNN variant consists of a convolutional layer, an average pooling layer, two fully connected layers (interspersed with dropout with a ratio 0.5), and an output layer. The input RGB image is resized to 20 × 20 × 3 pixels. The first layer is a convolutional layer (CONV) that filters the input with 120 kernels each of size 5 × 5 with a stride of 1 pixel. The convolutional layer produces 120 feature maps each of size 20 × 20 (with zero-paddings), followed by an average pooling layer that reduces the dimensionality of each feature map by a factor of 2. The output of the average pooling layer (i.e., L2) is then flattened and immediately followed by two fully connected layers and an output layer, which is a classification layer that has a number of units corresponding to the two categorical labels. The convolutional and fully connected layers (excluding the output utilizing softmax) are composed of rectified linear units (ReLUs). The prediction generated via the CNN variant differentiates good- from poor-quality embryos.
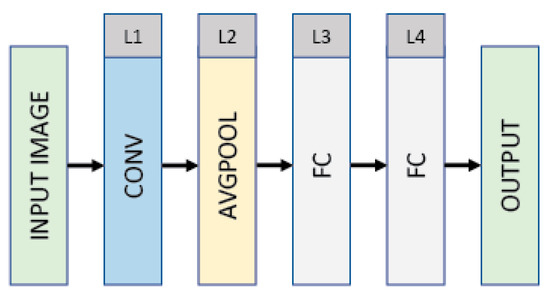
Figure 4.
The architecture of the CNN variant that is utilized via the proposed approach DL.
3.2. The First Boosted Deep Learning Approach (B1DL)
Figure 5 shows our first boosted deep learning approach (B1DL), which works as follows. Suppose that we are given a training set consisting of m labeled bovine embryo images, and a testing example of an unseen embryo image. We call our algorithm B1 (see Algorithm 2) to boost the prediction performance of the CNN variant via creating a better ensemble of several CNN variants. Initially, B1 takes as input the training set S (Line 1). In lines 2–4, each example of a jth ensemble is assigned an equal weight. Lines 5–20 aim to create a set of ensemble of CNN variants, where the size of the set is k. The number of epochs Λ is randomly generated (line 6). Training examples in S are selected without replacement based on weights in Dj,1. Then, Λ and S are provided as inputs to the CNN variant, to induce the first model of the jth ensemble, hj,1. Lines 8–19 aim to create T classifiers for a jth ensemble.
| Algorithm 2: Boost1 (B1). |
 |
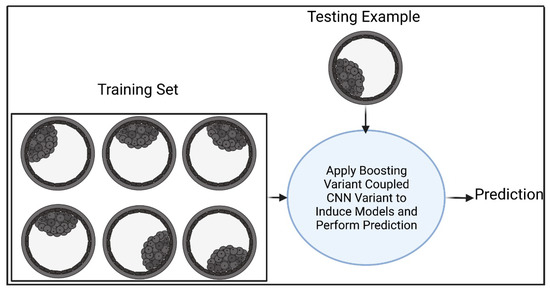
Figure 5.
The first boosted deep learning approach (B1DL) for predicting the quality of embryos. Figure created with BioRender.
For each t iteration, the error of the tth classifier of ensemble j on the training set S is calculated based on Dj,t (lines 9–10). The weight αj,t of classifier hj,t is calculated (line 11). For each training example xi in S, the weight update mechanism in lines 12–14 aims to increase the weight of incorrectly predicted example hj,t(xi) to focus on during the next iteration while decreasing the weight of correctly predicted example hj,t(xi). Zj,t is a normalization factor, such that Dj,t+1 becomes normally distributed. Line 15 selects m samples with replacement from S based on Dj,t+1, which are then provided with Λ as inputs to the CNN variant to learn a model hj,t+1. Such a process (i.e., lines 8–16) is repeated an additional t − 1 times. In lines 17–19, prediction of a given jth ensemble is performed via taking the sign of the weighted majority vote of T CNN variants. Lines 5–20 are repeated an additional k − 1 times to generate a k ensemble consisting of T CNN variants. Line 21 aims to select the ensemble that maximizes the area under the curve (AUC) during the training, where the AUC is a performance measure that takes predictions and actual training labels as input and returns the measured performance. For a given test example , the prediction is performed using the ensemble j* (see line 23).
3.3. The Second Boosted Deep Learning Approach (B2DL)
The B2DL approach aims to create a better set of classifiers and performs the same steps as B1DL. However, B2DL differs from B1DL in terms of the weight update mechanism of lines 12–14 in the B2 algorithm (Algorithm 3). As in [38], B2 aims to update the weights in Dj,t+1 using previous t classifiers. In particular, B2DL aims to increase the weight of an incorrectly predicted example xi to focus on in the next iteration if the majority of classifiers in the previous t iterations incorrectly predict xi. If the majority of classifiers in previous t iterations correctly predict xi, then its weight is decreased in the next iteration.
| Algorithm 3: Boost2 (B2). |
 |
4. Experiments and Results
In this section, we first describe the datasets utilized in this work. Then, the experimental methodology is presented. Lastly, the proposed approaches are compared against several baselines using several performance measures.
4.1. Dataset
The dataset consisted of 306 in vitro bovine embryo images. Of these, 112 were classified as good (i.e., grade 1), while the remaining images were classified as poor (i.e., grade 3). That is, a bovine embryo with grade 1 is considered a good enough embryo to be transferred or frozen for later use, while bovine images associated with grade 3 are considered not good enough to be transferred into a recipient. The dataset used in our study was downloaded from [2], which was produced via In Vitro Brasil (http://www.invitrobrasil.com/en/ (accessed on 14 March 2022)) according to the protocol described in [39]. The quality of bovine embryos was evaluated by three experienced embryologists utilizing the IETS standards [5]. As the grading might differ between embryologists, the modal value was used [2,40,41].
4.2. Experimental Methodology
In this study, we compared the proposed deep learning approaches against baseline convolutional neural network architectures, including LeNet and AlexNet. Details about the baselines are provided below. Also, a summary of all approaches is provided in Table 1.

Table 1.
Summary of the prediction algorithms used in our study.
4.2.1. LeNet
This baseline works under a supervised learning setting, providing a training set consisting of in vitro bovine embryo images as input to a CNN to obtain a model. Then, the resulted model is applied to test examples of unseen bovine embryo images, to generate predictions. The only difference between LeNet in this study and LeNet in Section 2.2 is the Softmax used in the output layer for the binary classification task (two instead of 10 possible categorical labels).
4.2.2. AlexNet
This baseline utilizes the CNN architecture, described in Section 2.3, which takes as input the set of in vitro bovine images, to generate a model. Then, the obtained model is used to classify unseen bovine embryo images. The only difference between AlexNet in this study and AlexNet in Section 2.3 is the Softmax used in the output layer for the binary classification task (two instead of 1000 possible categorical labels).
F1, accuracy (ACC), G-Mean (GM), and area under curve (AUC) performance measures were calculated based on the confusion matrix in Table 2 as follows [31,42]. As training deep learning is time-consuming and requires extensive computational resources [43,44], all prediction algorithms were evaluated using two-fold cross-validation as in [45,46,47], where training and test sets were allocated distinct folds. Specifically, in each iteration step j ∊ {1,2}, the test set took examples from fold j while the training set took all examples from the remaining folds. Then, learning was performed to induce the model and perform predictions on examples in test set, followed by calculating the performance results using F1, ACC, GM, and AUC. This process was repeated for one more iteration in which the four performance results were reported. Finally, we reported the mean for F1, ACC, GM, and AUC, which correspond to the performance results utilizing two-fold cross-validation. We designated these performance measures as MF1, MACC, MGM, and MAUC [44]. In addition, the standard deviation (SD) of the two testing folds was recorded. The software used in our work included TensorFlow [48], Keras [49], and reticulate [50]. The experimental results were analyzed using R [51].

Table 2.
Confusion matrix for the two-class classification problem.
4.3. Experimental Results
In this section, we evaluate all deep learning approaches, reporting two-fold cross-validation results based on in vitro bovine embryo images.
Predicting the Quality of Bovine Embryos
Table 3 provides a detailed overview of the structures of all approaches. The convolutional layer aims to extract special feature maps, followed by using either an average or max pooling layer for dimensionality reduction. The dropout layers were utilized to avoid overfitting during training [52,53]. A fully connected layer and an output layer were employed for the classification of bovine embryos. AlexNet utilizes a batch normalization layer to reduce the effects of covariate shift and accelerate the training process.

Table 3.
Detailed overview of Layer Type (and no. of parameters) for all models. DL is the proposed model. LeNet and AlexNet are the compared models. No. of parameters is omitted when no parameters were there.
Figure 6 reports the training accuracy curves for each approach. It can be seen that the training accuracy continues to increase as the number of epochs increases. While AlexNet generates the highest training accuracy, it does not outperform our approaches (see results in Table 4). Moreover, AlexNet in our study requires 24,773,378 parameters. The number of parameters in the architecture utilized in our approaches is 6,417,314. The total number of parameters in the LeNet architecture is 61,326.
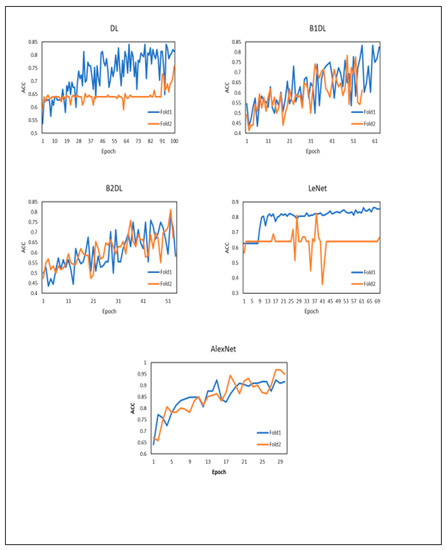
Figure 6.
Training accuracy plots for different deep learning approaches when running two-fold cross-validation. ACC denotes the accuracy.

Table 4.
Performance results of all prediction algorithms according to two-fold cross-validation on images pertaining to in vitro bovine embryos. The highest MF1, MACC, MGM, and MAUC values are indicated in bold. MF1 indicates the Mean F1. MACC indicates the mean accuracy. MGM indicates the mean G-Mean. MAUC indicates the mean area under the curve. SD indicates the standard deviation.
Table 4 reports the performance results of all studied prediction algorithms in this work. It can be seen from Table 4 that prediction algorithms utilizing our approaches generate better performance results when compared to the two baselines. Specifically, the prediction algorithm B2DL generates the highest MF1 of 0.863. Moreover, B2DL generates the highest MACC of 0.807. Considering performance metrics pertaining to the imbalanced classifications, B1DL performed better than all other prediction algorithms, achieving the highest MAUC of 0.773 and the highest MGM of 0.765. These results show the high performance of prediction algorithms exploiting the proposed deep learning approaches. For each prediction algorithm, Figure 7 ranks the total running times spent in the two-fold cross-validation technique. The results reported in Figure 7 show that the prediction algorithms DL and B2DL utilizing our approaches were faster than all other baseline algorithms. B1DL was slower than DL, B2DL, and LeNet. AlexNet was the slowest prediction algorithm.
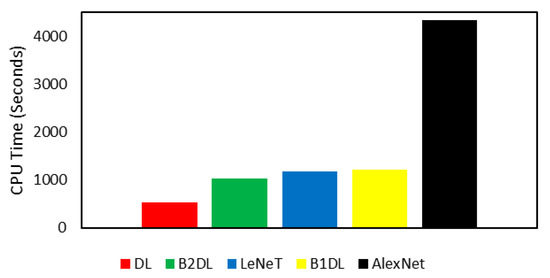
Figure 7.
Comparison of the total running times (in seconds) of all predictions of the quality of in vitro bovine embryos. The prediction algorithm on the far left is the fastest, while the prediction algorithm on the far right is the slowest.
We assessed the stability of the reported results, running cross-validation (CV) of prediction algorithms 10 times. Figure 8a–d and Figure 9a–d show the performance results obtained from 10 runs of cross-validation. Figure 8a,c and Figure 9a,c show that prediction algorithms utilizing B1DL and B2DL performed better than the baselines in terms of MF1, MACC, MGM, and MAUC. These results clearly demonstrate the predictive superiority of B1DL and B2DL.
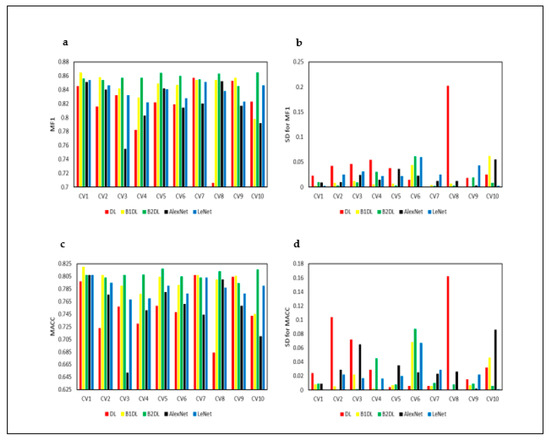
Figure 8.
Performance results of the studied prediction algorithms, applied to test examples pertaining to in vitro bovine embryos. These results are obtained by running CV 10 times. CVi is the ith cross-validation. MF1 is the mean F1. MACC is the mean accuracy. SD is the standard deviation.
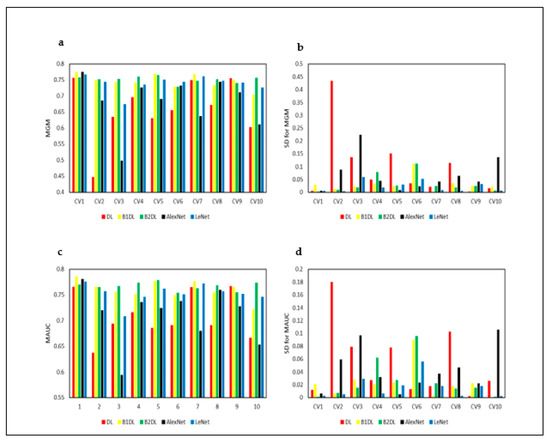
Figure 9.
Performance results of all prediction algorithms, applied to test examples pertaining to in vitro bovine embryos. These results are obtained by running CV 10 times. CVi is the ith cross-validation MGM is the mean G-Mean. MAUC is the mean AUC. SD is the standard deviation.
The nonparametric Friedman test was utilized to assess the statistical significance [54,55,56,57,58,59,60]. In Table 5, we report the average rankings of all pairs of prediction algorithms and the corresponding p-values, calculated based on MF1 in Figure 8a. Table 5 shows that B2DL outperformed DL, LeNet, and AlexNet, and the differences were statistically significant (p < 0.05). B1DL was also significantly better than DL and AlexNet (p < 0.05). The differences in prediction algorithms that were not significant (p ≥ 0.05) are shown in black.

Table 5.
p-values and average rankings of all pairs of the prediction algorithms in Figure 8a on the images pertaining to in vitro bovine embryos based on the Friedman test. The lower the rank of a prediction algorithm, the higher the performance. The difference between a pair of prediction algorithms with p < 0.001 (in red) is considered highly statistically significant. The difference between the two algorithms in a pair with p < 0.05 (in blue) is considered statistically significant.
5. Discussion
Our first deep learning approach (DL) aims to learn a model for classifying in vitro bovine embryos using a CNN variant, which includes a convolutional layer and pooling layer for the feature extraction and dimensionality reduction, fully connected layers, and an output layer for the classification. The resulting model is used to classify test examples of in vitro bovine embryo images. The second deep learning approach (B1DL) utilizes the CNN used in the DL approach, coupled with a boosting technique, to yield an ensemble of models, which are then used to classify test examples. The third deep learning approach (B2DL) behaves like the B1DL approach. However, B2DL has a different boosting technique in terms of the weight update mechanism.
Predicting the quality of bovine embryos is a challenging task. Making accurate predictions of the quality of bovine embryos could be specifically adapted to optimize the cryopreservation and avoid embryo selection in countries where it is not permitted, which in turn could help embryologists to enhance laboratory protocols, leading to successful pregnancies [20,22,61]. In this study, we aim to improve the prediction of the quality of bovine embryos via leveraging deep learning techniques that consist of a variant of CNN with or without boosting techniques. Experimental results show that our approaches perform better than baseline approaches.
For the optimization process during the training of LeNet and AlexNet, we used the Adam optimizer in this study with the default parameters (as suggested in [62]) to minimize the categorical cross-entropy loss function as in [63]. For our approaches, categorical cross-entropy is used as a loss function and the optimization is performed via RMSProp optimizer [64,65], with default parameters given in [49].
In this study, we ran the cross-validation 10 times to assess the stability of prediction algorithms. The reported p-values obtained from a Friedman test show that the prediction algorithms utilizing our approaches perform better than the baseline methods that utilize LeNet and AlexNet architectures.
The deep learning training phase requires additional time depending on the number of processing layers. The more layers a network has, the more computational running time is needed during the training. Therefore, we utilized deep learning consisting of fewer layers, because the proposed approaches work in an iterative manner, where DL is trained in each iteration.
While our boosted approaches require the invoking of CNN several times, our computational experiments show that the B2DL prediction algorithm is efficient as it requires less running time than the baseline methods. This is due to B2DL including just four layers compared to other baseline methods (i.e., LeNet and AlexNet) consisting of more than five. AlexNet was the slowest prediction algorithm.
6. Conclusions and Future Work
We propose three deep learning approaches to accurately predict the quality of in vitro bovine embryos. The first approach DL utilizes a convolutional neural network variant consisting of four layers, excluding the input and output layers. The DL approach aims to learn a model from a training set of images pertaining to in vitro bovine embryos. The resulting model is then applied to perform predictions on unseen test examples. The second approach, B1DL, is an extension of the DL approach. The only difference is that B1DL utilizes a variant of the boosting technique to select the best ensemble in terms of the maximum area under the curve during the training, from a set of several ensemble models. Then, the best ensemble is used to perform predictions on unseen text examples. The third approach, B2DL, is similar to B1DL. The only difference is that B2DL utilizes a boosting technique with a different weight update mechanism. These deep learning approaches are designed to improve the predictive performance. Experimental results on images pertaining to in vitro bovine embryos demonstrate that our approaches perform better than existing baseline methods and achieve statistically significant results.
In future work, we plan to (1) adopt the proposed approaches under the transfer learning setting; (2) incorporate the proposed approaches with generative adversarial networks to improve the prediction performance under the transfer learning setting [66,67,68]; and (3) evaluate the proposed deep learning approaches against others in the context of different medical imaging problems [69,70].
Author Contributions
T.T. and Z.W. planned the research. T.T. and Z.W. designed the algorithms. T.T. conducted the experimental work. T.T. and Z.W. discussed the results, wrote, and approved the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under grant no. D-289-611-1440. The authors, therefore, thank DSR for technical and financial support.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used in this study are available in [2].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jones, R.E.; Lopez, K.H. Human Reproductive Biology; Academic Press: New York, NY, USA, 2013. [Google Scholar]
- Rocha, J.C.; Passalia, F.J.; Matos, F.D.; Takahashi, M.B.; Maserati, M.P., Jr.; Alves, M.F.; De Almeida, T.G.; Cardoso, B.L.; Basso, A.C.; Nogueira, M.F.G. Automatized image processing of bovine blastocysts produced in vitro for quantitative variable determination. Sci. Data 2017, 4, 170192. [Google Scholar] [CrossRef] [PubMed]
- Sirard, M.A. The ovarian follicle of cows as a model for human. Anim. Models Hum. Reprod. 2017, 127, 127–144. [Google Scholar]
- Baerwald, A. Human antral folliculogenesis: What we have learned from the bovine and equine models. Anim. Reprod. 2009, 6, 20–29. [Google Scholar]
- Bo, G.; Mapletoft, R. Evaluation and classification of bovine embryos. Anim. Reprod. 2013, 10, 344–348. [Google Scholar]
- Alfuraiji, M.; Atkinson, T.; Broadbent, P.; Hutchinson, J. Superovulation in cattle using PMSG followed by PMSG-monoclonal antibodies. Anim. Reprod. Sci. 1993, 33, 99–109. [Google Scholar] [CrossRef]
- Behringer, R.; Gertsenstein, M.; Nagy, K.V.; Nagy, A. Manipulating the Mouse Embryo: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2014. [Google Scholar]
- Ferré, L.; Kjelland, M.; Strøbech, L.; Hyttel, P.; Mermillod, P.; Ross, P. Recent advances in bovine in vitro embryo production: Reproductive biotechnology history and methods. Animal 2020, 14, 991–1004. [Google Scholar] [CrossRef] [Green Version]
- Ker, J.; Wang, L.; Rao, J.; Lim, T. Deep Learning Applications in Medical Image Analysis. IEEE Access 2018, 6, 9375–9389. [Google Scholar] [CrossRef]
- Gibson, E.; Li, W.; Sudre, C.; Fidon, L.; Shakir, D.I.; Wang, G.; Eaton-Rosen, Z.; Gray, R.; Doel, T.; Hu, Y. NiftyNet: A deep-learning platform for medical imaging. Comput. Methods Programs Biomed. 2018, 158, 113–122. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-Van De Kaa, C.; Bult, P.; Van Ginneken, B.; Van Der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef] [Green Version]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Gargeya, R.; Leng, T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology 2017, 124, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, C.; Cheng, J.; Chen, X.; Wang, Z.J. A multi-scale data fusion framework for bone age assessment with convolutional neural networks. Comput. Biol. Med. 2019, 108, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Mohri, M.; Rostamizadeh, A.; Talwalkar, A. Foundations of Machine Learning; MIT Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Balaban, B.; Urman, B.; Sertac, A.; Alatas, C.; Aksoy, S.; Mercan, R. Blastocyst quality affects the success of blastocyst-stage embryo transfer. Fertil. Steril. 2000, 74, 282–287. [Google Scholar] [CrossRef]
- Lonergan, P. State-of-the-art embryo technologies in cattle. Soc. Reprod. Fertil. Suppl. 2007, 64, 315–325. [Google Scholar] [CrossRef]
- Voelkel, S.; Hu, Y. Direct transfer of frozen-thawed bovine embryos. Theriogenology 1992, 37, 23–37. [Google Scholar] [CrossRef]
- Hourvitz, A.; Lerner-Geva, L.; Elizur, S.E.; Baum, M.; Levron, J.; David, B.; Meirow, D.; Yaron, R.; Dor, J. Role of embryo quality in predicting early pregnancy loss following assisted reproductive technology. Reprod. Biomed. Online 2006, 13, 504–509. [Google Scholar] [CrossRef]
- Rocha, J.C.; Passalia, F.J.; Matos, F.D.; Takahashi, M.B.; de Souza Ciniciato, D.; Maserati, M.P.; Alves, M.F.; De Almeida, T.G.; Cardoso, B.L.; Basso, A.C. A method based on artificial intelligence to fully automatize the evaluation of bovine blastocyst images. Sci. Rep. 2017, 7, 7659. [Google Scholar] [CrossRef] [Green Version]
- Manna, C.; Nanni, L.; Lumini, A.; Pappalardo, S. Artificial intelligence techniques for embryo and oocyte classification. Reprod. Biomed. Online 2013, 26, 42–49. [Google Scholar] [CrossRef]
- Filho, E.S.; Noble, J.; Poli, M.; Griffiths, T.; Emerson, G.; Wells, D. A method for semi-automatic grading of human blastocyst microscope images. Hum. Reprod. 2012, 27, 2641–2648. [Google Scholar] [CrossRef] [Green Version]
- Tran, D.; Cooke, S.; Illingworth, P.; Gardner, D. Deep learning as a predictive tool for fetal heart pregnancy following time-lapse incubation and blastocyst transfer. Hum. Reprod. 2019, 34, 1011–1018. [Google Scholar] [CrossRef] [Green Version]
- Blank, C.; Wildeboer, R.R.; DeCroo, I.; Tilleman, K.; Weyers, B.; de Sutter, P.; Mischi, M.; Schoot, B.C. Prediction of implantation after blastocyst transfer in in vitro fertilization: A machine-learning perspective. Fertil. Steril. 2019, 111, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, Y.; Habara, T.; Hirata, R.; Hayashi, N. Feasibility of deep learning for predicting live birth from a blastocyst image in patients classified by age. Reprod. Med. Biol. 2019, 18, 190–203. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, X.; Zhang, Z.; Wu, F.; Zhao, L. Deep learning driven blockwise moving object detection with binary scene modeling. Neurocomputing 2015, 168, 454–463. [Google Scholar] [CrossRef]
- Dhungel, N.; Carneiro, G.; Bradley, A.P. A deep learning approach for the analysis of masses in mammograms with minimal user intervention. Med. Image Anal. 2017, 37, 114–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoaran, M.; Haghi, B.A.; Taghavi, M.; Farivar, M.; Emami-Neyestanak, A. Energy-efficient classification for resource-constrained biomedical applications. IEEE J. Emerg. Sel. Top. Circuits Syst. 2018, 8, 693–707. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Turki, T.; Taguchi, Y. Machine Learning Algorithms for Predicting Drugs–Tissues Relationships. Expert Syst. Appl. 2019, 127, 167–186. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Pool, J.; Tran, J.; Dally, W. Learning both weights and connections for efficient neural network. In Proceedings of the NIPS 2015, Montréal, QC, Canada, 7–10 December 2015; pp. 1135–1143. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Stateline, NV, USA, 3–8 December 2012; pp. 1097–1105. [Google Scholar]
- Juefei-Xu, F.; Naresh Boddeti, V.; Savvides, M. Local binary convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 19–28. [Google Scholar]
- Yang, H.-F.; Lin, K.; Chen, C.-S. Supervised learning of semantics-preserving hash via deep convolutional neural networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 437–451. [Google Scholar] [CrossRef] [Green Version]
- Turki, T.; Wang, J.T. Reverse engineering gene regulatory networks using sampling and boosting techniques. In Proceedings of the International Conference on Machine Learning and Data Mining in Pattern Recognition, New York, NY, USA, 15–19 July 2018; Springer: Berlin/Heidelberg, Germany; pp. 63–77. [Google Scholar]
- Sanches, B.V.; Lunardelli, P.A.; Tannura, J.H.; Cardoso, B.L.; Pereira, M.H.C.; Gaitkoski, D.; Basso, A.C.; Arnold, D.R.; Seneda, M.M. A new direct transfer protocol for cryopreserved IVF embryos. Theriogenology 2016, 85, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Cao, S.; Zheng, H.; Hu, D.; Lin, J.; Cui, B.; Lin, H.; Hu, R.; Wu, B.; Sun, Y. Genetic characterization of Chinese fir from six provinces in southern China and construction of a core collection. Sci. Rep. 2017, 7, 13814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, B.J.; Ishimoto, K.; Wheeler, R.J. Automated identification of flagella from videomicroscopy via the medial axis transform. Sci. Rep. 2019, 9, 5015. [Google Scholar] [CrossRef] [PubMed]
- Japkowicz, N.; Shah, M. Evaluating Learning Algorithms: A Classification Perspective; Cambridge University Press: New York City, NY, USA, 2011. [Google Scholar]
- Tang, J.; Deng, C.; Huang, G.-B. Extreme learning machine for multilayer perceptron. IEEE Trans. Neural Netw. Learn. Syst. 2015, 27, 809–821. [Google Scholar] [CrossRef]
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirinukunwattana, K.; Raza, S.e.A.; Tsang, Y.-W.; Snead, D.R.; Cree, I.A.; Rajpoot, N.M. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imaging 2016, 35, 1196–1206. [Google Scholar] [CrossRef] [Green Version]
- Kleesiek, J.; Urban, G.; Hubert, A.; Schwarz, D.; Maier-Hein, K.; Bendszus, M.; Biller, A. Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. NeuroImage 2016, 129, 460–469. [Google Scholar] [CrossRef]
- Andrew, W.; Greatwood, C.; Burghardt, T. Visual localisation and individual identification of Holstein Friesian cattle via deep learning. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2850–2859. [Google Scholar]
- Allaire, J.; Tang, Y. Tensorflow: R Interface to ‘TensorFlow’; R Package Version. 2018. Available online: https://cran.r-project.org/web/packages/tensorflow/index.html (accessed on 14 March 2022).
- Chollet, F.; Allaire, J. R Interface to Keras; GitHub: San Francisco, CA, USA, 2017. [Google Scholar]
- Allaire, J.; Ushey, K.; Tang, Y.; Eddelbuettel, D. Reticulate: Interface to ’Python’; R Package Version. 2018. Available online: https://cran.r-project.org/web/packages/reticulate/index.html (accessed on 14 March 2022).
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: http://www.R-project.org (accessed on 15 February 2021).
- Oh, S.L.; Ng, E.Y.; San Tan, R.; Acharya, U.R. Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats. Comput. Biol. Med. 2018, 102, 278–287. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Raghavendra, U.; Tan, J.H.; Adam, M.; Gertych, A.; Hagiwara, Y. Automated identification of shockable and non-shockable life-threatening ventricular arrhythmias using convolutional neural network. Future Gener. Comput. Syst. 2018, 79, 952–959. [Google Scholar] [CrossRef]
- Shang, L.; Lu, Z.; Li, H. Neural Responding Machine for Short-Text Conversation. In Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics (ACL), Beijing, China, 16–21 August 2015; pp. 1577–1586. [Google Scholar]
- Brzezinski, D.; Stefanowski, J. Reacting to different types of concept drift: The accuracy updated ensemble algorithm. IEEE Trans. Neural Netw. Learn. Syst. 2014, 25, 81–94. [Google Scholar] [CrossRef] [Green Version]
- Calvo, B.; Santafé Rodrigo, G. scmamp: Statistical comparison of multiple algorithms in multiple problems. R J. 2016, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- García, S.; Fernández, A.; Luengo, J.; Herrera, F. Advanced nonparametric tests for multiple comparisons in the design of experiments in computational intelligence and data mining: Experimental analysis of power. Inf. Sci. 2010, 180, 2044–2064. [Google Scholar] [CrossRef]
- Demšar, J. Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Rodríguez-Fdez, I.; Canosa, A.; Mucientes, M.; Bugarín, A. STAC: A web platform for the comparison of algorithms using statistical tests. In Proceedings of the 2015 IEEE International Conference on Fuzzy Systems, Istanbul, Turkey, 2–5 August 2015; pp. 1–8. [Google Scholar]
- Howell, D.C. Fundamental Statistics for the Behavioral Sciences; PSY 200 (300) Quantitative Methods in Psychology; Wadsworth Cengage Learnin: Mason, OH, USA, 2010. [Google Scholar]
- Cervera, R.; Garcia-Ximénez, F. Vitrification of zona-free rabbit expanded or hatching blastocysts: A possible model for human blastocysts. Hum. Reprod. 2003, 18, 2151–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Anthimopoulos, M.; Christodoulidis, S.; Ebner, L.; Christe, A.; Mougiakakou, S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans. Med. Imaging 2016, 35, 1207–1216. [Google Scholar] [CrossRef]
- Vadicamo, L.; Carrara, F.; Cimino, A.; Cresci, S.; Dell’Orletta, F.; Falchi, F.; Tesconi, M. Cross-media learning for image sentiment analysis in the wild. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 308–317. [Google Scholar]
- Tieleman, T.; Hinton, G. Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude. Coursera Neural Netw. Mach. Learn. 2012, 4, 26–31. [Google Scholar]
- Bengio, Y.; Goodfellow, I.J.; Courville, A. Deep learning. Nature 2015, 521, 436–444. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the NIPS 2014, Montreal, QC, Canada, 8–13 December 2014; pp. 2672–2680. [Google Scholar]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).