Emerging Applications of Bio-Inspired Algorithms in Image Segmentation
Abstract
1. Introduction
- Presenting four bio-inspired-algorithm-based image-segmentation techniques not provided in previous surveys.
- Indicating the well-used AI algorithms in solving image segmentation to help researchers choose the best segmentation technique.
- Emphasizing the algorithms that are not mainly used in this regard to open new opportunities in handling and enhancing image segmentation.
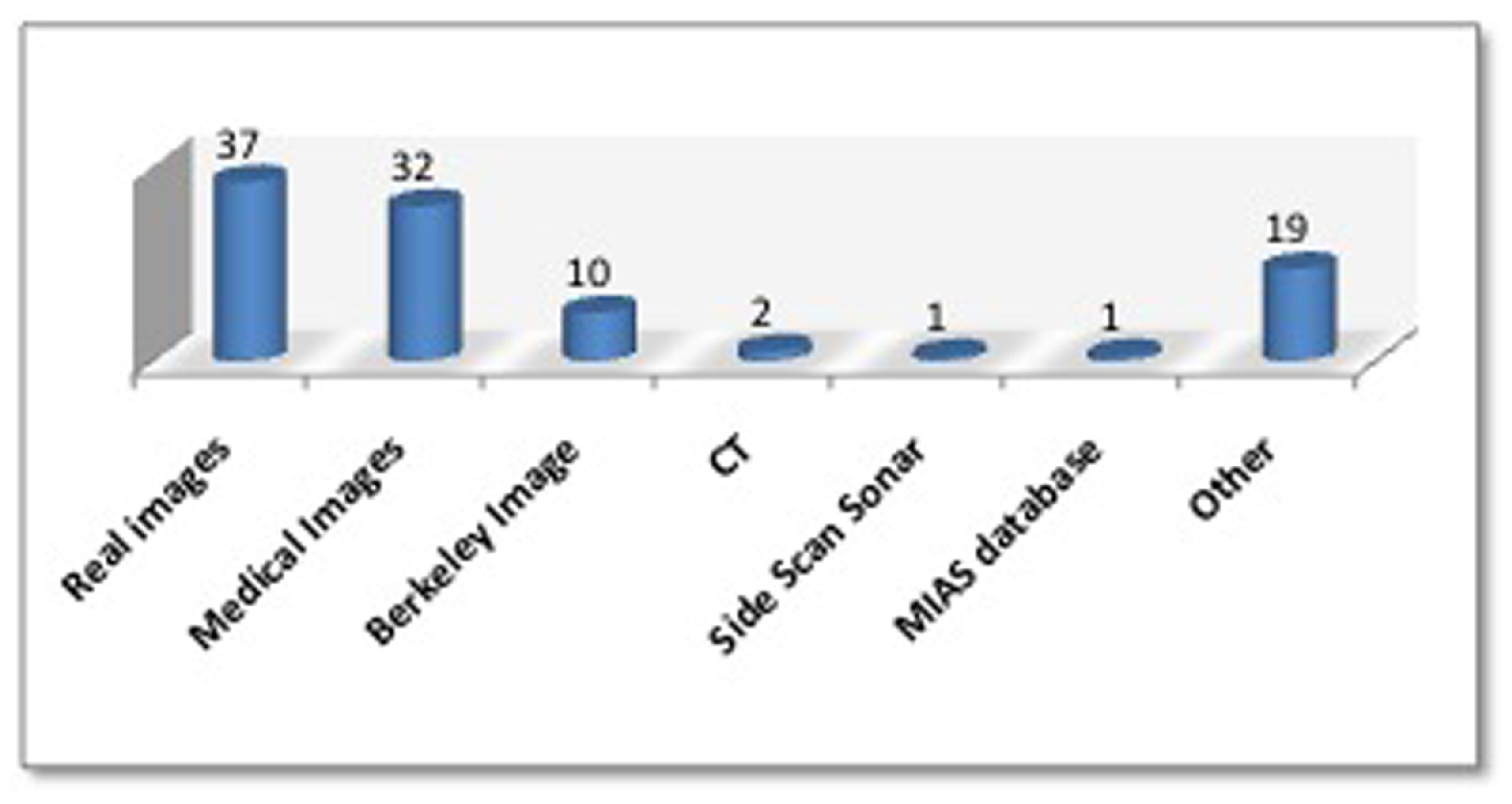
- Presenting various databases dedicated for the use in image segmentation to be served as a repository for future research.
- Investigating the parameter setting and recommending the most-used values for the non-expert AI researchers working in image segmentation.
- Suggesting new directions and future works for exploration to expand this research field.
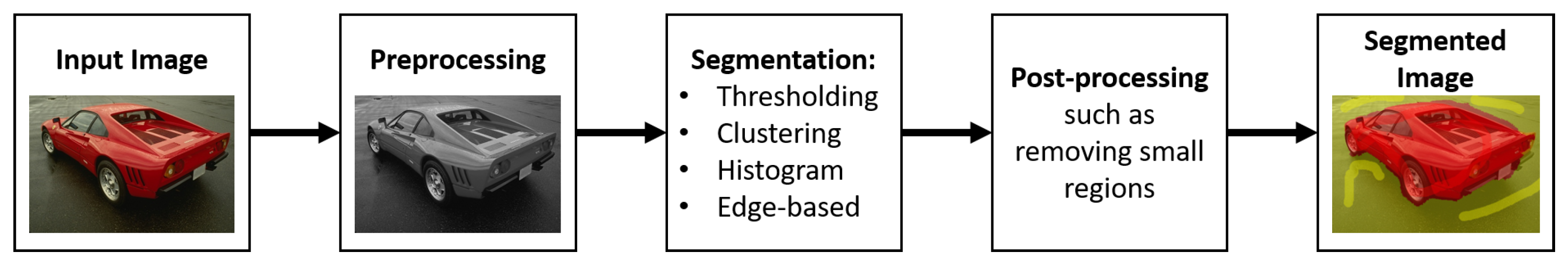
2. Image Segmentation
3. Bio-Inspired Algorithms
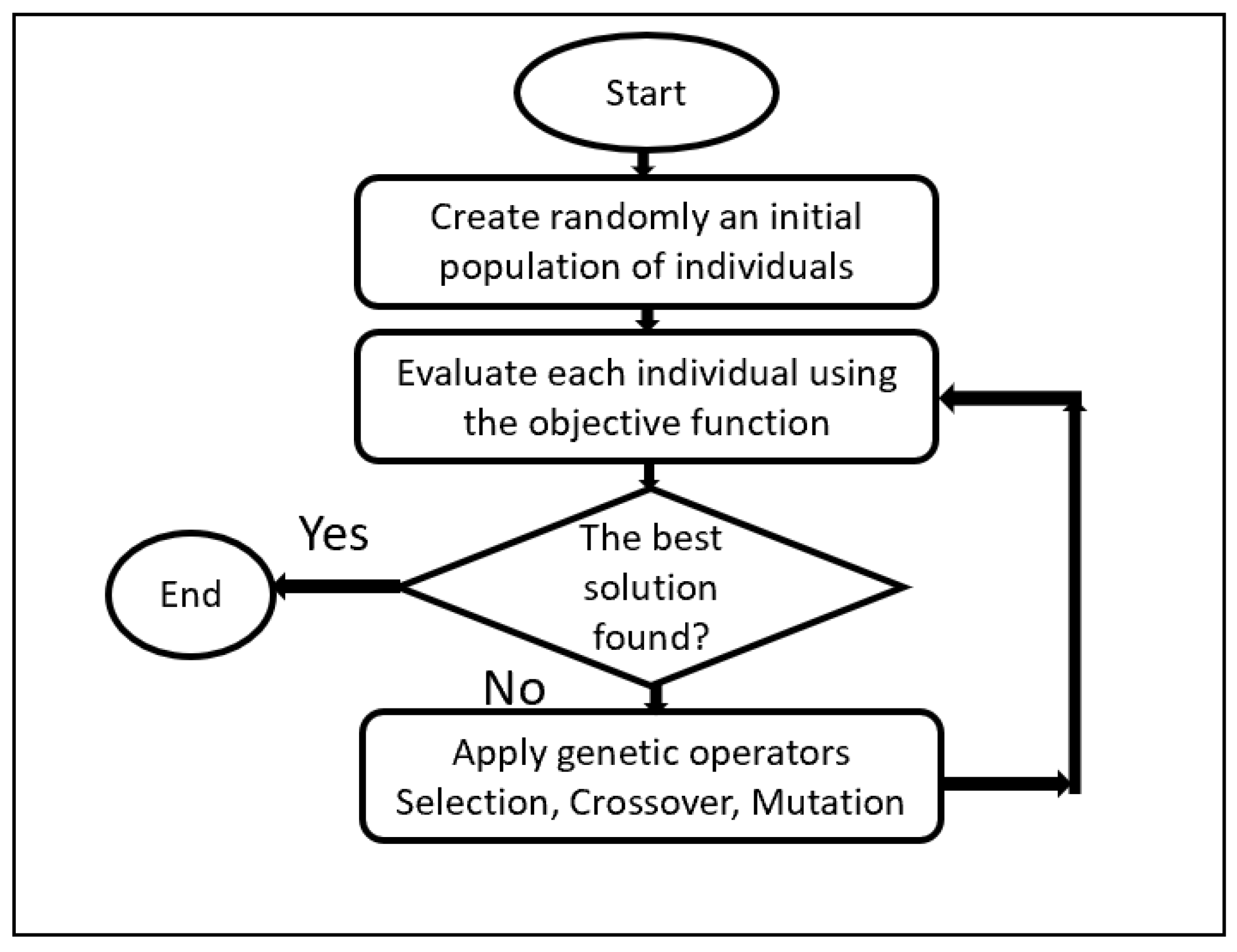
3.1. Genetic Algorithm (GA)
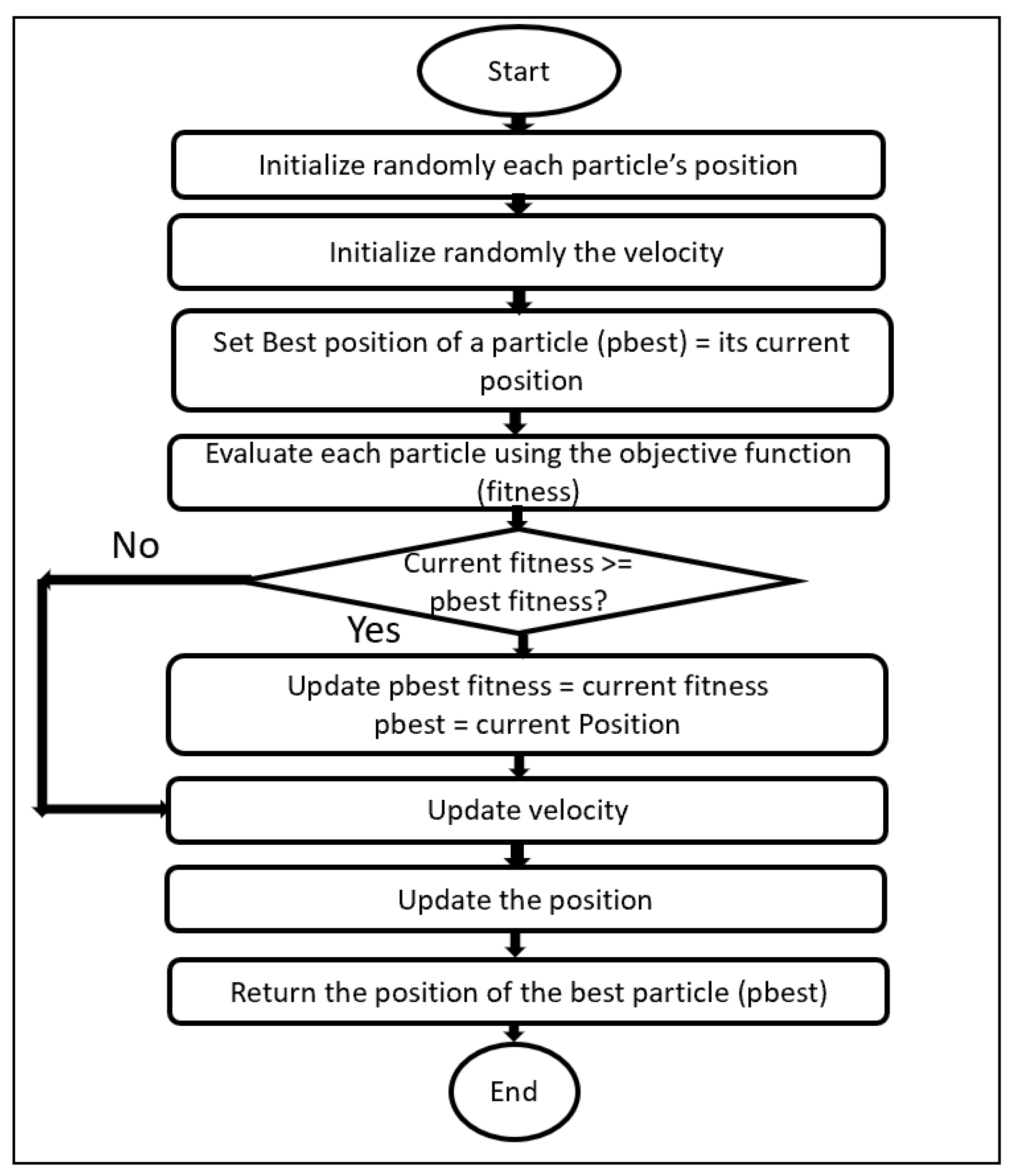
3.2. Particle Swarm Optimization (PSO)
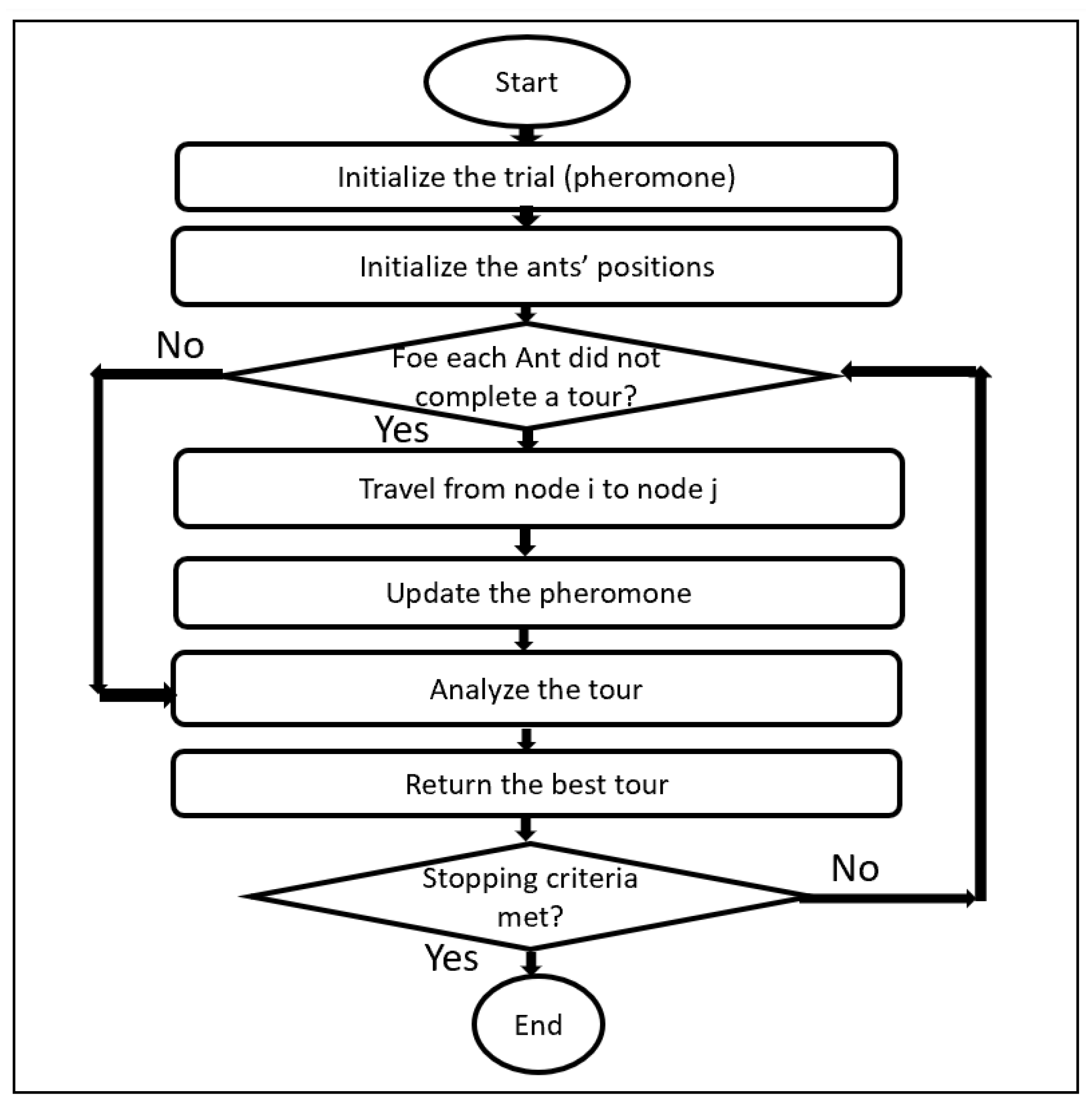
3.3. Ant Colony Optimization (ACO)
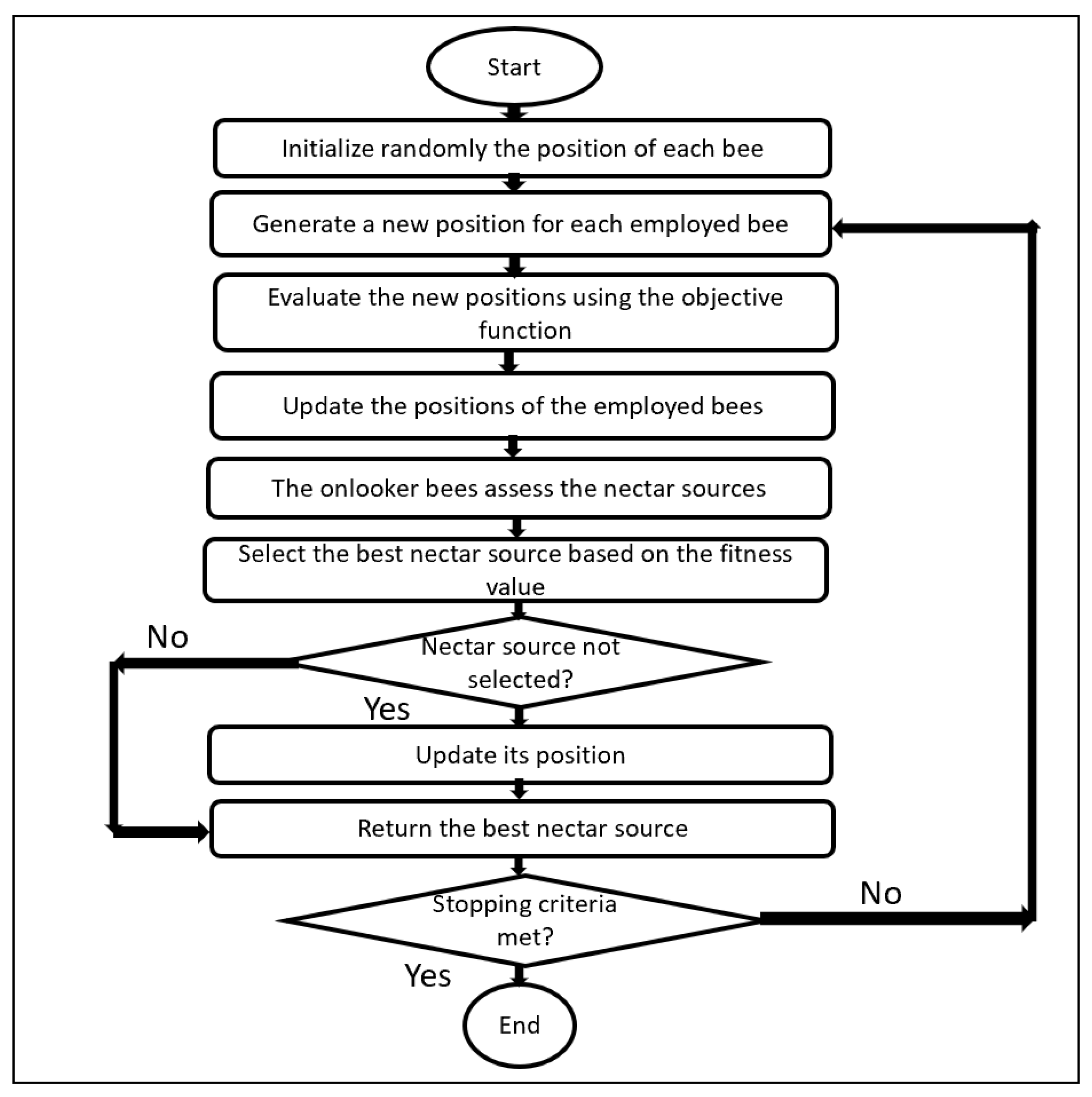
3.4. Artificial Bee Colony (ABC)
3.5. Adaptation of the Bio-Inspired Algorithms
- Defining the candidate solution.
- Defining the fitness function.
3.5.1. Thresholding Techniques
3.5.2. Clustering Techniques
4. Literature Review
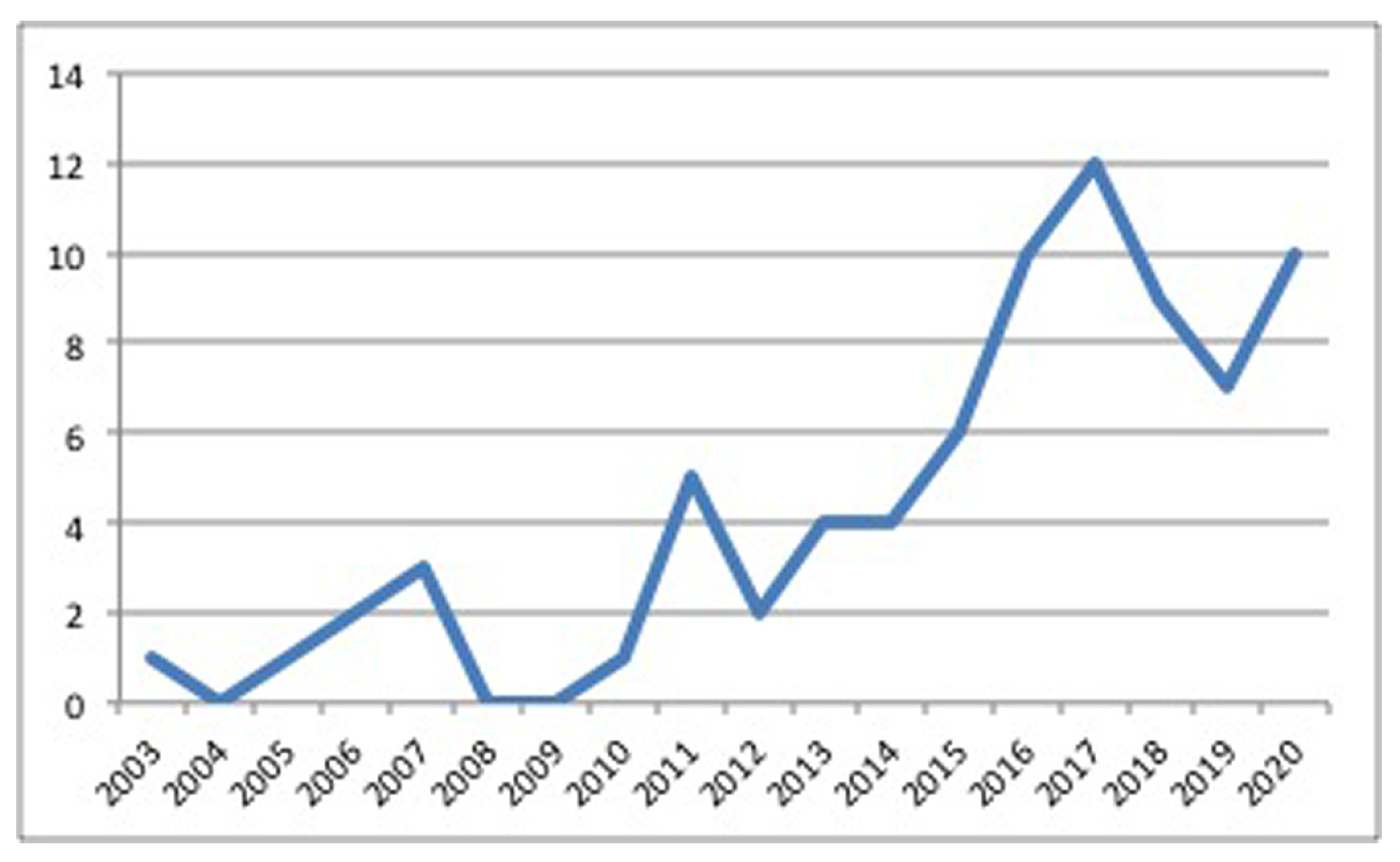
4.1. Sources and Search Method
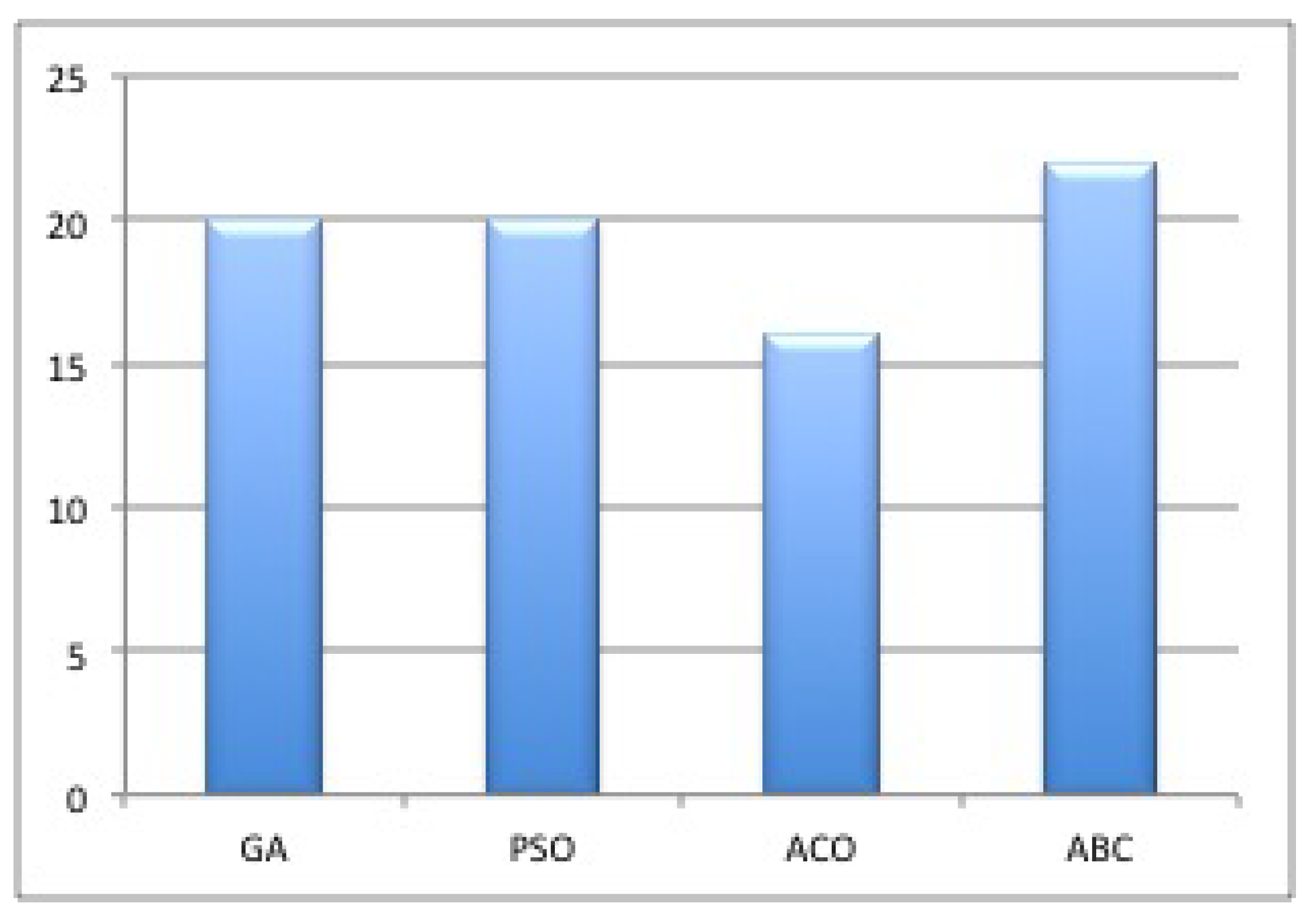
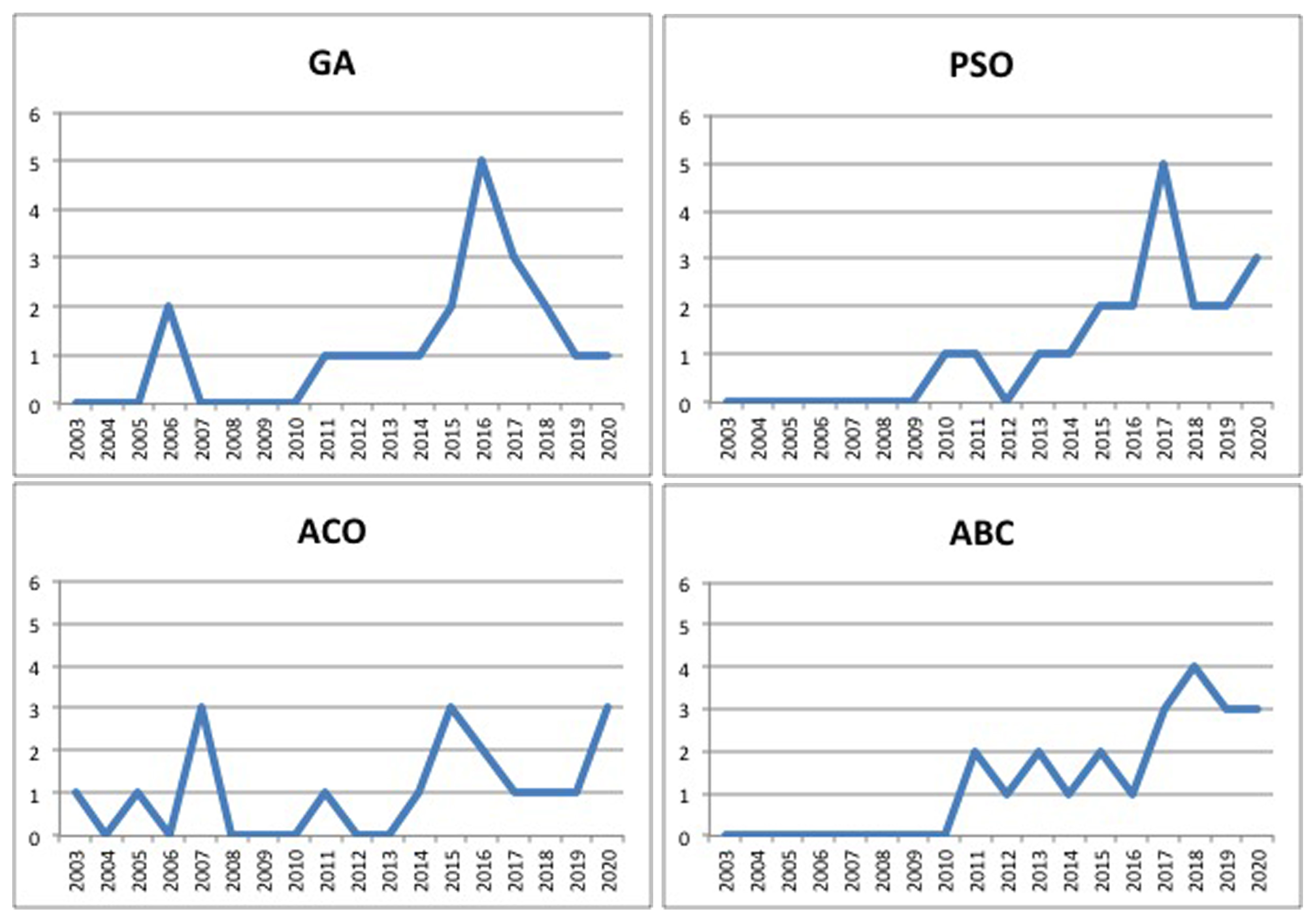
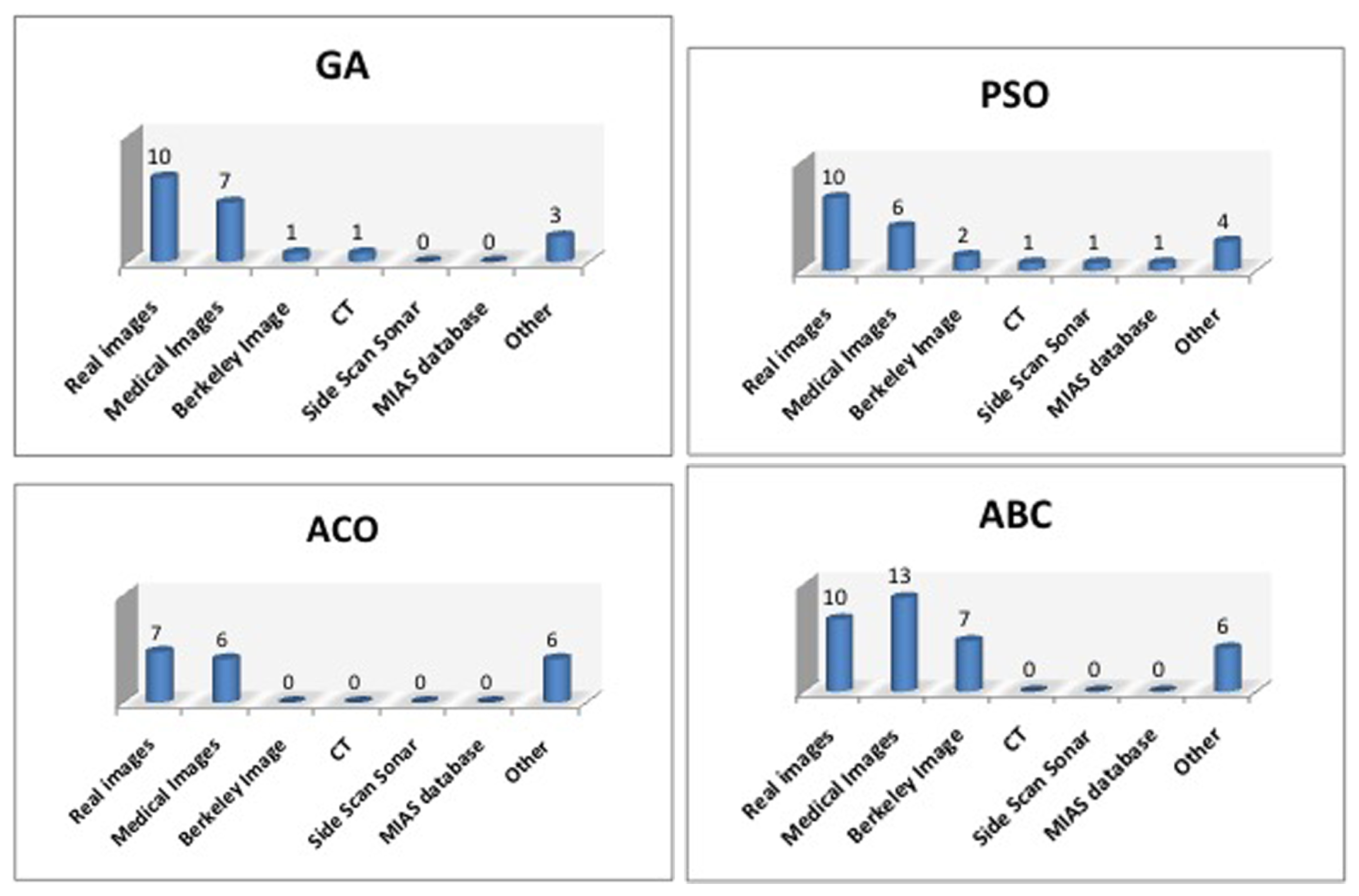
4.2. Image-Segmentation-Based Bio-Inspired Algorithms
4.3. Image Segmentation Based on Genetic Algorithms (GA)
| Population Size (N), Crossover Probability (), Mutation Probability (), Iteration Numver (Iter) | |||
|---|---|---|---|
| Reference | Dataset | Parameter Setting | Compared Methods |
| [29] | Cardiac images | Not provided | None |
| [30] | MRI brain images | Exhaustive search method | |
| Basic GA | |||
| [31] | Science Photo Library | Not provided | Sorting GA (SGA) |
| Adaptive GA (AGA) | |||
| Non-dominated sorting GA (NSGA) | |||
| [33] | CHASE database | Not provided | None |
| [34] | Lena, Cameraman, House | GSA with multi-threshold | |
| Berkeley Segmentation dataset | , | Three GSA variants with multi-level threshold | |
| [35] | MRI and CT | and 50 | Chan and Vese algorithm |
| and | |||
| [36] | MRI brain tumor | , | None |
| [37] | Grape, Peppers, Brain | 2D thresholding methods | |
| Light microscopy and others | |||
| [38] | 22 benchmark images | Classical thresholding method | |
| [39] | Plant leaf images | None | K-means |
| [40] | Skin lesions demoscopic images | None | K-means and -K-means |
| [41] | Simulated thermal images | , | Otsu, Iterative Thresholding |
| K-means, Hamadani | |||
| Seeded Region Growing | |||
| [42] | Plant dataset | No Applied | None |
| [43] | A set of 12 images | , | 6 swarm methods |
| [44] | Arbitrary gray images | None | |
| [45] | House | Otsu’s approach | |
| Table lamp | [19] | ||
| Ceiling light | |||
| [46] | Rice Image | None | |
| Cameraman | |||
| Lifting body | |||
| Fishman | |||
| [47] | Lena | FHNN | |
| [48] | Car | , | None |
| RGB Images | and not provided | ||
| [49] | MRI images | Not provided | None |
4.4. Image Segmentation Based on Particle Swarm Optimization (PSO)
4.5. Image Segmentation Based on Ant Colony Optimization (ACO)
4.6. Image Segmentation Based on Artificial Bee Colony (ABC)
5. Discussion
5.1. Segmentation Techniques
5.2. Bioinspired Techniques
5.3. Parameter Settings
5.4. Datasets
5.5. Evaluation Metrics
- Accuracy: measures how close the results are to the desired value calculated as the ratio of the correct predictions to the total sample.
- Error rate: is the inaccuracy of the predicted output values of the categorical dataset.
- Sensitivity: is the amount of positives that are correctly recognized.
- Precision: is the fraction of the related instances within the retrieved instances.
- Recall: is the fraction of all the quantities of the related instances that are actually retrieved.
- Specificity: is the amount of negatives that are recognized correctly.
- F measure: is a measure of accuracy calculated by the precision and recall of the experiment, which is also called the Dice index or the Dice similarity coefficient. It indicates no spatial overlap of the binary segmentation results when it equals zero and indicates complete overlap when it equals one.
- Peak Signal-to-Noise Ratio (PSNR): is related to the ratio of the highest possible value of a signal and the value of noise that affects signal representation. In the case of image segmentation, the signal refers to the original data, and the noise refers to the error introduced by compression.
- The Jaccard Similarity Coefficient (The Jaccard Index): measures the similarity and diversity of the finite sample sets calculated by dividing the size of the intersection by the size of the union.
- Structural-Similarity-Index Measure: measures the perceptual similarity between two images.
- Probabilistic Rand Index: measures the similarity between two sets during the process of data clustering.
- True Positive: is a test result where the experiment correctly predicts the positive result.
- True Negative: is a test result where the experiment correctly predicts the negative result.
- False Positive: is a test result where the experiment incorrectly predicts the positive result.
- False Negative: is a test result where the experiment incorrectly predicts the negative result.
- Variation of Information: is a simple linear expression that measures the distance between two clusters of mutual information.
- Local Consistency Error (LCE): measures the relative consistency between two different segmentation solutions using the average minimal refinement error of all pixels.
- Global Consistency Error (GCE): measures the relative consistency between two different segmentation solutions using minimum cumulative refinement errors over all pixels in each coupling of clusters, then it finds the average of the result over all clusters.
- Boundary Displacement Error (BDE): gauges the average displacement error of the boundary pixels and the nearest boundary pixels in the other segmentation.
- Mean Square Error (MSE): is a positive value that measures the average squared difference between the estimated values and the real values: the smaller values of MSE are better.
- Elapsed time: is the amount of time consumed from the beginning of an experiment to its finish.
5.6. Tools
6. Conclusions and Suggestions
- Encourage the hybridization of the segmentation method with the bio-inspired algorithms.
- Modify the bio-inspired algorithms (combined with the segmentation methods) to improve their performance. This can be done by enhancing the movement strategy, enhancing the communication between individuals, and/or improving the evaluation process, etc.
- Combine the bio-inspired algorithms with other well-known segmentation methods such as the split and merge approach, region growing, and the Markov random model.
- Employ a second bio-inspired algorithm to enhance the performance of the principal bio-inspired technique that will be combined with a segmentation method.
- Treat the image-segmentation problem as an optimization problem for parameter setting.
- Investigate the recent bio-inspired algorithms such as the lion optimization method, the firefly algorithm, the camel algorithm, the elephant search algorithm, and the wolf optimization algorithm.
- Investigate the use of the bio-inspired image-segmentation approaches with other datasets that have not been considered such as urban-facade image segmentation.
- Propose a new clustering technique for image segmentation including the projection-pursuit clustering method to find regions into an image.
- Compare the investigated methods to the deep-learning methods. These methods have achieved very good results, so they provide a baseline for comparison against other algorithms.
- Encourage research in the image-segmentation-based medical field.
- Apply the proposed techniques on video processing.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| GA | Genetic algorithm |
| PSO | Particle swarm optimization |
| ABC | Artificial bee colony optimization |
| ACO | Ant colony optimization |
| PDE | Partial differential equation |
| ANN | Artificial neural network |
| FCM | Fuzzy C-means |
| KFCM | Kernel-based fuzzy C-means |
| RGB | Red green blue |
| MRI | Magnetic resonance imaging |
| SSIM | Structural-similarity-index measure |
| PSNR | Peak signal-to-noise ratio |
| MSE | Mean square error |
| MRF | Markov random field |
References
- Omarov, B.; Saparkhojayev, N.; Shekerbekova, S.; Akhmetova, O.; Sakypbekova, M.; Kamalova, G.; Alimzhanova, Z.; Tukenova, L.; Akanova, Z. Artificial Intelligence in Medicine: Real Time Electronic Stethoscope for Heart Diseases Detection. Comput. Mater. Contin. 2022, 70, 2815–2833. [Google Scholar] [CrossRef]
- Eberhart, R.C.; Shi, Y.; Kennedy, J. Swarm Intelligence; Elsevier: Amsterdam, The Netherlands, 1995. [Google Scholar]
- Holland, J. Adaptation in Natural and Artificial Systems; University of Michigan Press: Ann Arbor, MI, USA, 1975. [Google Scholar]
- Karaboga, D. An Idea Based on Honey Bee Swarm for Numerical Optimization; Technical Report, Technical Report-tr06; Erciyes University, Engineering Faculty, Computer Engineering Department: Kayseri, Turkey, 2005. [Google Scholar]
- Dorigo, M.; Gambardella, L.M. Ant colony system: A cooperative learning approach to the traveling salesman problem. IEEE Trans. Evol. Comput. 1997, 1, 53–66. [Google Scholar] [CrossRef]
- Khan, A.; Ravi, S. Image segmentation methods: A comparative study. Int. J. Soft Comput. Eng. IJSCE 2013, 3, 84–93. [Google Scholar]
- Sonawane, M.; Dhawale, C. A brief survey on image segmentation methods. Int. J. Comput. Appl. 2015, 975, 8887. [Google Scholar]
- Liang, Y.; Zhang, M.; Browne, W.N. Image segmentation: A survey of methods based on evolutionary computation. In Asia-Pacific Conference on Simulated Evolution and Learning; Springer: Berlin/Heidelberg, Germany, 2014; pp. 847–859. [Google Scholar]
- Zaitoun, N.M.; Aqel, M.J. Survey on image segmentation techniques. Procedia Comput. Sci. 2015, 65, 797–806. [Google Scholar] [CrossRef]
- Yuheng, S.; Hao, Y. Image Segmentation Algorithms Overview. arXiv 2017, arXiv:1707.02051. [Google Scholar]
- Chauhan, A.S.; Silakari, S.; Dixit, M. Image segmentation methods: A survey approach. In Proceedings of the IEEE 2014 Fourth International Conference on Communication Systems and Network Technologies (CSNT), Bhopal, India, 7–9 April 2014; pp. 929–933. [Google Scholar]
- Chouhan, S.S.; Kaul, A.; Singh, U.P. Image Segmentation Using Computational Intelligence Techniques: Review. Arch. Comput. Methods Eng. 2018, 26, 533–596. [Google Scholar] [CrossRef]
- Chouhan, S.S.; Kaul, A.; Singh, U.P. Soft computing approaches for image segmentation: A survey. Multimed. Tools Appl. 2018, 77, 28483–28537. [Google Scholar] [CrossRef]
- Rew, J.; Kim, H.; Hwang, E. Hybrid segmentation scheme for skin features extraction using dermoscopy images. Comput. Mater. Contin. 2021, 69, 801–817. [Google Scholar] [CrossRef]
- Khan, K.; Khan, R.U.; Albattah, W.; Nayab, D.; Qamar, A.M.; Habib, S.; Islam, M. Crowd Counting Using End-to-End Semantic Image Segmentation. Electronics 2021, 10, 1293. [Google Scholar] [CrossRef]
- Huang, H.Y.; Liu, Z.H. Stereo Matching with Spatiotemporal Disparity Refinement Using Simple Linear Iterative Clustering Segmentation. Electronics 2021, 10, 717. [Google Scholar] [CrossRef]
- Hassanien, A.E.; Emary, E. Swarm Intelligence: Principles, Advances, and Applications; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Berro, A.; Marie-Sainte, S.L.; Ruiz-Gazen, A. Genetic algorithms and particle swarm optimization for exploratory projection pursuit. Ann. Math. Artif. Intell. 2010, 60, 153–178. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Yue, X.; Zhang, H. Modified hybrid bat algorithm with genetic crossover operation and smart inertia weight for multilevel image segmentation. Appl. Soft Comput. 2020, 90, 106157. [Google Scholar] [CrossRef]
- Srikanth, M.V.; Prasad, V.V.K.D.V.; Prasad, K.S. An Improved Firefly Algorithm-Based 2-D Image Thresholding for Brain Image Fusion. Int. J. Cogn. Informatics Nat. Intell. (IJCINI) 2020, 14, 60–96. [Google Scholar] [CrossRef]
- Rahaman, J.; Sing, M. An efficient multilevel thresholding based satellite image segmentation approach using a new adaptive cuckoo search algorithm. Expert Syst. Appl. 2021, 174, 114633. [Google Scholar] [CrossRef]
- Chakraborty, F.; Durgapur, N.; India, D. Elephant Herding Optimization for Multi-Level Image Thresholding. Int. J. Appl. Metaheuristic Comput. 2020, 11, 167–176. [Google Scholar] [CrossRef]
- Boulanouar, S.; Lamiche, C. A New Hybrid Image Segmentation Method Based on Fuzzy C-Mean and Modified Bat Algorithm. Univ. Bahrain Sci. J. 2020, 9, 677–687. [Google Scholar] [CrossRef]
- Hrosik, R.C.; Tuba, E.; Dolicanin, E.; Jovanovic, R.; Tuba, M. Brain Image Segmentation Based on Firefly Algorithm Combined with K-means Clustering. Stud. Inform. Control 2019, 28, 167–176. [Google Scholar] [CrossRef]
- Wachs-Lopes, G.; Santos, R.; Saito, N.; Rodrigues, P. Recent nature-Inspired algorithms for medical image segmentation based on tsallis statistics. Commun. Nonlinear Sci. Numer. Simul. 2020, 88, 105256. [Google Scholar] [CrossRef]
- Xu, L.; Jia, H.; Lang, C.; Peng, X.; Sun, K. A Novel Method for Multilevel Color Image Segmentation Based on Dragonfly Algorithm and Differential Evolution. IEEE Access 2019, 7, 19502–19538. [Google Scholar] [CrossRef]
- Gonzalez, C.I.; Melin, P.; Castro, J.R.; Castillo, O.; Mendoza, O. Optimization of interval type-2 fuzzy systems for image edge detection. Appl. Soft Comput. 2016, 47, 631–643. [Google Scholar] [CrossRef]
- Singh, V.; Misra, A.K. Cardiac image segmentation using Simulated Genetic algorithm. In Proceedings of the 2015 International Conference on Advances in Computer Engineering and Applications, Ghaziabad, India, 19–20 March 2015; pp. 1024–1027. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, F.; Li, P. Medical image segmentation based on 2D maximum fuzzy entropy and improved genetic algorithm. In Proceedings of the 2015 8th International Congress on Image and Signal Processing (CISP), Shenyang, China, 14–16 October 2015; pp. 1583–1587. [Google Scholar] [CrossRef]
- Zhu, W.; Shen, Y. A segmentation approach for tissue images using non-dominated sorting GA. In Proceedings of the 2016 10th IEEE International Conference on Anti-counterfeiting, Security, and Identification (ASID), Xiamen, China, 23–25 September 2016; pp. 1–5. [Google Scholar] [CrossRef]
- Lu, S.; Xu, C.; Zhong, R.Y. An active RFID tag-enabled locating approach with multipath effect elimination in AGV. IEEE Trans. Autom. Sci. Eng. 2016, 13, 1333–1342. [Google Scholar] [CrossRef]
- Kaur, A.; Kaur, P. An integrated approach for Diabetic Retinopathy exudate segmentation by using Genetic Algorithm and Switching Median Filter. In Proceedings of the 2016 International Conference on Image, Vision and Computing (ICIVC), Xi’an, China, 26–28 July 2016; pp. 119–123. [Google Scholar] [CrossRef]
- Sun, G.; Zhang, A.; Yao, Y.; Wang, Z. A novel hybrid algorithm of gravitational search algorithm with genetic algorithm for multi-level thresholding. Appl. Soft Comput. 2016, 46, 703–730. [Google Scholar] [CrossRef]
- Ghosh, P.; Mitchell, M.; Tanyi, J.A.; Hung, A.Y. Incorporating priors for medical image segmentation using a genetic algorithm. Neurocomputing 2016, 195, 181–194. [Google Scholar] [CrossRef]
- Chandra, G.R.; Rao, K.R.H. Tumor Detection in Brain Using Genetic Algorithm. In Procedia Computer Science; Elsevier B.V.: Amsterdam, The Netherlands, 2016; Volume 79, pp. 449–457. [Google Scholar] [CrossRef]
- Abdel-Khalek, S.; Ishak, A.B.; Omer, O.A.; Obada, A.S.F. A two-dimensional image segmentation method based on genetic algorithm and entropy. Optik 2017, 131, 414–422. [Google Scholar] [CrossRef]
- Ishak, A.B. A two-dimensional multilevel thresholding method for image segmentation. Appl. Soft Comput. 2017, 52, 306–322. [Google Scholar] [CrossRef]
- Singh, V.; Misra, A.K. Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf. Process. Agric. 2017, 4, 41–49. [Google Scholar] [CrossRef]
- Ashour, A.S.; Hawas, A.R.; Guo, Y.; Wahba, M.A. A novel optimized neutrosophic k-means using genetic algorithm for skin lesion detection in dermoscopy images. Signal Image Video Process. 2018, 12, 1311–1318. [Google Scholar] [CrossRef]
- Gao, B.; Li, X.; Woo, W.L.; Tian, G.Y. Physics-based image segmentation using first order statistical properties and genetic algorithm for inductive thermography imaging. IEEE Trans. Image Process. 2018, 27, 2160–2175. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Lali, M.I.U.; Sharif, M.; Javed, K.; Aurangzeb, K.; Haider, S.I.; Altamrah, A.S.; Akram, T. An Optimized Method for Segmentation and Classification of Apple Diseases Based on Strong Correlation and Genetic Algorithm Based Feature Selection. IEEE Access 2019, 7, 46261–46277. [Google Scholar] [CrossRef]
- Abd Elaziz, M.; Ewees, A.A.; Oliva, D. Hyper-heuristic method for multilevel thresholding image segmentation. Expert Syst. Appl. 2020, 146, 113201. [Google Scholar] [CrossRef]
- Kanungo, P.; Nanda, P.; Samal, U. Image segmentation using thresholding and Genetic Algorithm. In Proceedings of the Conference on Soft Computing Technique for Engineering Applications, Rourkela, India, 24–26 March 2006. [Google Scholar]
- Kanungo, P.; Nanda, P. Parallel Genetic Algorithm Based Thresholding for Image Segmentation. In Proceedings of the National Seminar on IT and Soft Computing ITSC06, Nagpur, India, 17–18 November 2006. [Google Scholar]
- Banimelhem, O.; Yahya, A. Multi-thresholding image segmentation using Genetic Algorithm. In Proceedings of the International Conference on Image Processing, Computer Vision, and Pattern Recognition (IPCV), Las Vegas, NV, USA, 18–21 July 2011; pp. 1–6. [Google Scholar]
- Halder, A.; Pramanik, S. An unsupervised dynamic image segmentation using fuzzy hopfield neural network based genetic algorithm. arXiv 2012, arXiv:1205.6572. [Google Scholar]
- Hole, M.K.R.; Gulhane, V.S.; Shellokar, N.D. Application of Genetic Algorithm for Image Enhancement and Segmentation. Int. J. Adv. Res. Comput. Eng. Technol. (IJARCET) 2013, 2, 1342. [Google Scholar]
- Dilpreet, K.; Yadwinder, K. Intelligent medical image segmentation using FCM, GA and PSO. Int. J. Comput. Sci. Inf. Technol. 2014, 5, 6089–6093. [Google Scholar]
- Feng, Z.; Zhang, B. Fuzzy clustering image segmentation based on particle swarm optimization. Telkomnika (Telecommun. Comput. Electron. Control) 2015, 13, 128–136. [Google Scholar] [CrossRef][Green Version]
- Mekhmoukh, A.; Mokrani, K. Improved Fuzzy C-Means based Particle Swarm Optimization (PSO) initialization and outlier rejection with level set methods for MR brain image segmentation. Comput. Methods Programs Biomed. 2015, 122, 266–281. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, X.; Zhang, H.; Hu, J.; Jian, X. Side scan sonar image segmentation based on neutrosophic set and quantum-behaved particle swarm optimization algorithm. Mar. Geophys. Res. 2016, 37, 229–241. [Google Scholar] [CrossRef]
- Hu, Y.; Yu, M.; Du, J. An improved image segmentation approach using FGFCM with an edges-based neighbor selection strategy and PSO. In Proceedings of the 2017 36th Chinese Control Conference (CCC), Dalian, China, 26–28 July 2017; pp. 10951–10955. [Google Scholar] [CrossRef]
- Na, L.; Yan, J.; Shu, L. Application of PSO algorithm with dynamic inertia weight in medical image thresholding segmentation. In Proceedings of the 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom), Dalian, China, 12–15 October 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Taie, S.; Ghonaim, W. A New Image Segmentation Algorithm Based on Particle Swarm Optimization and Rough Set. J. Comput. 2017, 13, 130–138. [Google Scholar] [CrossRef]
- Suresh, S.; Lal, S. Multilevel thresholding based on Chaotic Darwinian Particle Swarm Optimization for segmentation of satellite images. Appl. Soft Comput. 2017, 55, 503–522. [Google Scholar] [CrossRef]
- Li, Y.; Bai, X.; Jiao, L.; Xue, Y. Partitioned-cooperative quantum-behaved particle swarm optimization based on multilevel thresholding applied to medical image segmentation. Appl. Soft Comput. 2017, 56, 345–356. [Google Scholar] [CrossRef]
- Tian, D.; Shi, Z. MPSO: Modified particle swarm optimization and its applications. Swarm Evol. Comput. 2018, 41, 49–68. [Google Scholar] [CrossRef]
- Gautam, K.; Singhai, R. Color Image Segmentation Using Particle Swarm Optimization in Lab Color Space. Int. J. Eng. Dev. Res. IJEDR 2018, 6, 373–377. [Google Scholar]
- Sharif, M.; Amin, J.; Raza, M.; Mussarat, Y.; Satapathy, S. An Integrated Design of Particle Swarm Optimization (PSO) with Fusion of Features for Detection of Brain Tumor. Pattern Recognit. Lett. 2019, 129, 150–157. [Google Scholar] [CrossRef]
- Astuti, N.R.D.P.; Mardhia, M.M. Multilevel thresholding hyperspectral image segmentation based on independent component analysis and swarm optimization methods. Int. J. Adv. Intell. Inform. 2019, 5, 66–75. [Google Scholar] [CrossRef]
- Sengar, S.S.; Mukhopadhyay, S. Motion segmentation-based surveillance video compression using adaptive particle swarm optimization. Neural Comput. Appl. 2020, 32, 11443–11457. [Google Scholar] [CrossRef]
- Di Martino, F.; Sessa, S. PSO image thresholding on images compressed via fuzzy transforms. Inf. Sci. 2020, 506, 308–324. [Google Scholar] [CrossRef]
- Farshi, T.R.; Drake, J.H.; Özcan, E. A Multimodal Particle Swarm Optimization-based Approach for Image Segmentation. Expert Syst. Appl. 2020, 149, 113233. [Google Scholar] [CrossRef]
- Mohsen, F.; Hadhoud, M.M.; Moustafa, K.; Ameen, K. A new image segmentation method based on particle swarm optimization. Int. Arab J. Inf. Technol. 2010, 9, 487–493. [Google Scholar]
- Palus, H.; Bereska, D. Region-based Colour Image Segmentation. In Proceedings of the 5th Workshop Farbbildverarbeitung Ilmenau, Ilmenau, Germany, 7–8 October 1999; Volume 67, p. 74. [Google Scholar]
- Mohsen, F.M.; Hadhoud, M.M.; Amin, K. A new optimization-based image segmentation method by particle swarm optimization. IJACSA Int. J. Adv. Comput. Sci. Appl. Spec. Issue Image Process. Anal. 2011. [Google Scholar] [CrossRef]
- Raju, N.G.S.; Rao, P.A.N. Particle Swarm Optimization Methods for Image Segmentation Applied in Mammography. Int. J. Eng. Res. Appl. 2013, 3, 1572–1579. [Google Scholar]
- Tandan, A.; Raja, R.; Chouhan, Y. Image Segmentation Based on Particle Swarm Optimization Technique. Int. J. Sci. Eng. Technol. Res. (IJSETR) 2014, 3, 257–260. [Google Scholar]
- Dewan, S.; Bajaj, S.; Prakash, S. Using Ant’s Colony Algorithm for improved segmentation for number plate recognition. In Proceedings of the 2015 IEEE/ACIS 14th International Conference on Computer and Information Science (ICIS), Las Vegas, NV, USA, 28 June–1 July 2015; pp. 313–318. [Google Scholar]
- Aslam, A.; Khan, E.; Beg, M.S. Multi—Threading based Implementation of Ant-Colony Optimization Algorithm for Image Edge Detection. In Proceedings of the 2015 Annual IEEE India Conference (INDICON), New Delhi, India, 17–20 December 2015; Volume 151, pp. 10–17. [Google Scholar]
- Ye, Z.; Wang, M.; Jin, H.; Liu, W.; Lai, X. An Image Thresholding Approach Based on Ant Colony Optimization Algorithm Combined with Genetic Algorithm. Int. J. Intell. Syst. Appl. 2015, 7, 8–15. [Google Scholar] [CrossRef]
- Arnay, R.; Fumero, F.; Sigut, J. Ant Colony Optimization-based method for optic cup segmentation in retinal images. Appl. Soft Comput. J. 2016, 52, 409–417. [Google Scholar] [CrossRef]
- Zou, G. Ant colony clustering algorithm and improved markov random fusion algorithm in image segmentation of brain images. Int. J. Bioautom. 2016, 20, 505, Technical Report 4. [Google Scholar]
- Selmani, A.; Seddik, H.; Braiek, E.B. A novel ant colonies approach to medical image segmentation. In Proceedings of the 2017 IEEE 14th International Multi-Conference on Systems, Signals & Devices (SSD), Marrakech, Morocco, 28–31 March 2017; pp. 22–26. [Google Scholar]
- Al-Ruzouq, R.; Shanableh, A.; Gibril, M.B.A.; Al-Mansoori, S. Image segmentation parameter selection and ant colony optimization for date palm tree detection and mapping from very-high-spatial-resolution aerial imagery. Remote Sens. 2018, 10, 1413. [Google Scholar] [CrossRef]
- Singh, R.; Vashishath, M.; Kumar, S. Ant colony optimization technique for edge detection using fuzzy triangular membership function. Int. J. Syst. Assur. Eng. Manag. 2019, 10, 91–96. [Google Scholar] [CrossRef]
- Khudov, H.; Ruban, I.; Makoveichuk, O.; Pevtsov, H.; Khudov, V.; Khizhnyak, I.; Fryz, S.; Podlipaiev, V.; Polonskyi, Y.; Khudov, R. Development of methods for determining the contours of objects for a complex structured color image based on the ant colony optimization algorithm. EUREKA Phys. Eng. 2020, 2020, 34–47. [Google Scholar] [CrossRef]
- Devarajan, D.; Ramesh, S.M.; Gomathy, B. A metaheuristic segmentation framework for detection of retinal disorders from fundus images using a hybrid ant colony optimization. Soft Comput. 2020, 24, 13347–13356. [Google Scholar] [CrossRef]
- Sandhya, G.; Kande, G.B.; Savithri, T.S. Q4: Segmentation of Magnetic Resonance Brain Images Using the Advanced Ant Colony Optimization Technique. J. Biomim. Biomater. Biomed. Eng. 2020, 44, 37–49. [Google Scholar] [CrossRef]
- Gonzalez, R.C.; Woods, R.E. Digital Image Processing; Prentice Hall: Harlow, UK, 2007. [Google Scholar]
- Maini, R.; Sohal, J. Performance evaluation of Prewitt edge detector for noisy images. GVIP J. 2006, 6, 39–46. [Google Scholar]
- Roberts, L.G. Machine Perception of Three Dimensional Solids. In Optical and Electro-Optical Information Processing; MIT Press: Cambridge, MA, USA, 1965; pp. 159–197. [Google Scholar]
- Ouadfel, S.; Batouche, M. MRF based image segmentation using Ant Colony System. ELCVIA Electron. Lett. Comput. Vis. Image Anal. 2003, 2, 12–24. [Google Scholar] [CrossRef]
- Ouadfel, S.; Batouche, M.; Talhi, S. Ant Colonies For MRF Based Image Segmentation. In Proceedings of the 3rd International Conference: Sciences of Electronic, Technologies of Information and Telecommunications, Sousse, Tunisia, 27–31 March 2005. [Google Scholar]
- Zhao, B.; Zhu, Z.; Mao, E.; Song, Z. Image segmentation based on ant colony optimization and K-means clustering. In Proceedings of the 2007 IEEE International Conference on Automation and Logistics, Jinan, China, 18–21 August 2007; pp. 18–21. [Google Scholar]
- Laptik, R.; Navakauskas, D. Application of ant colony optimization for image segmentation. Electron. Electr. Eng. J. 2007, 80, 13–18. [Google Scholar]
- Han, Y.; Shi, P. An improved ant colony algorithm for fuzzy clustering in image segmentation. Neurocomput. J. 2007, 70, 665–671. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, Z. Ant Colony Optimization for Image Segmentation. In Ant Colony Optimization; Ostfeld, A., Ed.; IntechOpen: Rijeka, Croatia, 2011; Chapter 17. [Google Scholar] [CrossRef]
- Xumin, L.; Shi, N.; Cailing, L. Image segmentation algorithm based on improved ant colony algorithm. Int. J. Signal Process. Image Process. Pattern Recognit. 2014, 7, 433–442. [Google Scholar]
- Bouaziz, A.; Draa, A.; Chikhi, S. Artificial bees for multilevel thresholding of iris images. Swarm Evol. Comput. 2015, 21, 32–40. [Google Scholar] [CrossRef]
- Sağ, T.; Çunkaş, M. Color image segmentation based on multiobjective artificial bee colony optimization. Appl. Soft Comput. 2015, 34, 389–401. [Google Scholar] [CrossRef]
- Bose, A. Fuzzy-based artificial bee colony optimization for gray image. Signal Image Video Process. 2016, 10, 1089–1096. [Google Scholar] [CrossRef]
- Li, L.; Sun, L.; Guo, J.; Han, C.; Zhou, J.; Li, S. A quick artificial bee colony algorithm for image thresholding. Information 2017, 8, 16. [Google Scholar] [CrossRef]
- Huo, F.; Liu, Y.; Wang, D.; Sun, B. Bloch quantum artificial bee colony algorithm and its application in image threshold segmentation. Signal Image Video Process. 2017, 11, 1585–1592. [Google Scholar] [CrossRef]
- Fu, Z.; Liu, Y.; Hu, H.; Wu, D.; Gao, H. An efficient method of white blood cells detection based on artificial bee colony algorithm. In Proceedings of the 2017 IEEE 29th Chinese Control And Decision Conference (CCDC), Chongqing, China, 28–30 May 2017; pp. 3266–3271. [Google Scholar]
- Feng, Y.; Yin, H.; Lu, H.; Cao, L.; Bai, J. FCM-based quantum artificial bee colony algorithm for image segmentation. In Proceedings of the 10th International Conference on Internet Multimedia Computing and Service, Nanjing, China, 17–19 August 2018; p. 6. [Google Scholar]
- Zhang, S.; Jiang, W.; Satoh, S. Multilevel thresholding color image segmentation using a modified artificial bee colony algorithm. IEICE Trans. Inf. Syst. 2018, E101D, 2064–2071. [Google Scholar] [CrossRef]
- Lingappa, H.K.; Suresh, H.N.; Manvi, S.K. Medical image segmentation based on extreme learning machine algorithm in Kernel Fuzzy C-Means using artificial bee colony method. Int. J. Intell. Eng. Syst. 2018, 11, 128–136. [Google Scholar] [CrossRef]
- Banharnsakun, A. Artificial bee colony algorithm for enhancing image edge detection. Evol. Syst. 2018, 10, 679–687. [Google Scholar] [CrossRef]
- Ma, L.; Wang, X.; Shen, H.; Huang, M.; Ma, L.; Wang, X.; Shen, H.; Huang, M. A novel artificial bee colony optimiser with dynamic population size for multi-level threshold image segmentation. Int. J. Bio-Inspired Comput. 2019, 13, 32–44, Technical Report 1. [Google Scholar] [CrossRef]
- Gupta, S.; Deep, K. Hybrid sine cosine artificial bee colony algorithm for global optimization and image segmentation. Neural Comput. Appl. 2019, 6, 9521–9543. [Google Scholar] [CrossRef]
- Huo, F.; Sun, X. Multilevel image threshold segmentation using an improved Bloch quantum artificial bee colony algorithm. Multimed. Tools Appl. 2020, 79, 2447–2471. [Google Scholar] [CrossRef]
- Hancer, E. Artificial Bee Colony: Theory, Literature Review, and Application in Image Segmentation. In Recent Advances on Memetic Algorithms and its Applications in Image Processing. Studies in Computational Intelligence; Springer: Singapore, 2020; Volume 873, pp. 47–67. [Google Scholar] [CrossRef]
- Alrosan, A.; Alomoush, W.; Norwawi, N.; Alswaitti, M.; Makhadmeh, S.N. An improved artificial bee colony algorithm based on mean best-guided approach for continuous optimization problems and real brain MRI images segmentation. Neural Comput. Appl. 2020, 33, 1671–1697. [Google Scholar] [CrossRef]
- Zhi, H.; Liu, S. Gray image segmentation based on fuzzy c-means and artificial bee colony optimization. J. Intell. Fuzzy Syst. 2020, 38, 3647–3655. [Google Scholar] [CrossRef]
- Pare, S.; Kumar, A.; Singh, G.K.; Bajaj, V. Image Segmentation Using Multilevel Thresholding: A Research Review. Iran. J. Sci. Technol.—Trans. Electr. Eng. 2020, 44, 1–29. [Google Scholar] [CrossRef]
- Peng, L.; Hua, Z. Parameter Selection for Ant Colony Algorithm Based on Bacterial Foraging Algorithm. Math. Probl. Eng. 2016, 2016, 6469721. [Google Scholar] [CrossRef]
| Population Size (N), Cognitive and Social, Parameters () and (), Maximum and Minimum Velocity (, ) | |||
|---|---|---|---|
| Inertia (), Dimension of the Search Space (D), Range of Gray Levels in Image (), Iteration Number () | |||
| Reference | Dataset | Parameter Setting | Compared Methods |
| [50] | Not provided | , | Fuzzy clustering |
| , | Multi-threshold Otsu | ||
| , | |||
| [51] | Brain Cancer Medical Images | Fuzzy C-mean (FCM) | |
| Synthetic Images | FCM-S1 | ||
| Simulated Images | KPCM | ||
| IKPCM | |||
| RFCM | |||
| [52] | Berkeley | 2D D-L MPSO | |
| BSDS300 | 1D D-L MPSO | ||
| 2D D-L classic PSO | |||
| 1D Renyi entropy MPSO | |||
| [52] | Side Scan Sonar | 2D-Otsu, MRF | |
| FCM | |||
| Level set, fractal theory | |||
| [53] | Wheel image | Fast generalized FCM | |
| Airplane | |||
| MRI images | |||
| [54] | MRI images | PSO | |
| , | |||
| , | |||
| [55] | MRI images | Bat algorithm | |
| Cameraman | |||
| [56] | Satellite images | , | None |
| , | |||
| [57] | Stomach CT | , | None |
| [58] | Lena & Pepper | , | None |
| , | |||
| [59] | view color images | Not applied | None |
| [60] | Medical images | Not Applied | None |
| from RIDER and BRATS | |||
| [61] | Aviris Indian Pines hyperspectral image | Not provided | PSO, DPSO, FODPSO |
| [62] | Hallway | Cuckoo search | |
| Foreman | Flower pollination, bat | ||
| Container | bird swarm | ||
| Coastguard | Gbest-guided gravitational search | ||
| [63] | Lena | DPSO | |
| Jet | CDPSO | ||
| Mandrill | |||
| Berkeley Segmentation Dataset | |||
| Image Processing Standard Database | |||
| [64] | 20 colour images | Not Applied | FCM |
| Lena | SFFCM | ||
| [65] | Cameraman | Region-based segmentation | |
| [66] | |||
| [67] | House | Normal region-based | |
| Cameraman | segmentation | ||
| Peppers | K-means algorithm | ||
| [68] | MIAS database (*) | Not provided | PSO, Darwinian PSO (DPSO) |
| Fractional-order DPSO (FO-DPSO) | |||
| [69] | Arbitrary RGB Images | Not provided | None |
| Number of Iterations (), Number of Ants (N) | |||
|---|---|---|---|
| Evaporation of the Pheromone Trail (), Initial Pheromone Quantity () | |||
| Maximum Pheromone Quantity (), the Relative Importance of the Residual Pheromones () | |||
| the Relative Importance of the Pheromone Trails (), the Pheromone Decay Coefficient () | |||
| Reference | Dataset | Parameter Setting | Compared Methods |
| [70] | Vehicle image | Not provided | SOBEL operator [81] |
| Prewitt operator [82] | |||
| ResultACOoberts’s | |||
| operator [83] | |||
| [71] | Shapes | Conventional ACO | |
| Lena | |||
| Mandril | (i.e., is the number of pixels) | ||
| Cameraman | |||
| Chairs | |||
| Pepper | |||
| [72] | Person | 2D Otsu | |
| Rice | Traditional GA | ||
| Man | |||
| Tank | |||
| [73] | RIM-ONE dataset | Thresholding | |
| R-bend | |||
| ASM | |||
| Regression | |||
| Superpixel | |||
| [74] | Medical MRI images | Not provided | FCM |
| Markov random field (MRF) | |||
| [75] | MRI-guided prostate biopsies | Not provided | Manual method |
| [76] | Very-high-spatial-resolution | Not provided | Chi-square |
| (VHSR) aerial imagery | CFS | ||
| provided by the | Gain ratio | ||
| Ajman municipality | Information gain | ||
| SVM | |||
| Principal component | |||
| analysis (PCA) | |||
| [77] | Infrared image of | FCM baed ACO | |
| ship | |||
| [78] | Structured color image | Canny method | |
| K-means method (, ) | |||
| Random forest method | |||
| [79] | DRIVE database | Not provided | Bayesian set models |
| STARE database | Standalone ANN models | ||
| [80] | BrainWeb database | Not provided | FCM |
| Expectation-maximization (EM) | |||
| Improved bacterial foraging | |||
| Algorithm (IBFA) | |||
| Improved PSO | |||
| Optimization (IPSO) | |||
| [84] | Cerebral MRI | Simulated annealing (SA) | |
| Resonance (MR) (*) | GA | ||
| House | |||
| [85] | Cerebral MRI | Simulated annealing (SA) | |
| Resonance (MR) (*) | GA | ||
| Muscle cells image | ACS-MRF | ||
| House | |||
| [86] | Banana | None | |
| RGB images | |||
| [87] | Two-dimensional | None | |
| electrophoresis gel images | |||
| [88] | RGB images | Sobel approach | |
| Canny approach | |||
| Watershed approaches | |||
| [89] | Heart ventricle image | Original ACO | |
| GA | |||
| [90] | Pepper | Original ACO | |
| Number of Iterations (), Number of Ants (N), Evaporation of the Pheromone Trail (), Initial Pheromone Quantity () | |||
|---|---|---|---|
| Maximum Pheromone Quantity (), the Relative Importance of the Residual Pheromones () | |||
| the Relative Importance of the Pheromone Trails (), the Pheromone Decay Coefficient () | |||
| Reference | Dataset | Parameter Setting | Compared Methods |
| [70] | Vehicle image | Not provided | SOBEL operator [81] |
| Prewitt operator [82] | |||
| ResultACOoberts’s | |||
| operator [83] | |||
| [71] | Shapes | Conventional ACO | |
| Lena | |||
| Mandril | (i.e., is the number of pixels) | ||
| Cameraman | |||
| Chairs | |||
| Pepper | |||
| [72] | Person | 2D Otsu | |
| Rice | Traditional GA | ||
| Man | |||
| Tank | |||
| [73] | RIM-ONE dataset | Thresholding | |
| R-bend | |||
| ASM | |||
| Regression | |||
| Superpixel | |||
| [74] | Medical MRI images | Not provided | FCM |
| MRF | |||
| [75] | MRI-guided prostate biopsies | Not provided | Manual method |
| [76] | Very-high-spatial-resolution | Not provided | Chi-square |
| (VHSR) aerial imagery | CFS | ||
| provided by the | Gain ratio | ||
| Ajman municipality | Information gain | ||
| SVM | |||
| Principal component | |||
| analysis (PCA) | |||
| [77] | Infrared image of | FCM based ACO | |
| ship | |||
| [78] | Structured color image | Canny method | |
| K-means method (, ) | |||
| Random forest method | |||
| [79] | DRIVE database | Not provided | Bayesian set models |
| STARE database | Standalone ANN models | ||
| [80] | BrainWeb database | Not provided | FCM |
| Expectation-maximization (EM) | |||
| Improved bacterial foraging | |||
| algorithm (IBFA) | |||
| Improved particle swarm | |||
| optimization (IPSO) | |||
| [84] | Cerebral MRI | Simulated annealing (SA) | |
| resonance (MR) (*) | GA | ||
| House | |||
| [85] | Cerebral MRI | Simulated annealing (SA) | |
| Resonance (MR) (*) | GA | ||
| Muscle cells image | ACS-MRF | ||
| House | |||
| [86] | Banana | None | |
| RGB images | |||
| [87] | Two-dimensional | None | |
| electrophoresis gel images | |||
| [88] | RGB images | Sobel approach | |
| Canny approach | |||
| Watershed approaches | |||
| [89] | Heart ventricle image | Original ACO | |
| GA | |||
| [90] | Pepper | Original ACO | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larabi-Marie-Sainte, S.; Alskireen, R.; Alhalawani, S. Emerging Applications of Bio-Inspired Algorithms in Image Segmentation. Electronics 2021, 10, 3116. https://doi.org/10.3390/electronics10243116
Larabi-Marie-Sainte S, Alskireen R, Alhalawani S. Emerging Applications of Bio-Inspired Algorithms in Image Segmentation. Electronics. 2021; 10(24):3116. https://doi.org/10.3390/electronics10243116
Chicago/Turabian StyleLarabi-Marie-Sainte, Souad, Reham Alskireen, and Sawsan Alhalawani. 2021. "Emerging Applications of Bio-Inspired Algorithms in Image Segmentation" Electronics 10, no. 24: 3116. https://doi.org/10.3390/electronics10243116
APA StyleLarabi-Marie-Sainte, S., Alskireen, R., & Alhalawani, S. (2021). Emerging Applications of Bio-Inspired Algorithms in Image Segmentation. Electronics, 10(24), 3116. https://doi.org/10.3390/electronics10243116






