4.1. Comparison of RMU-Net with other Deep Learning Segmentation Models
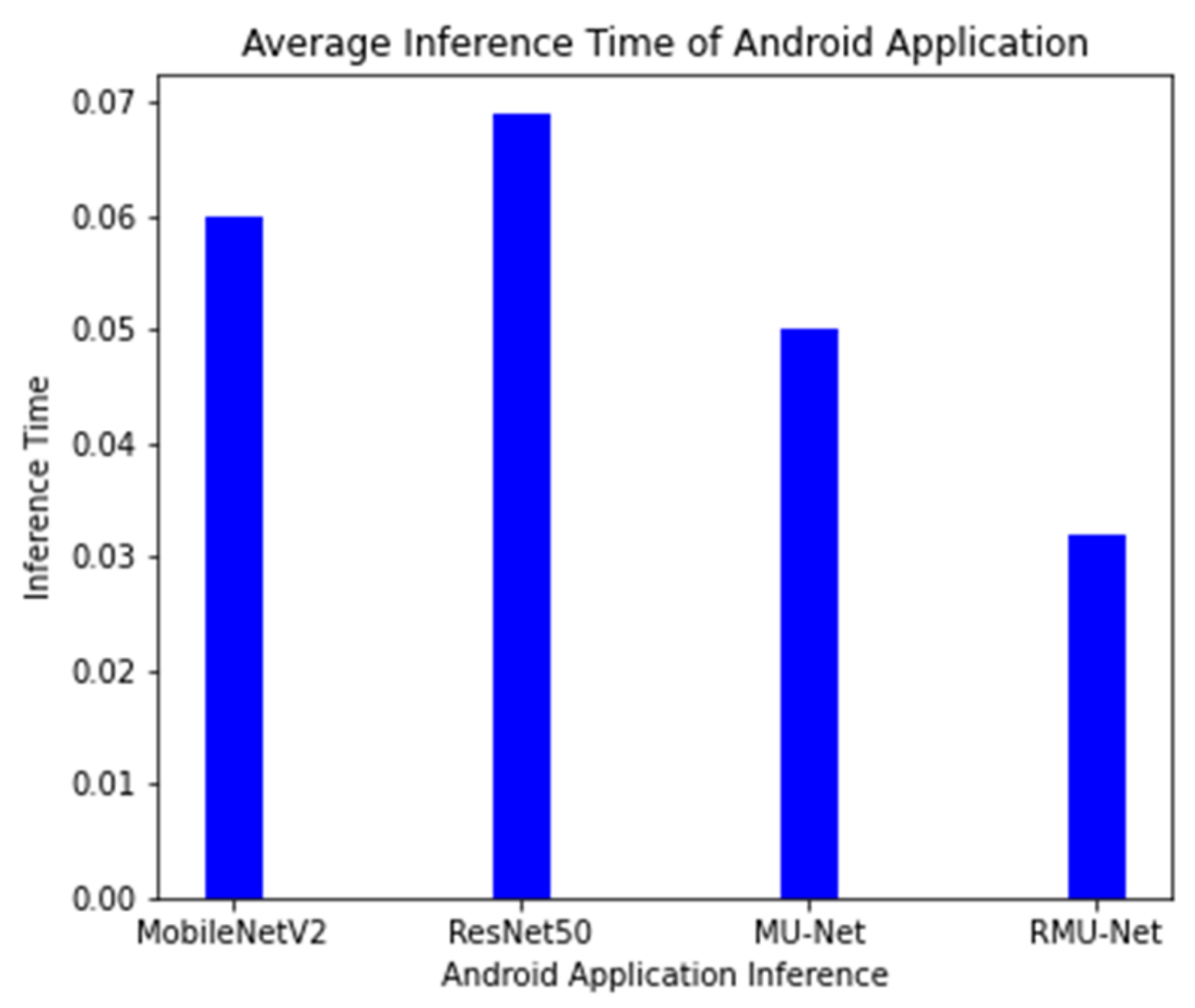
On an industrial machine, the proposed model is tested, and the cost of computation is compared with the current system. RMU-Net is a deep, lightweight neural network designed with convolution depth-wise. In U-Net, the depth-wise convolution used is much faster than standard convolution. On the central processor unit (CPU) platform, separation convolution is generally quicker than traditional convolution. On both GPU and CPU platforms, the proposed RMU-Net performs well in segmentation time. However, the number of parameters impacts the number of computer resources used and the time it takes to train. RMU-Net time is also assessed on various hardware platforms, including two GPU platforms (GTX 1080Ti and GTX 745), a CPU platform (Intel i7), and an embedded platform. The suggested RMU-Net performs well in segmentation time on the GPU platform with two h 47 min. However, the CPU takes 47 h, while the embedded system takes 32 h to complete the process. A lighter weight model with a limited number of parameters and model size is the proposed technique. The results obtained from different segmentation models are shown in
Table 11, which includes a brief explanation, trainable parameters, and the models’ size.
In the article [
29], Lucas Fidon introduced a 3D U-Net model to segment brain tumors. The author used the same model as before, but he experimented with non-standard loss functions such as the Wasserstein loss function. Ranger, a non-standard optimizer, was adopted for optimization. Ranger is a more generalized version of Adam that works well with small batches and noisy labels. To find the best results, three deep learning models were trained using different optimizers. The BraTS 2020 dataset was used to test the model. The model had dice scores of 88.9%, 84.1%, 81.4% for the whole tumor, tumor core, and enhanced tumor, respectively, and Hausdorff distances of 6.4, 19.4, and 15.8 for the entire tumor, tumor core, and enhanced tumor. Another automatic brain tumor segmentation approach was proposed by Yixin Wang et al. [
30] with modality-pairing learning methods. To extract complex information from several modalities, different layer connections were used. An average ensemble of all the models was used to obtain results, along with post-processing methods. The model performed well on the BraTS 2020 dataset, with dice scores of 89.1%, 84.2%, and 81.6% for the entire tumor, tumor core, and enhanced tumor, respectively. Haozhe jia et al. [
31] used H
2NF-Net for the segmentation of brain tumor from multi-modal MRI images. To separate the distinct parts of the tumor, the author employed a single, cascaded network and concatenated the pre-predictions to reach the final segmentation result. BraTS 2020 training and validation datasets were used to train and evaluate the model. The model attained dice scores of 78.75%, 91.29%, and 85.46% for the enhanced tumor, whole tumor, and tumor core, respectively, and Hausdorff distances of 26.57, 4.18, and 4.97 by integrating the single and cascaded networks.
A modified nnU-Net was proposed by [
32] for the segmentation of brain tumors with data augmentation, post-processing, and region-based training. The model showed improved results with several minor modifications and achieved first place in the BraTS 2020 dataset challenge. The dice scores of the model were 88.95%, 85.06%, and 82.03% for the whole tumor, tumor core, and enhanced tumor, respectively. Wenbo Zhang et al. [
33] used a multi-encoder framework for brain tumor segmentation. In addition, the author created a new loss function called categorical loss and assigned various weights to different segmented regions. The model was evaluated using the BraTS 2020 dataset. The method achieved promising results with dice scores of 70.24%, 88.26%, and 73.86% for the entire tumor, tumor core, and enhanced tumor. A deep neural network architecture for brain tumor segmentation [
34] is proposed to cascade three deep learning models. The output feature map of the previous stage was used in the next step as input. The dataset used for this study was the publicly available BraTS 2020 dataset. The model achieved dice scores of 88.58%, 82.97%, and 79% for the whole tumor, core tumor, and enhanced tumor. Another modified architecture of U-Net was proposed by Parvez Ahmad et al. [
35] for automatic brain tumor segmentation. The author extracts multi-contextual features by using dense connections between encoder and decoder. In addition, local and global information was also extracted with residual inception blocks. The author validated the model on the BraTS 2020 dataset. The dice scores for the whole tumor, tumor core, and enhanced tumor were 89.12%, 84.74%, and 79.12%, respectively.
Henry et al. [
36] trained multiple U-Net network-like models with stochastic weights and deep vision on a Multi-modal BraTS 2020 training dataset to make the process automated and standardized. Two different models were trained separately, and feature maps from both models were concatenated. The BraTS 2020 test dataset was used for testing the model that achieved dice scores of 81%, 91%, and 95% for the enhanced tumor, whole tumor, and tumor core. Carlo Russo [
37] used spherical space transformed input data to extract better features than standard feature extraction methods. The spherical coordinate transformation was used as pre-processing to improve the accuracy for brain tumor segmentation on the BraTS 2020 dataset. The model achieved dice scores for the whole tumor, tumor core, and enhanced tumor of 86.87%, 80.66%, and 78.98%. In article [
38], the author trained a two-dimensional network for the three-dimensional segmentation of a brain tumor. EfficientNet was used as the encoder part that achieved promising results compared to previous work with dice scores of 69.59%, 80.86%, and 75.20% for the enhanced tumor, whole tumor, and tumor core.
A multi-step deep neural network [
39] was proposed, which takes the hierarchical structure of the brain tumor and segments the substructures. Deep supervision along with data augmentation techniques was used to overcome the gradient vanishing and overfitting. The model has evaluated the BraTS 2019 dataset with dice scores of 88.6%, 81.3%, 77.1% for the whole tumor, tumor core, and enhanced tumor. Wang et al. [
40] proposed a 3D U-net based deep earning model using brain-wise normalization and a patching method for brain tumor segmentation. The model was tested on the BraTS 2019 challenge dataset. Dice scores of the enhanced tumor, tumor core and whole tumor are 77.8%, 79.8%, and 85.5%. In [
41], a CNN model was trained on high-contrast images to improve the segmentation results of the sub-regions. A Generative Adversarial Network is used for synthesizing high-contrast images. The experiments were conducted on the BraTS 2019 dataset, showing that the high-contrast images have more segmentation accuracy. The dice scores of the synthetic images are 76.65%, 89.65%, and 79.01% for the ET, WT, and TC, respectively.
An automated three-dimensional [
42] deep model for the segmentation of gliomas in 3D pre-operative MRI scans was proposed—the model segments the tumor and its subregions. One deep learning model learns the local features of the input data, and another model extracts the global features from the whole image. The output from both the models is ensembled to develop a more accurate learning process. The model is trained on the BraTS 2019 dataset, which gives promising segmentation results. A comparison of 3D semantic segmentation [
43], convolutional neural network, and encoder–decoder architecture is used to improve the performance of the segmentation results. The method is evaluated on the BraTS 2019 dataset, which achieved dice scores for the ET, WT, and TC classes of 82.6%, 88.2%, and 83.7%, respectively. The segmentation results of the testing dataset were 0.82, 0.72, and 0.70 for the whole tumor, tumor core, and enhanced tumor.
A two-step approach [
44] for brain tumor segmentation was proposed using two different 3D U-net models. First, the tumor is located using 3D U-net, and the second model segments the detected tumor into sub regions. The segmentation results of the ET, WT, and TC classes are 62.1, 84.4, and 72.8, respectively. An automated 2D [
45] brain tumor segmentation method is proposed. The network architecture used in this work was a modified U-net architecture for improving the segmentation results. To address the class imbalance problem, weighted cross-entropy and the generalized dice score were used as loss functions. The proposed segmentation system has been tested on the BraTS 2018 dataset, which achieved dice scores of 78.3%, 86.8%, and 80.5%, respectively. Another modified 3D U-net architecture [
46] was introduced with the augmentation technique to handle MRI input data. The quality of the tumor segmentation was enhanced with context obtained from models of same network. A cascade of CNN networks [
47] for the segmentation of brain tumors using MRI images was introduced that is a trade-off between computational cost and model complexity. Experiments with the BraTS dataset showed that the model achieved dice scores for WT, ET, and TC of 90.5%, 78.6%, and 83.8%, respectively.
A similar model was proposed by AMADEUS et al. [
49], which introduced an approach that can be used in low resource devices such as mobile devices. Two different models were introduced in which both models used three convolutional layers to decrease the computational cost. Batch normalization, residual layers, and depthwise separable convolution layers were used to preserve the features and reduce the number of operations. These models were tested on ImageNet, CIFAR 10, CIFAR 100, and some other datasets. The input size of the datasets was 32 × 32 with total trainable parameters of 907,449. Whereas, in the proposed model, we applied the modified architecture of MobileNetV2 with additional residual blocks as an encoder, which was integrated with the U-Net decoder for the segmentation task. The input size is 224 × 224 with a total of 4.6 million trainable parameters.
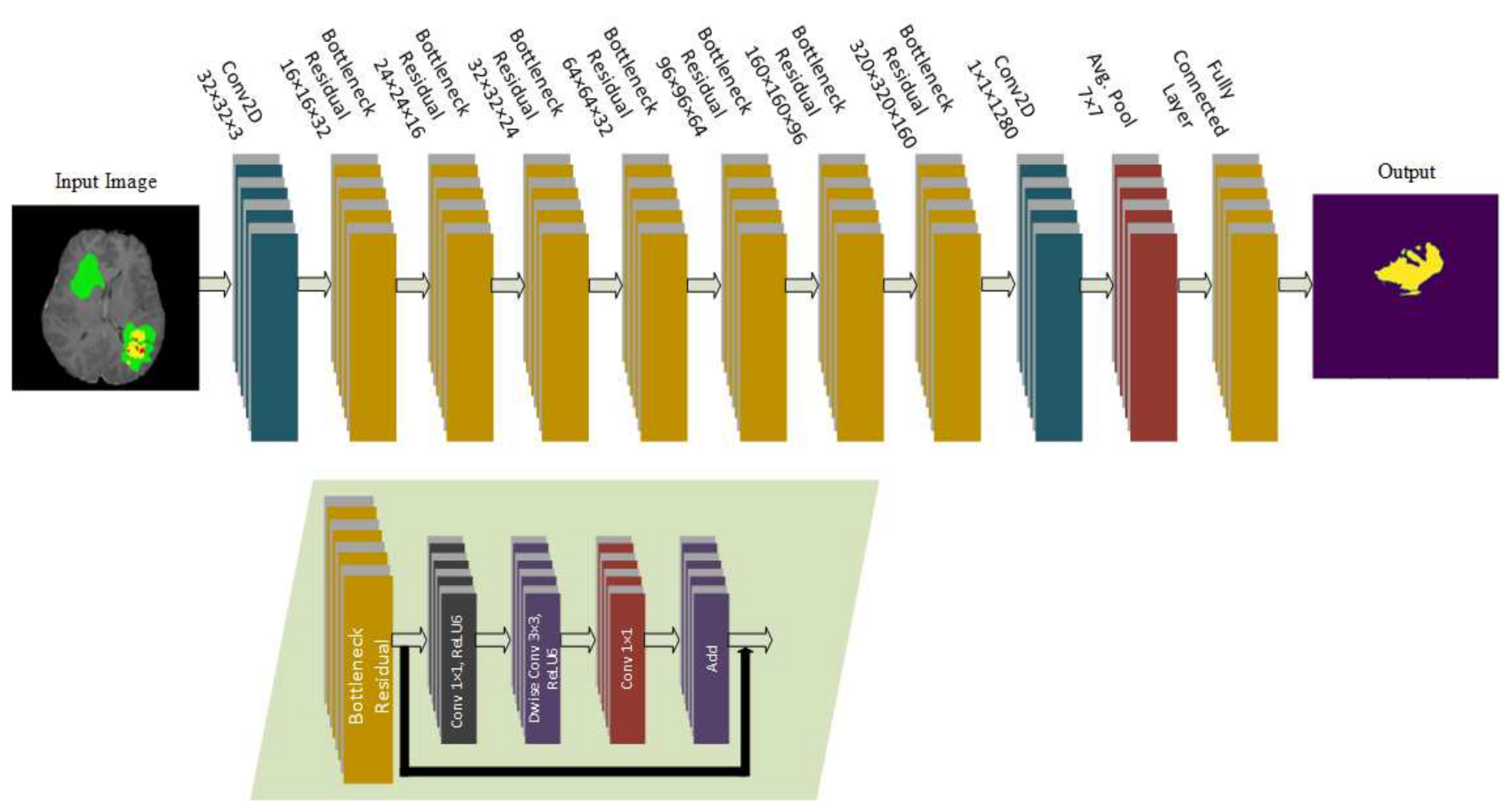
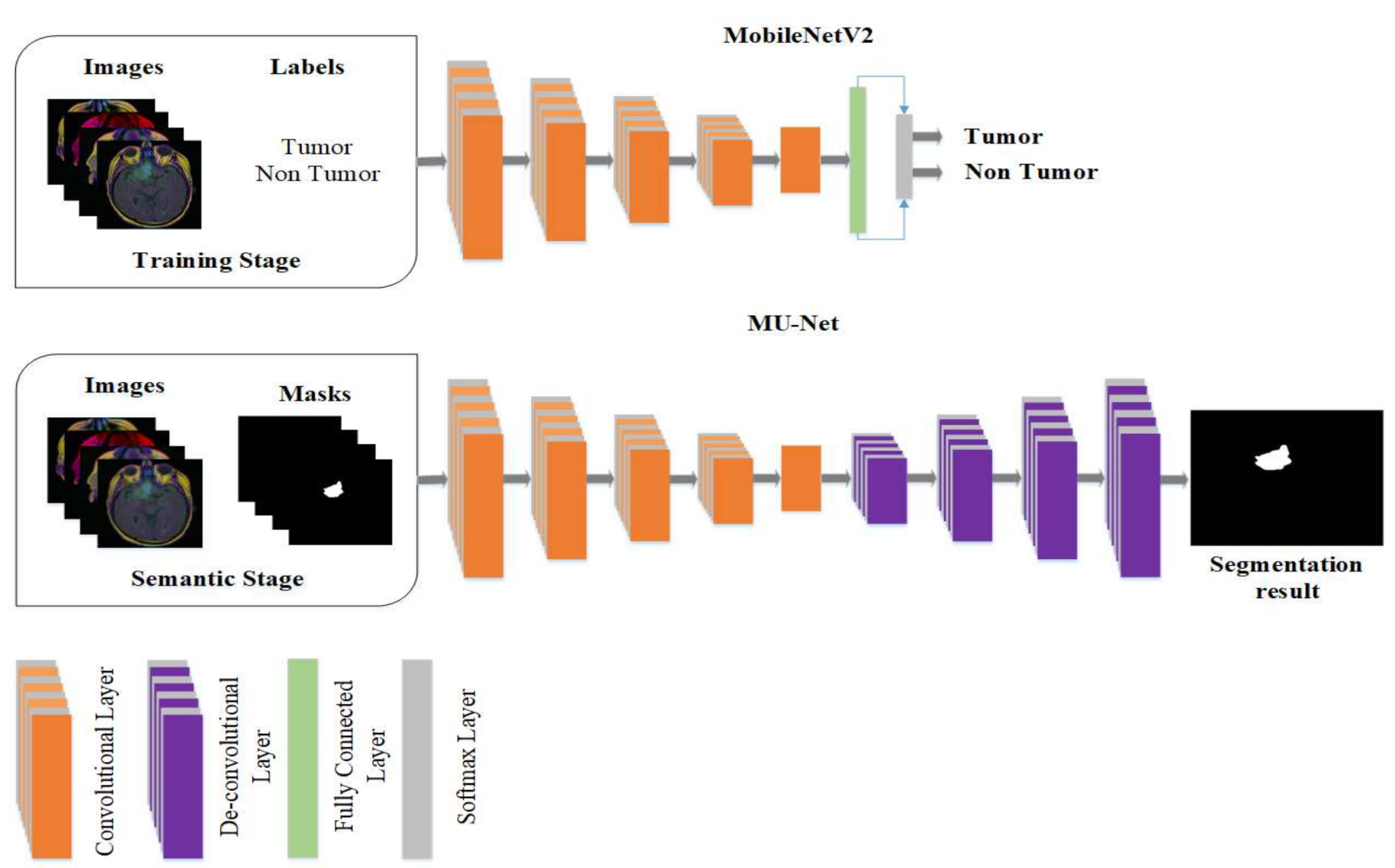
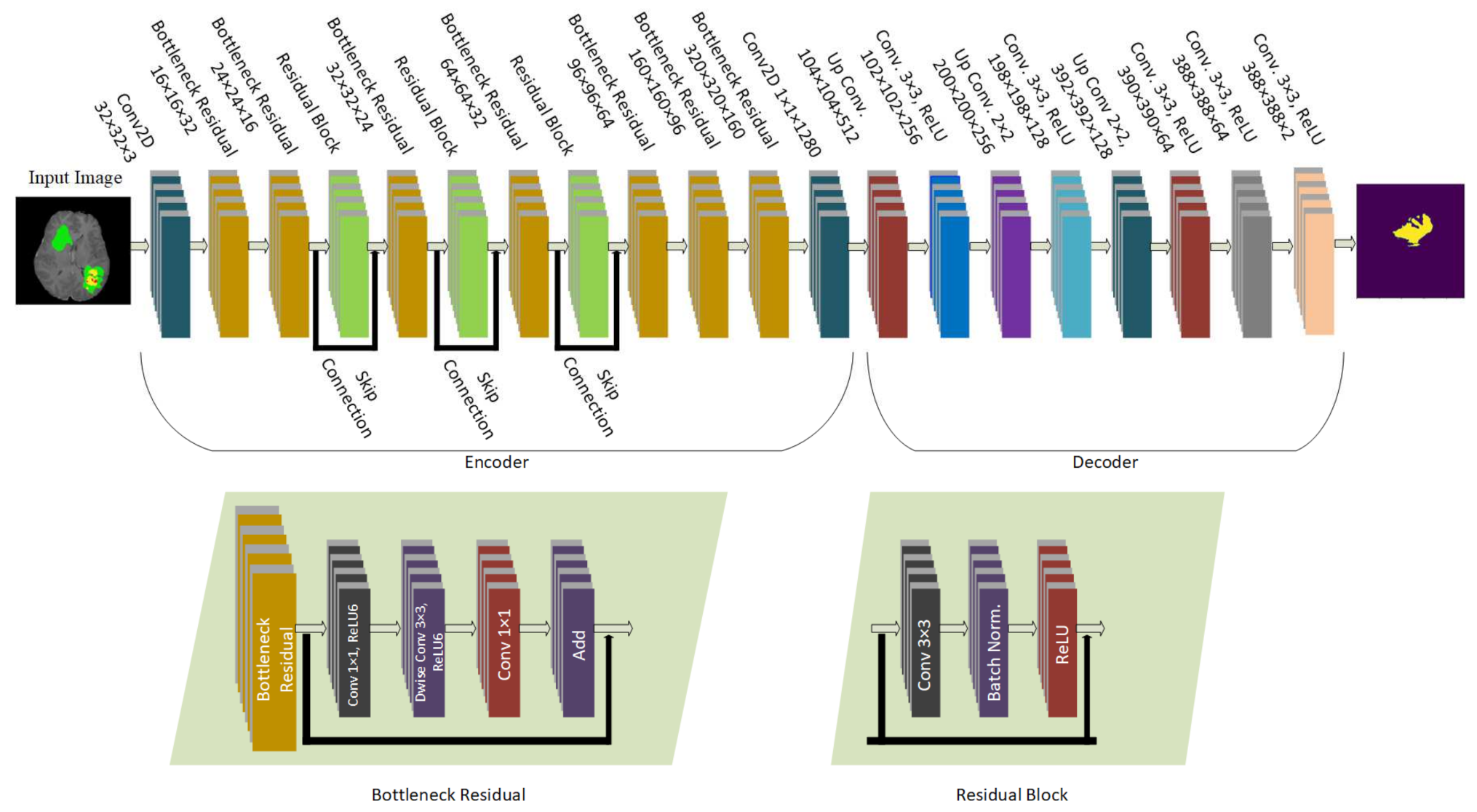
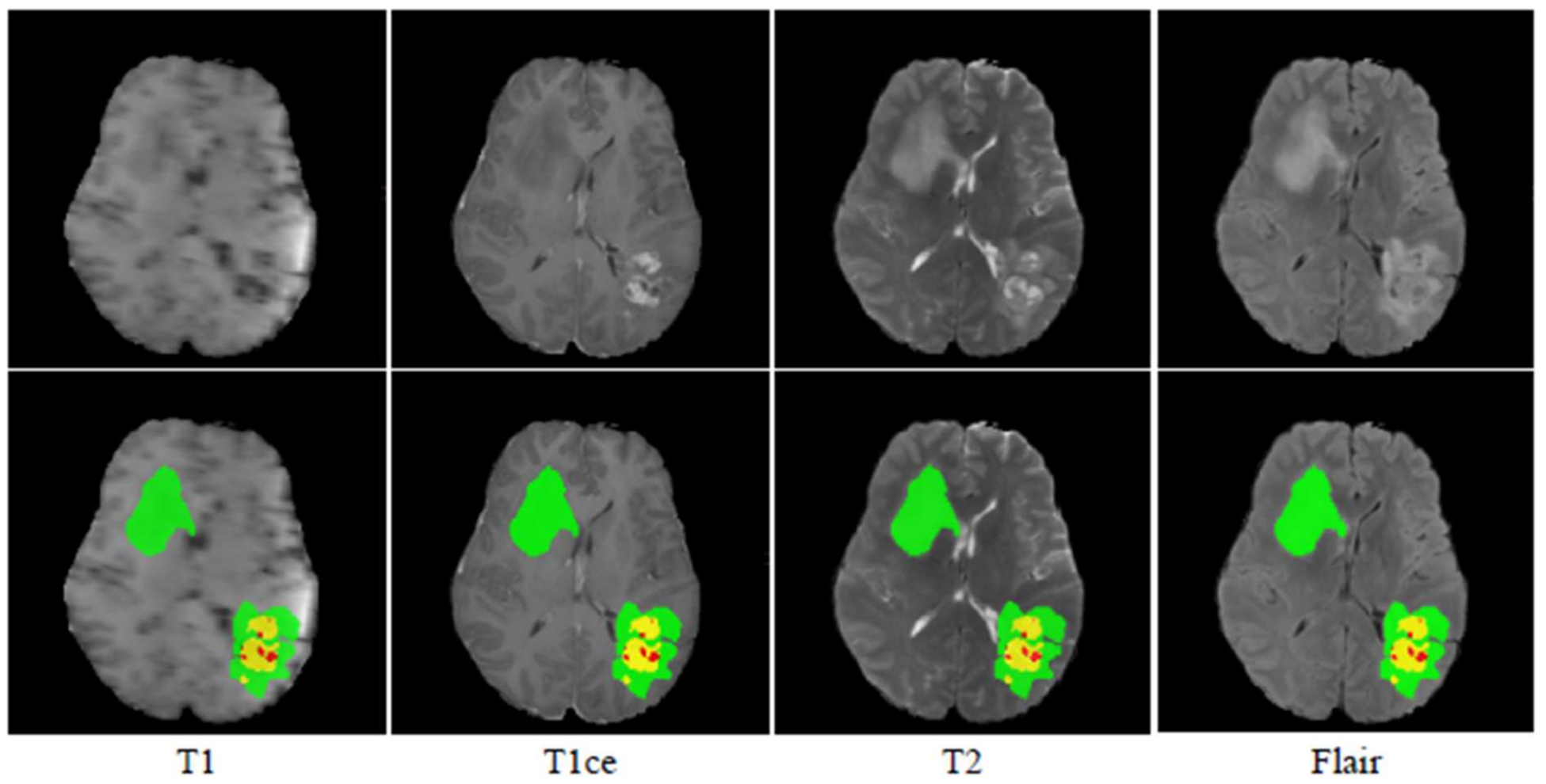
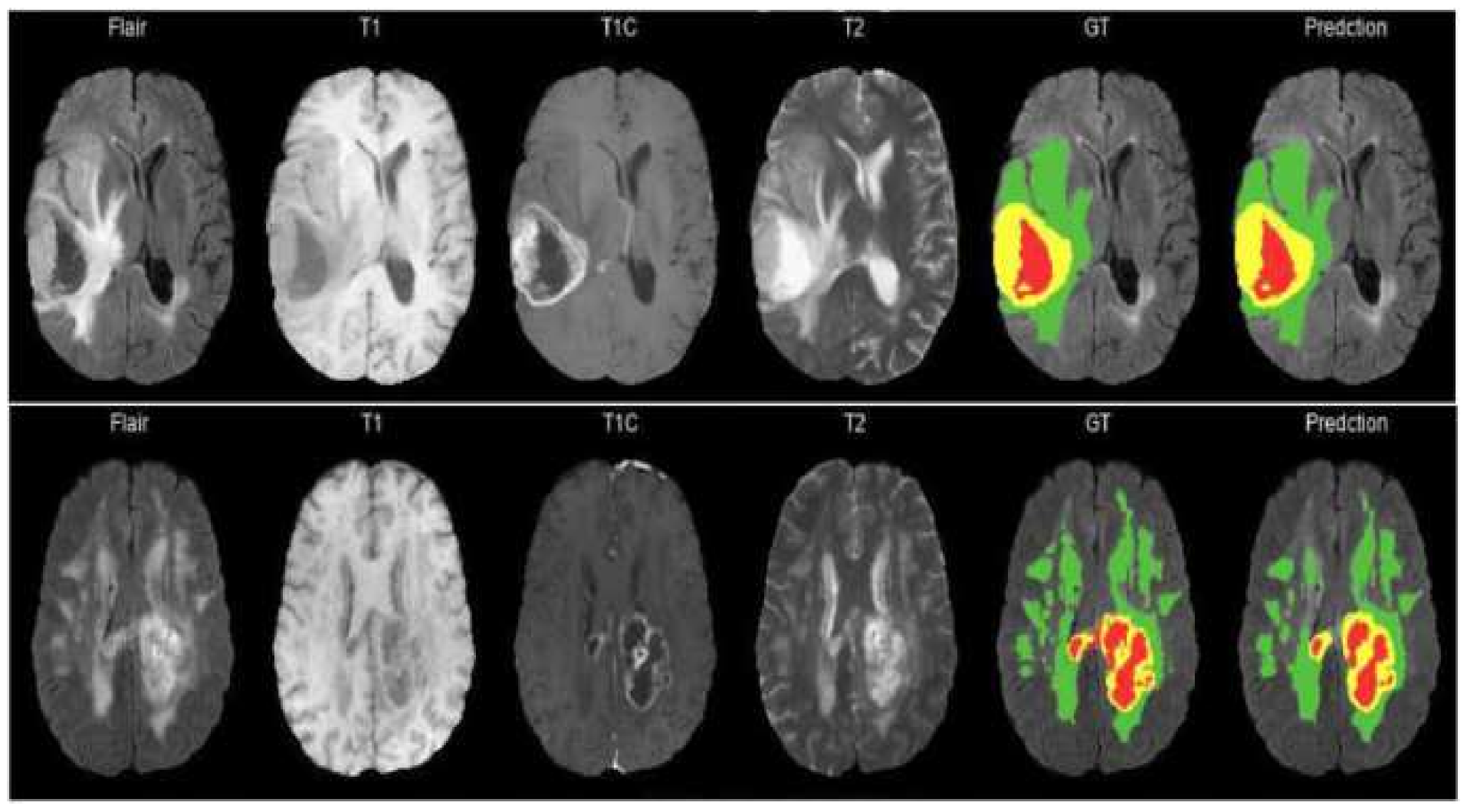
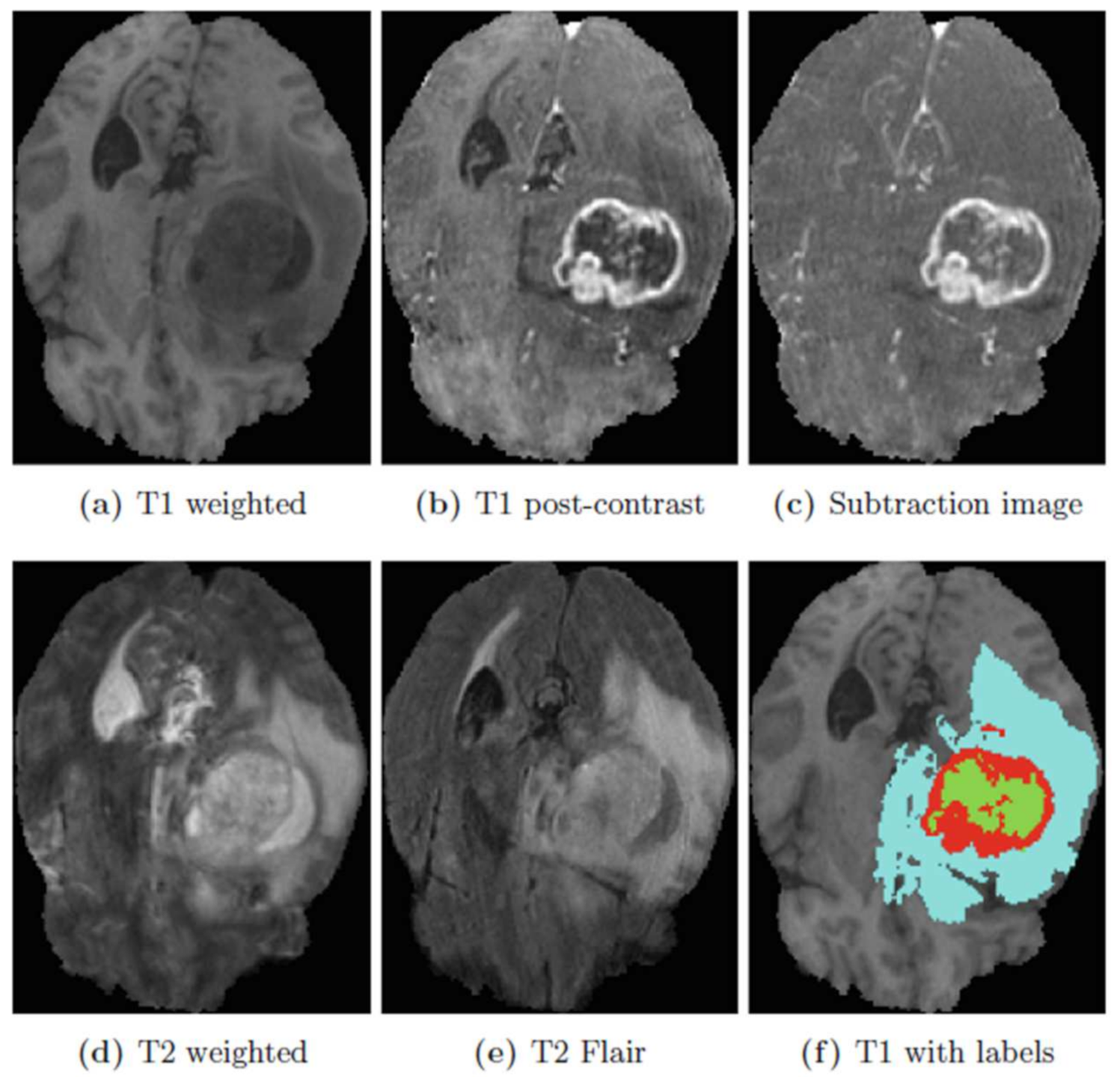
The model proposed in this study used an encoder–decoder architecture for brain tumor segmentation. The encoder part used modified MobileNetV2 for extracting features from MRI images. These feature maps were given as input to the decoder part of U-Net [
8] for the segmentation task. The model is evaluated using the BraTS 2020 dataset. The model contains 4.6M parameters along with a model size of 53MB. Experiments show that the model achieved the dice coefficient scores for WT, TC, and ET as 91.35%, 88.13%, and 83.26% on BraTS 2020, 91.76%, 91.23%, and 83.19% on BraTS 2019, 90.80%, 86.75%, and 79.36% on BraTS 2018 datasets, respectively. Thus, RMU-Net is an improved method for brain tumor segmentation with fewer computational parameters while maintaining high accuracy.