Compounds in Indonesian Ginger Rhizome Extracts and Their Potential for Anti-Skin Aging Based on Molecular Docking
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation and Sample Extraction
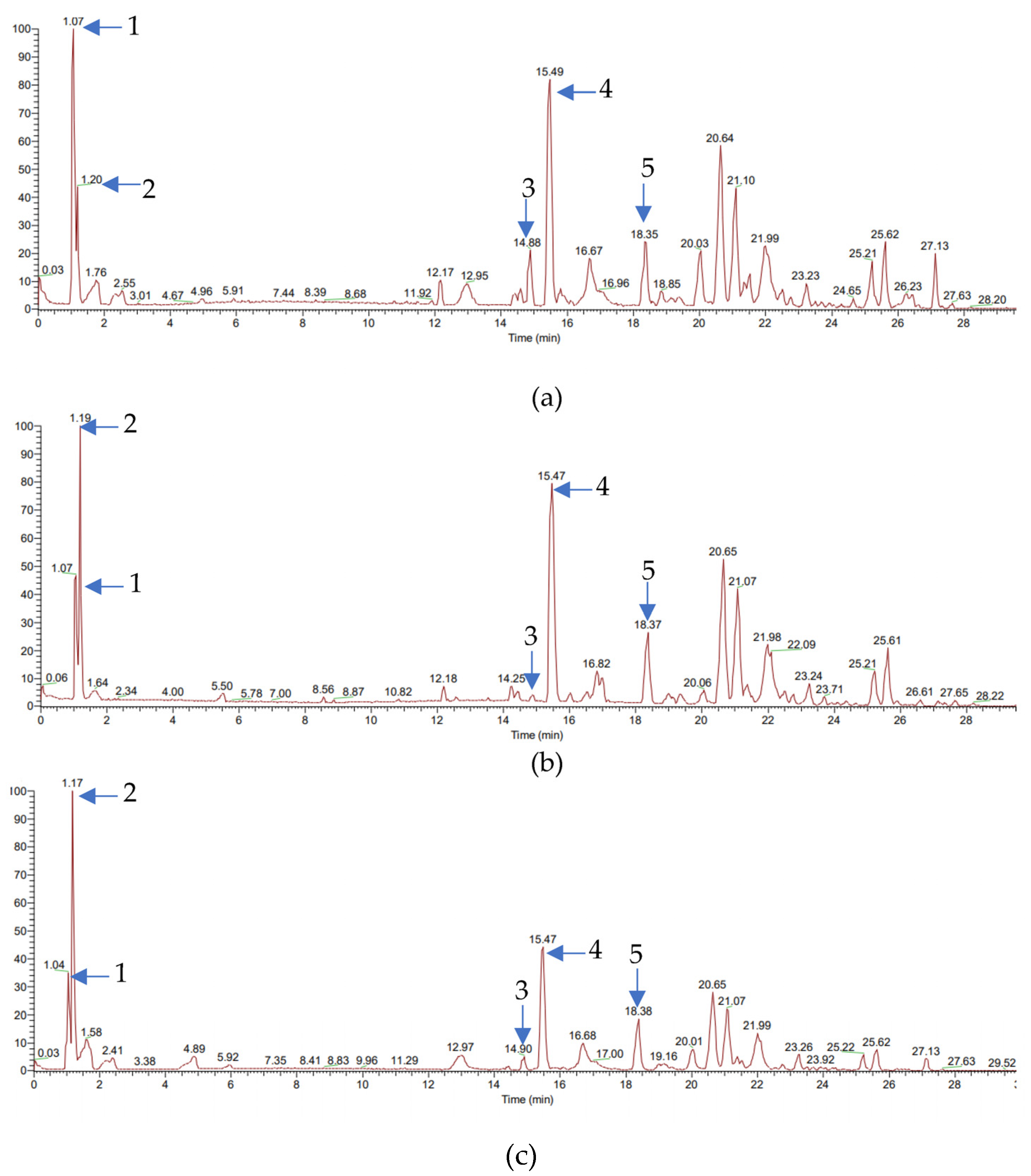
2.2. Identification Compounds in Indonesian Ginger Rhizome Extracts Using Liquid Chromatography–Mass Spectrometry/Mass Spectrometry (LC–MS/MS)
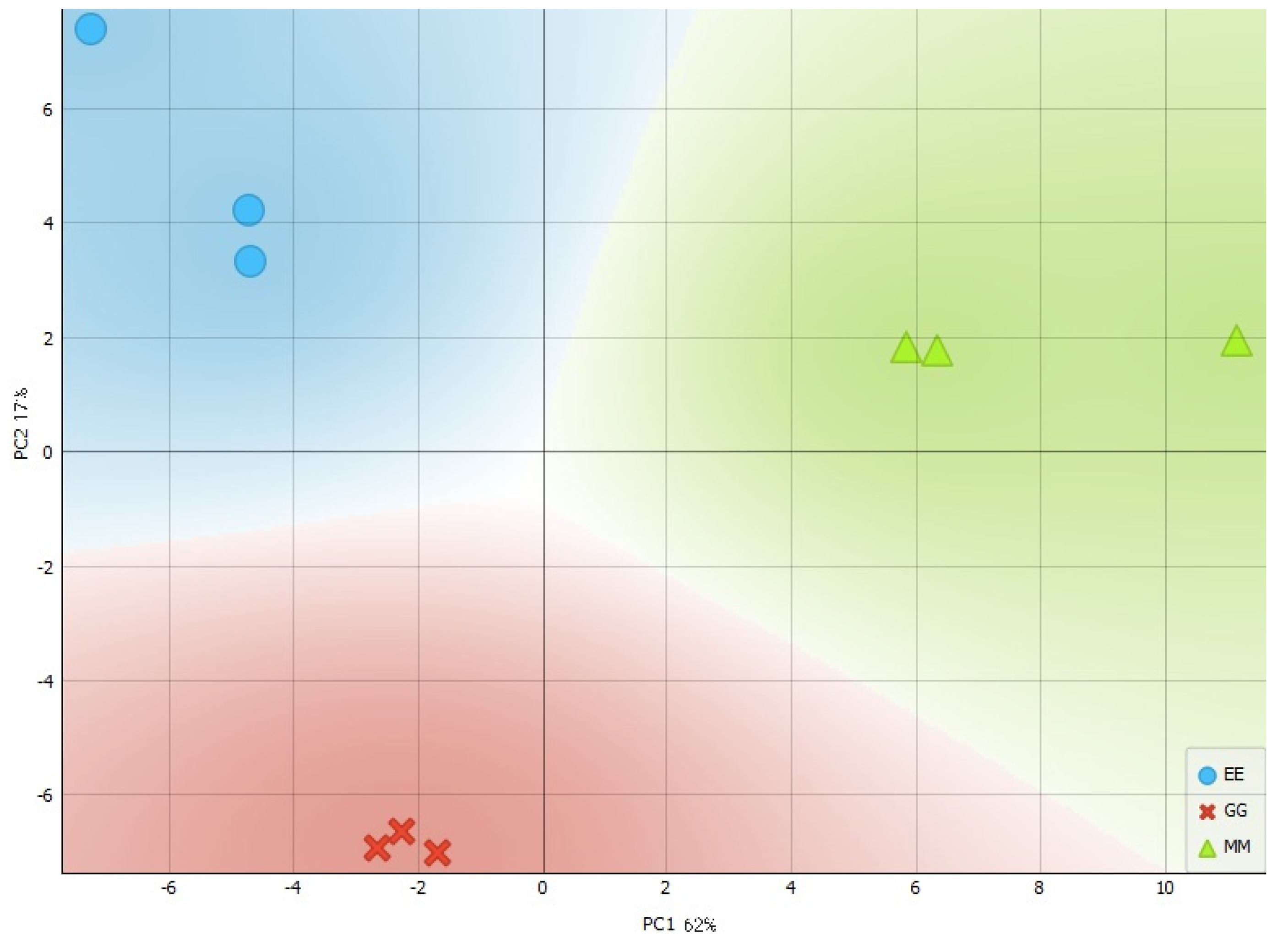
2.3. Statistical Analysis
2.4. Ligands and Receptors Preparation for Molecular Docking
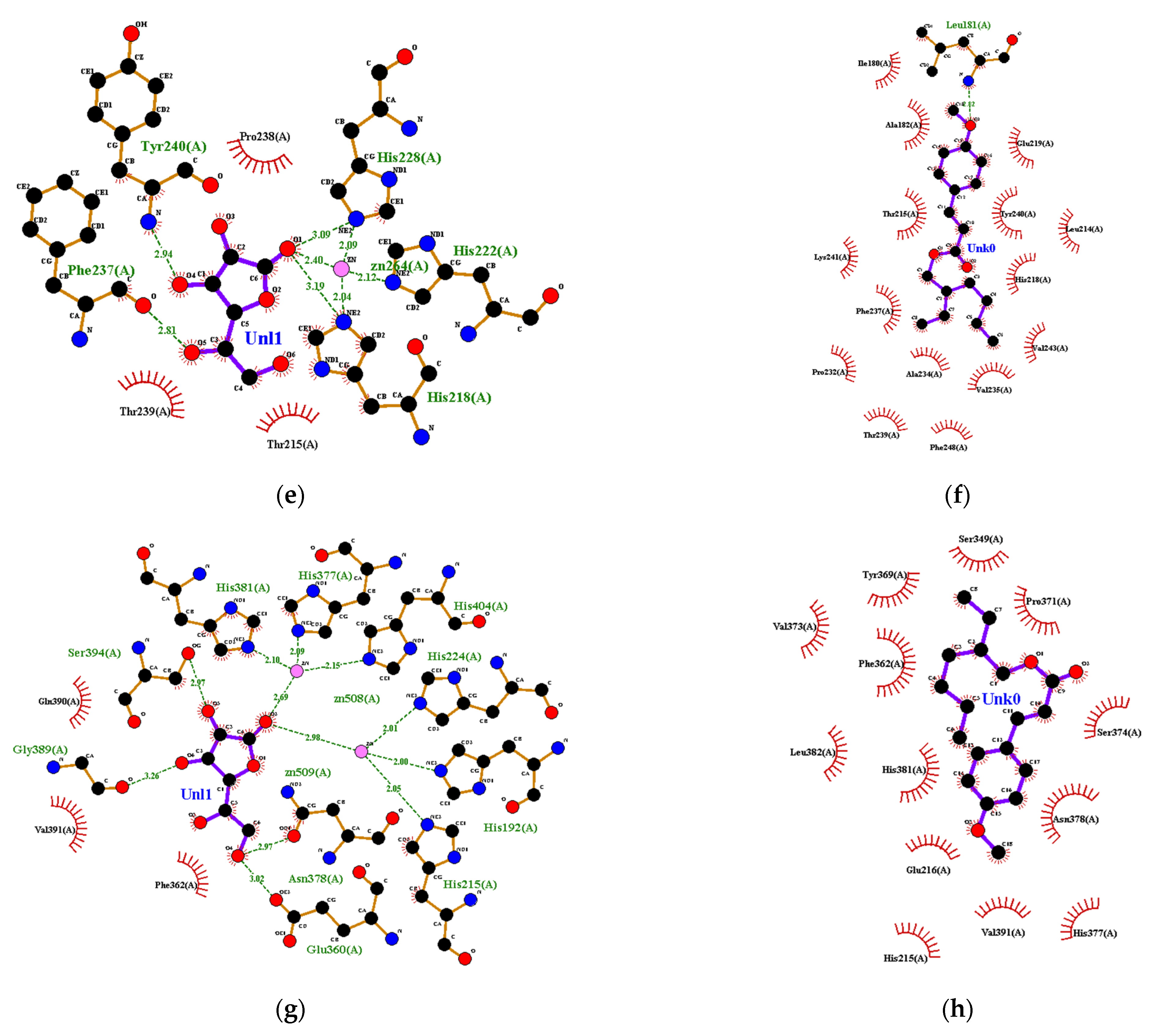
2.5. Molecular Docking Simulation, Visualization, and admetSAR Analysis
3. Results
3.1. Compounds in Indonesian Ginger Rhizome Extracts
3.2. Principal Component Analysis
3.3. Molecular Docking for Anti-Skin Aging Activity Evaluation
3.4. Visualization and Determination of Physicochemical Properties of Ligands Based on admetSAR Parameters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rose, M.R.; Flatt, T.; Graves, J.L.; Greer, L.F.; Martinez, D.E.; Matos, M.; Mueller, L.D.; Shmookler Reis, R.J.; Shahrestani, P. What is aging? Front. Genet. 2012, 3, 2011–2013. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A. Complex systems dynamics in aging: New evidence, continuing questions. Biogerontology 2016, 17, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Liu, Y.; Wong, N.K.; Xiao, J.; So, K.F. Oxidative Stress in Stem Cell Aging. Cell Transplant. 2017, 26, 1483–1495. [Google Scholar] [CrossRef]
- Risques, R.A.; Kennedy, S.R. Aging and the rise of somatic cancer-associated mutations in normal tissues. PLoS Genet. 2018, 14, e1007108. [Google Scholar] [CrossRef] [PubMed]
- Sirard, M.A. Why is the human ovary aging faster than other organs? Women Health 2022, 62, 577–579. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Vierkötter, A.; Schikowski, T.; Hüls, A.; Ding, A.; Matsui, M.S.; Deng, B.; Ma, C.; Ren, A.; Zhang, J.; et al. Epidemiological evidence that indoor air pollution from cooking with solid fuels accelerates skin aging in Chinese women. J. Dermatol. Sci. 2014, 79, 148–154. [Google Scholar] [CrossRef]
- Tottoli, E.M.; Dorati, R.; Genta, I.; Chiesa, E.; Pisani, S.; Conti, B. Skin wound healing process and new emerging technologies for skin wound care and regeneration. Pharmaceutics 2020, 12, 735. [Google Scholar] [CrossRef]
- Keaney, T.C. Aging in the Male Face: Intrinsic and Extrinsic Factors. Dermatol. Surg. 2016, 42, 797–803. [Google Scholar] [CrossRef]
- Rabe, J.H.; Mamelak, A.J.; McElgunn, P.J.S.; Morison, W.L.; Sauder, D.N. Photoaging: Mechanisms and repair. J. Am. Acad. Dermatol. 2006, 55, 1–19. [Google Scholar] [CrossRef]
- Harman, D. Forum Mini Review. Antioxid. Redox Signal. 2003, 5, 557–561. [Google Scholar] [CrossRef]
- Lee, E.J.; Kim, J.Y.; Oh, S.H. Advanced glycation end products (AGEs) promote melanogenesis through receptor for AGEs. Sci. Rep. 2016, 6, 27848. [Google Scholar] [CrossRef] [PubMed]
- Lete, I.; Allué, J. The Effectiveness of Ginger in the Prevention of Nausea and Vomiting during Pregnancy and Chemotherapy. Integr. Med. Insights 2016, 11, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Mahassni, S.H.; Bukhari, O.A. Journal of Nutrition & Intermediary Metabolism Bene fi cial e ff ects of an aqueous ginger extract on the immune system cells and antibodies, hematology, and thyroid hormones in male smokers and. J. Nutr. Intermed. Metab. 2019, 15, 10–17. [Google Scholar] [CrossRef]
- Singh, G.; Kapoor, I.P.S.; Singh, P.; de Heluani, C.S.; de Lampasona, M.P.; Catalan, C.A.N. Chemistry, antioxidant and antimicrobial investigations on essential oil and oleoresins of Zingiber officinale. Food Chem. Toxicol. 2008, 46, 3295–3302. [Google Scholar] [CrossRef]
- Höferl, M.; Stoilova, I.; Wanner, J.; Schmidt, E.; Jirovetz, L.; Trifonova, D.; Stanchev, V.; Krastanov, A. Composition and Comprehensive Antioxidant Activity of Ginger (Zingiber officinale) Essential Oil from Ecuador. Nat. Prod. Commun. 2015, 10, 1085–1090. [Google Scholar] [CrossRef]
- Feng, J.; Du, Z.; Zhang, L.; Luo, W.; Zheng, Y.; Chen, D.; Pan, W.; Yang, Z.; Lin, L.; Xi, L. Chemical Composition and Skin Protective Effects of Essential Oil Obtained from Ginger (Zingiber officinale Roscoe). J. Essent. Oil-Bear. Plants 2018, 21, 1542–1549. [Google Scholar] [CrossRef]
- Lestari, A.R.; Batubara, I.; Wahyudi, S.T.; Ilmiawati, A.; Achmadi, S.S. Bioactive Compounds in Garlic (Allium sativum) and Black Garlic as Antigout Agents, Using Computer Simulation. Life 2022, 12, 1131. [Google Scholar] [CrossRef]
- Rampogu, S.; Baek, A.; Gajula, R.G.; Zeb, A.; Bavi, R.S.; Kumar, R.; Kim, Y.; Kwon, Y.J.; Lee, K.W. Ginger (Zingiber officinale) phytochemicals-gingerenone-A and shogaol inhibit SaHPPK: Molecular docking, molecular dynamics simulations and in vitro approaches. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 16. [Google Scholar] [CrossRef]
- Moitessier, N.; Englebienne, P.; Lee, D.; Lawandi, J.; Corbeil, C.R. Towards the development of universal, fast and highly accurate docking/scoring methods: A long way to go. Br. J. Pharmacol. 2008, 153, 7–26. [Google Scholar] [CrossRef]
- Bailey, D.; Brown, D. High-throughput chemistry and structure-based design: Survival of the smartest. Drug Discov. Today 2001, 6, 57–59. [Google Scholar] [CrossRef]
- McConkey, B.J.; Sobolev, V.; Edelman, M. The performance of current methods in ligand-protein docking. Curr. Sci. 2002, 83, 845–856. [Google Scholar]
- Agarwal, S.; Mehrotra, R. An Overview of Molecular Simulation. JSM Chem. 2016, 4, 1024–1028. [Google Scholar]
- Lee, H.S.; Jo, S.; Lim, H.S.; Im, W. Application of binding free energy calculations to prediction of binding modes and affinities of MDM2 and MDMX inhibitors. J. Chem. Inf. Model. 2012, 52, 1821–1832. [Google Scholar] [CrossRef] [PubMed]
- Arakaki, P.A.; Marques, M.R.; Santos, M.C.L.G. MMP-1 polymorphism and its relationship to pathological processes. J. Biosci. 2009, 34, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.; Buhren, B.; Schrumpf, H.; Wohlrab, J.; Gerber, P. Clinical applications of hyaluronidase. Adv. Exp. Med. Biol. 2019, 1148, 255–277. [Google Scholar] [PubMed]
- Wittenauer, J.; MäcKle, S.; Sußmann, D.; Schweiggert-Weisz, U.; Carle, R. Inhibitory effects of polyphenols from grape pomace extract on collagenase and elastase activity. Fitoterapia 2015, 101, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Eun Lee, K.; Bharadwaj, S.; Yadava, U.; Gu Kang, S. Evaluation of caffeine as inhibitor against collagenase, elastase and tyrosinase using in silico and in vitro approach. J. Enzyme Inhib. Med. Chem. 2019, 34, 927–936. [Google Scholar] [CrossRef]
- Agricultural Technology Study Center. BPTP Sumatera Utara. In Petunjuk Teknis Budidaya Tanaman Jahe.; Ministry of Agricultural Republic of Indonesia: Jakarta, Indonesia, 2012; pp. 25–27. Available online: http://repository.pertanian.go.id/handle/123456789/8585 (accessed on 23 December 2021).
- Kherif, F.; Latypova, A. Principal component analysis. Mach. Learn. Methods Appl. Brain Disord. 2019, 1, 209–225. [Google Scholar] [CrossRef]
- Marlin, M.; Maharijaya, A.; Purwito, A.; Sobir, S. Secondary metabolites change under vernalization and its relation to flowering competency in shallot (Allium cepa var. aggregatum). Rasayan J. Chem. 2019, 12, 1418–1425. [Google Scholar] [CrossRef]
- Ravetti, S.; Clemente, C.; Brignone, S.; Hergert, L.; Allemandi, D.; Palma, S. Ascorbic acid in skin health. Cosmetics 2019, 6, 58. [Google Scholar] [CrossRef]
- Carp, H. A systematic review of dydrogesterone for the treatment of threatened miscarriage. Gynecol. Endocrinol. 2012, 28, 983–990. [Google Scholar] [CrossRef] [PubMed]
- Dugasani, S.; Pichika, M.R.; Nadarajah, V.D.; Balijepalli, M.K.; Tandra, S.; Korlakunta, J.N. Comparative antioxidant and anti-inflammatory effects of [6]-gingerol, [8]-gingerol, [10]-gingerol and [6]-shogaol. J. Ethnopharmacol. 2010, 127, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Ehrt, C.; Brinkjost, T.; Koch, O. A benchmark driven guide to binding site comparison: An exhaustive evaluation using tailor-made data sets (ProSPECCTs). PLoS Comput. Biol. 2018, 14, e1006483. [Google Scholar] [CrossRef] [PubMed]
- Chao, K.L.; Muthukumar, L.; Herzberg, O. Structure of human hyaluronidase-1, a hyaluronan hydrolyzing enzyme involved in tumor growth and angiogenesis. Biochemistry 2007, 46, 6911–6920. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.B.; Liu, J.Z.; Zhang, S.E.; Du, X.; Nie, F.; Tian, J.Y.; Ye, F.; Huang, K.; Hu, J.P.; Li, Y.; et al. 3-Phenylpropanoic acid-based phosphotyrosine (pTyr) mimetics: Hit evolution to a novel orally active protein tyrosine phosphatase 1B (PTP1B) inhibitor. ChemMedChem 2014, 9, 918–921. [Google Scholar] [CrossRef]
- Chhabra, N.; Aseri, M.; Padmanabhan, D. A review of drug isomerism and its significance. Int. J. Appl. Basic Med. Res. 2013, 3, 16–18. [Google Scholar] [CrossRef]
- Rahman, A.J.; Sharma, D.; Kumar, D.; Pathak, M.; Singh, A.; Kumar, V.; Chawla, R.; Ojha, H. Spectroscopic and molecular modelling study of binding mechanism of bovine serum albumin with phosmet. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2021, 244, 118803. [Google Scholar] [CrossRef]
- Benet, L.Z.; Hosey, C.M.; Ursu, O.; Oprea, T.I. BDDCS, the Rule of 5 and drugability. Adv. Drug Deliv. Rev. 2016, 101, 89–98. [Google Scholar] [CrossRef]
- Doak, B.C.; Over, B.; Giordanetto, F.; Kihlberg, J. Oral druggable space beyond the rule of 5: Insights from drugs and clinical candidates. Chem. Biol. 2014, 21, 1115–1142. [Google Scholar] [CrossRef]
- Mishra, A.; Dixit, S.; Ratan, V.; Srivastava, M.; Trivedi, S.; Srivastava, Y.K. Identification and in silico screening of biologically active secondary metabolites isolated from Trichoderma harzianum. Ann. Phytomed. An Int. J. 2018, 7. [Google Scholar] [CrossRef]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. AdmetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [Google Scholar] [CrossRef] [PubMed]
- Andrews, P.R.; Craik, D.J.; Martin, J.L. Functional Group Contributions to Drug-Receptor Interactions. J. Med. Chem. 1984, 27, 1648–1657. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Han, J.; Jiang, C.; Zhang, Y. Biomarkers, oxidative stress and autophagy in skin aging. Ageing Res. Rev. 2020, 59, 101036. [Google Scholar] [CrossRef] [PubMed]
- Yuriev, E.; Holien, J.; Ramsland, P.A. Improvements, trends, and new ideas in molecular docking: 2012–2013 in review. J. Mol. Recognit. 2015, 28, 581–604. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, C.R.; Williams, C.I.; Labute, P. Variability in docking success rates due to dataset preparation. J. Comput. Aided. Mol. Des. 2012, 26, 775–786. [Google Scholar] [CrossRef]
- Chen, F.; Liu, H.; Sun, H.; Pan, P.; Li, Y.; Li, D.; Hou, T. Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein-protein binding free energies and re-rank binding poses generated by protein-protein docking. Phys. Chem. Chem. Phys. 2016, 18, 22129–22139. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided-Drug Des. 2012, 7, 146–157. [Google Scholar] [CrossRef]
- Vakser, I.A. Challenges in protein docking. Curr. Opin. Struct. Biol. 2020, 64, 160–165. [Google Scholar] [CrossRef]


| Receptor | Coordinates | ||
|---|---|---|---|
| X | Y | Z | |
| Collagenase (2TCL) | 73.986 | 8.655 | 9.279 |
| Hyaluronidase (2PE4) | 41.877 | −22.142 | −16.287 |
| Elastase (3F19) | −11.338 | −4.481 | −18.305 |
| Tyrosinase (5M8R) | −30.322 | −4.944 | −24.773 |
| Ligand | Affinity Energy (kcal/mol) | |||
|---|---|---|---|---|
| 2TCL 2 | 2PE4 3 | 3F19 4 | 5M8R 5 | |
| Ascorbic acid | −6.4 | −5.6 | −6.1 | −6.0 |
| d-Corlin | −14.6 | −13.9 | −16.0 | −13.4 |
| DG 1 | −10.6 | −11.4 | −13.0 | −10.6 |
| Catechin | −8.8 | −7.9 | −9.7 | −8.4 |
| 6-Gingerol | −6.1 | −6.7 | −8.0 | −6.8 |
| 8-Shogaol | −6.0 | −6.9 | −8.0 | −6.4 |
| 4-Shogaol | −6.1 | −6.8 | −7.6 | −6.3 |
| 10-Shogaol | −5.5 | −7.0 | −7.7 | −6.6 |
| 6-Paradol | −5.7 | −6.8 | −7.9 | −5.5 |
| Octinoxate | −6.1 | −7.2 | −7.8 | −6.3 |
| 6-Gingerdione | −5.9 | −6.4 | −7.9 | −6.6 |
| p-Cymene | −5.9 | −6.6 | −6.5 | −5.8 |
| Ethyl cinnamate | −6.1 | −6.1 | −7.0 | −5.5 |
| Ligand | Lipinski’s Rules | admetSAR Parameters | Enzyme Inhibitor | ||
|---|---|---|---|---|---|
| a, b, c, d, e | Mutagenicity | Carcinogenicity | Eye Irritation | ||
| Catechin | Pass | + | - | + | 0.47 |
| 6-Paradol | Pass | - | - | + | 0.18 |
| 6-Gingerol | Pass | - | - | + | 0.38 |
| EC 1 | Pass | - | - | + | −0.45 |
| p-Cymene | Pass | - | - | + | −0.78 |
| Octinoxate | Pass | - | - | - | 0.02 |
| 8-Shogaol | Pass | + | - | + | 0.27 |
| 6G 2 | Pass | + | - | + | 0.21 |
| 4-Shogaol | Pass | - | - | + | 0.27 |
| 10-Shogaol | Pass | + | - | + | 0.25 |
| DG 3 | Pass | - | - | - | 0.56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asoka, S.F.; Batubara, I.; Lestari, A.R.; Wahyuni, W.T.; Wahyudi, S.T. Compounds in Indonesian Ginger Rhizome Extracts and Their Potential for Anti-Skin Aging Based on Molecular Docking. Cosmetics 2022, 9, 128. https://doi.org/10.3390/cosmetics9060128
Asoka SF, Batubara I, Lestari AR, Wahyuni WT, Wahyudi ST. Compounds in Indonesian Ginger Rhizome Extracts and Their Potential for Anti-Skin Aging Based on Molecular Docking. Cosmetics. 2022; 9(6):128. https://doi.org/10.3390/cosmetics9060128
Chicago/Turabian StyleAsoka, Shadila F., Irmanida Batubara, Ayu Rahmania Lestari, Wulan Tri Wahyuni, and Setyanto Tri Wahyudi. 2022. "Compounds in Indonesian Ginger Rhizome Extracts and Their Potential for Anti-Skin Aging Based on Molecular Docking" Cosmetics 9, no. 6: 128. https://doi.org/10.3390/cosmetics9060128
APA StyleAsoka, S. F., Batubara, I., Lestari, A. R., Wahyuni, W. T., & Wahyudi, S. T. (2022). Compounds in Indonesian Ginger Rhizome Extracts and Their Potential for Anti-Skin Aging Based on Molecular Docking. Cosmetics, 9(6), 128. https://doi.org/10.3390/cosmetics9060128








