1. Introduction
Skin cancer represents one of the most prevalent malignancies worldwide, with melanoma accounting for approximately 75% of skin cancer-related deaths despite comprising fewer than 5% of cases. Early detection dramatically improves survival rates from 14% to over 99%, highlighting the urgent need for accurate and accessible diagnostic tools. While deep learning has shown promise in dermatological diagnosis, existing approaches lack clinical explainability and deployable interfaces that bridge the gap between research innovation and practical healthcare applications.
Traditional dermatoscopic image assessment faces significant challenges, particularly in regions with limited expert availability and variability in clinical experience. Tschandl et al. noted that neural network training for automated diagnosis is limited by small, non-diverse dermatoscopic datasets [
1]. The emergence of artificial intelligence, particularly convolutional neural networks (CNNs), has opened new possibilities for automated diagnosis. Esteva et al. demonstrated that deep neural networks could achieve dermatologist-level performance using a dataset of 129,450 clinical images encompassing 2032 diseases, with CNNs matching 21 board-certified dermatologists across critical binary classification tasks [
2].
The HAM10000 dataset by Tschandl et al. comprises 10,015 high-resolution dermatoscopic images from diverse populations, encompassing seven diagnostic categories: melanocytic nevi (66.9%), melanoma (11.1%), benign keratosis-like lesions (11.0%), basal cell carcinoma (5.1%), actinic keratoses (3.3%), vascular lesions (1.4%), and dermatofibroma (1.1%). Over 50% of lesions were confirmed histopathologically, with the remaining cases validated by expert consensus, follow-up examination, or confocal microscopy [
1].
Significant progress has been achieved through deep learning architectures. Wu et al. showed that CNN architectures, such as VGGNet, GoogleNet, ResNet, and variants, have been successfully applied to skin cancer classification [
3]. Ahmad et al. achieved accuracies of 99.3% and 91.5% on the HAM10000 and ISIC2018 datasets using deep learning with explainable AI, data augmentation, fine-tuned pre-trained models, and the Butterfly Optimization Algorithm for feature selection [
4]. Recent innovations include Perez et al.’s data augmentation, achieving an AUC of 0.882 [
5], Himel et al.’s Vision Transformer, achieving 96.15% accuracy [
6], Liu et al.’s SkinNet, achieving 86.7% accuracy with an AUC of 0.96 [
7], Munjal et al.’s SkinSage XAI, achieving 96% accuracy and 99.83% AUC [
8], and Cino et al.’s test-time augmentation, achieving 97.58% balanced accuracy [
9].
Network-level fusion architectures have emerged as promising approaches. Arshad et al. proposed a novel architecture, integrating multiple deep models through depth concatenation, achieving 91.3% and 90.7% accuracies on the HAM10000 and ISIC2018 datasets while maintaining lower computational complexity [
10]. Hussein et al. introduced hybrid quantum deep learning utilizing HQCNN, BiLSTM, and MobileNetV2, achieving 97.7% training and 89.3% test accuracy [
11]. Krishna et al.’s LesionAid utilized ViTGANs for class imbalance, achieving 99.2% training and 97.4% validation accuracy [
12]. Wang et al.’s boundary-aware transformers integrated CNNs with transformer architectures for segmentation [
13].
Multimodal approaches have shown significant promise. Tang and Lasser’s asymmetric multimodal fusion method reduced parameters from 58.49 M to 32.48 M without compromising performance [
14]. Hasan and Rifat’s hybrid ensemble framework achieved a partial AUC of 0.1755 with an above 80% true positive rate [
15]. Thomas demonstrated that combining image features with patient metadata enhanced classification across six deep neural networks [
16], while Tran Van and Le achieved 95.73% accuracy through cross-attention fusion [
17]. Advanced frameworks include Wang et al.’s self-supervised multi-modality learning [
18], Yu et al.’s MDSIS-Net [
19], and Christopoulos et al.’s SLIMP nested contrastive learning [
20].
Recent contributions by Aksoy et al. evaluated seven deep learning models, with DenseNet169 achieving 85% accuracy and successful web-based deployment featuring visualization and automated medical knowledge extraction [
21,
22]. Yan et al.’s PanDerm foundation model, pre-trained on over 2 million images, achieved state-of-the-art performance across 28 benchmarks, outperforming clinicians by 10.2% in early-stage melanoma detection [
23]. Vision–language models, like Kamal and Oates’ MedGrad E-CLIP, enhanced diagnostic transparency through weighted entropy mechanisms [
24].
Clinical accessibility has been addressed through smartphone-based approaches. Oyedeji et al. investigated 13 deep learning models using clinical images, with DenseNet-161 achieving 79.40% binary accuracy and EfficientNet-B7 reaching 85.80% [
25]. For clinical deployment, explainability frameworks have been essential. Wu et al. emphasized the importance of explainable AI [
3], while Ahmad et al., Munjal et al., and Cino et al. implemented Grad-CAM, LIME, and t-SNE visualizations [
4,
8,
9]. Advanced approaches include Patrício et al.’s concept-based explanations [
26], Ieracitano et al.’s TIxAI trustworthiness index [
27], and Metta et al.’s ABELE explainer [
28].
Comprehensive evaluations have been conducted across ISIC datasets. Yao achieved a validation AUC greater than 94% using EfficientNet B6 and VGG 16 with GBDT ensemble learning [
29]. Paccotacya-Yanque et al. compared seven explainability methods, identifying essential properties of fidelity, meaningfulness, and effectiveness [
30].
Despite significant advances in deep learning-based skin lesion classification, important gaps remain in the literature. While explainability techniques like Grad-CAM and LIME have been extensively explored for providing pixel-level attribution and feature importance visualization, XRAI has not been implemented for skin lesion analysis. XRAI represents a significant advancement over traditional explainability methods by providing superior region-based explanations that generate more coherent and spatially connected visual explanations compared to conventional pixel-level attribution methods. Unlike Grad-CAM, which often produces scattered heatmaps, or LIME, which segments images into superpixels that may not align with clinical reasoning patterns, XRAI creates contiguous regions that better correspond to how dermatologists naturally examine lesions, focusing on unified areas of clinical significance rather than individual pixels or arbitrary segments. This region-based approach aligns more closely with dermatological reasoning patterns, where clinicians assess lesions based on coherent morphological features, such as asymmetry, border irregularity, color variation, and diameter changes. Furthermore, clinically deployable interfaces that provide both diagnostic capabilities and practical patient guidance are lacking in the existing literature.
These limitations have been addressed in this study through two key novel contributions. First, the first implementation of XRAI explainability for skin lesion classification has been presented, providing more coherent and spatially connected explanations compared to traditional pixel-level attribution methods. Second, the first deployable web-based clinical interface has been developed, which combines diagnostic predictions with visual explanations and evidence-based cosmetic recommendations for benign conditions, bridging the gap between AI research and practical clinical utility.
2. Methodology
2.1. Data Acquisition and Preprocessing
This study utilized the HAM10000 dataset (“Human Against Machine with 10,000 training images”), a comprehensive collection of dermatoscopic images specifically designed for machine learning applications in dermatological diagnosis. The dataset was originally developed for the International Skin Imaging Collaboration (ISIC) 2018 challenge and represents one of the largest publicly available collections of annotated skin lesion images for academic research purposes.
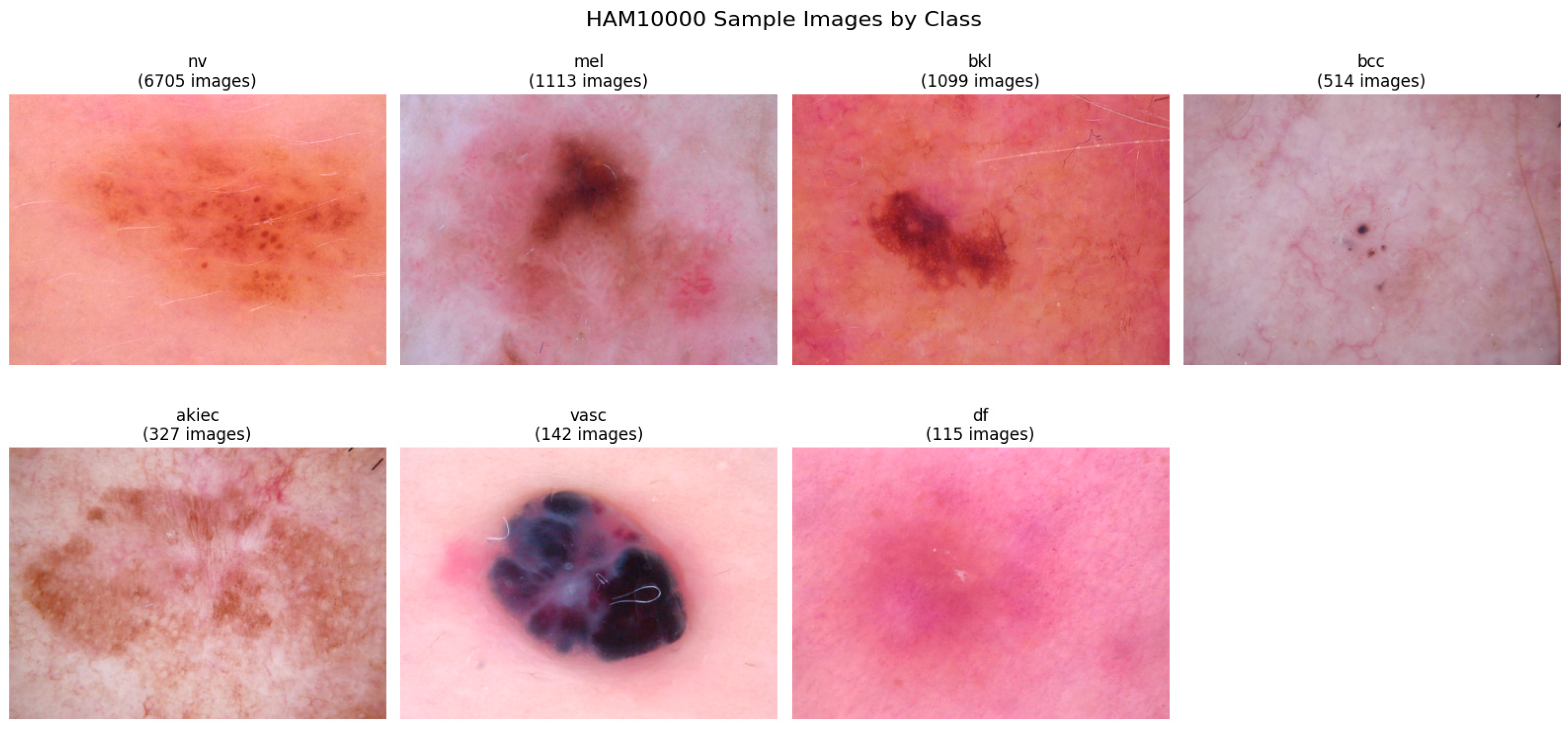
The HAM10000 dataset comprises 10,015 high-resolution dermatoscopic images collected from diverse populations using multiple acquisition modalities and imaging devices. This multi-source approach ensures enhanced generalizability and robustness compared to single-institution datasets. The images encompass seven distinct diagnostic categories representing the most clinically significant types of pigmented skin lesions encountered in dermatological practice.
As illustrated in
Table 1, the dataset exhibits significant class imbalance, with melanocytic nevi representing approximately two-thirds of all samples, while rare conditions, such as dermatofibroma and vascular lesions, constitute less than three percent combined. This distribution reflects real-world clinical prevalence patterns but necessitates specialized handling techniques during model training to prevent bias toward majority classes.
The diagnostic ground truth for the HAM10000 dataset was established through multiple validation methodologies, ensuring high confidence in label accuracy. More than fifty percent of the lesions received histopathological confirmation, which represents the gold standard for dermatological diagnosis. The remaining cases were validated through expert consensus among board-certified dermatologists, follow-up examination protocols, or confirmation via in vivo confocal microscopy techniques.
This multimodal validation approach provides robust ground truth labels while accommodating the practical constraints of clinical practice, where not all lesions undergo invasive histopathological examination. The dataset includes lesions with multiple images, trackable through unique lesion identifiers, allowing for comprehensive analysis of individual cases across different imaging conditions and time points.
All dermatoscopic images underwent systematic preprocessing to ensure consistency and optimal model performance. The raw images, originally stored in JPEG format across two directory partitions, were first validated for accessibility and quality. The images were resized to a standardized input resolution of 224 × 224 pixels using high-quality interpolation algorithms to maintain visual fidelity while ensuring computational efficiency.
To optimize feature extraction and model convergence, custom normalization parameters were calculated specifically for the HAM10000 dataset rather than relying on generic ImageNet statistics. Through stratified sampling of 1996 images across all diagnostic categories, the following normalization parameters were derived:
Channel-wise means: [0.763, 0.544, 0.568] for RGB channels, respectively;
Channel-wise standard deviations: [0.141, 0.152, 0.169] for RGB channels, respectively.
These parameters reflect the unique color characteristics of dermatoscopic imagery, which typically exhibit higher red channel intensity and distinct color distributions compared to natural images used in ImageNet pretraining.
Beyond image data, the HAM10000 dataset provides comprehensive metadata, including patient demographics and lesion characteristics. This multimodal information was systematically processed and integrated to enhance diagnostic accuracy.
Age information was normalized using z-score standardization to account for the broad age distribution. Missing age values were imputed with population mean values. Gender information was encoded using binary representation (male = 1, female = 0) with appropriate handling for unspecified cases. Lesion location data encompassed twelve distinct body regions, including face, scalp, neck, trunk, extremities, back, abdomen, and chest. This categorical information was transformed using one-hot encoding to preserve spatial relationships.
The integration of demographic and anatomical metadata creates a comprehensive multimodal dataset that mirrors real-world clinical decision-making processes, where dermatologists consider patient characteristics alongside visual examination findings.
To ensure robust model evaluation and prevent data leakage, the dataset was partitioned using stratified random sampling to maintain proportional class representation across training, validation, and test sets. The final partitioning scheme allocated seventy percent of samples for training (7010 images), fifteen percent for validation (1502 images), and fifteen percent for final testing (1503 images).
This partitioning strategy ensured adequate representation of minority classes in each subset while providing sufficient training data for complex model architectures. The stratified approach maintained the original class distribution across all partitions, preventing potential bias that could arise from uneven class allocation.
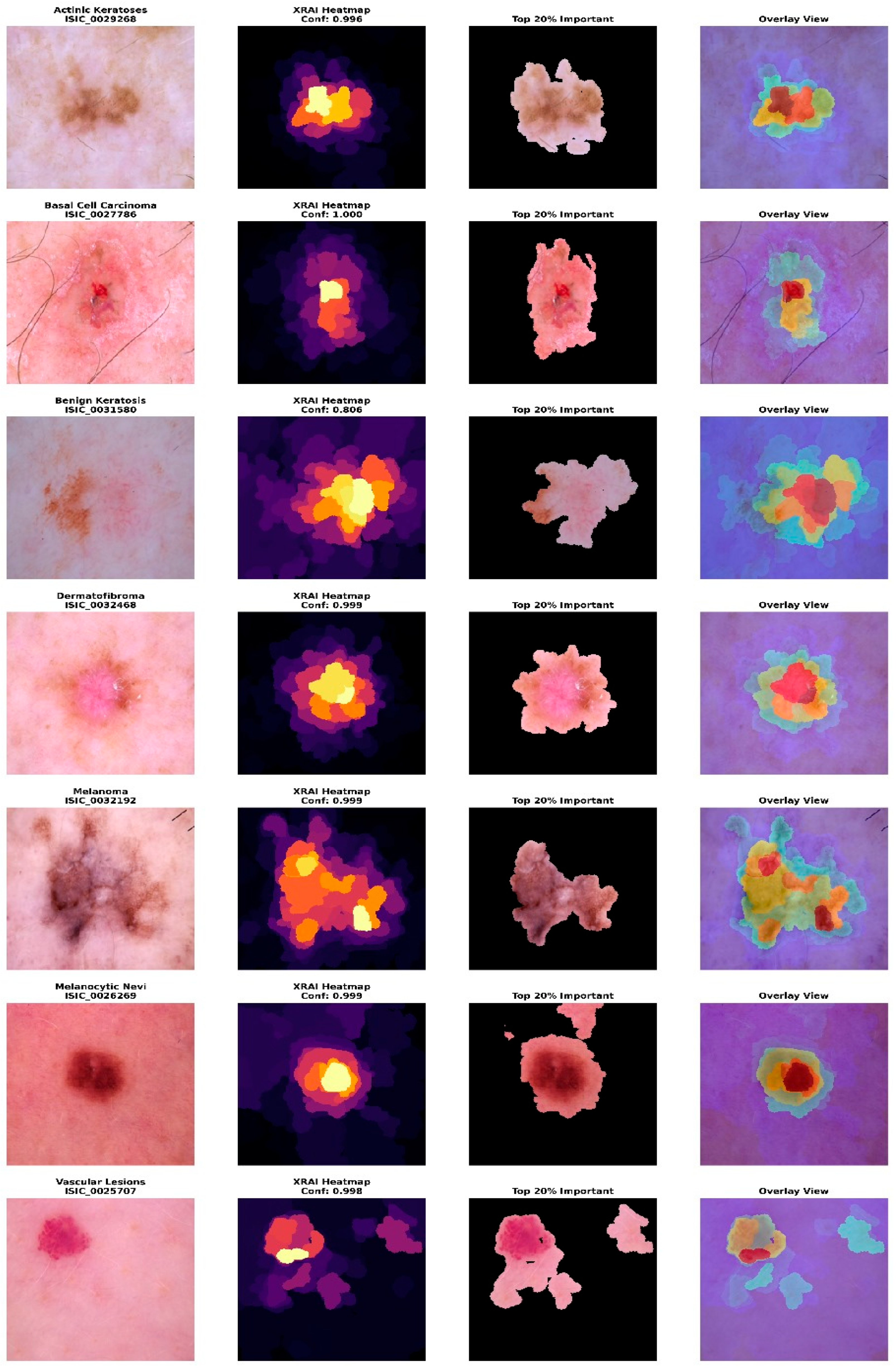
Figure 1 presents representative samples from each diagnostic category, illustrating the visual diversity and characteristic features of different lesion types within the dataset. The samples demonstrate the challenging nature of automated skin lesion classification, with subtle visual differences between benign and malignant conditions requiring sophisticated pattern recognition capabilities.
2.2. Model Architectures and Training Methodology
This study employed a comprehensive multi-architecture strategy to evaluate different deep learning approaches for skin lesion classification. Three distinct convolutional neural network architectures were implemented and compared: DenseNet-121, EfficientNet-B3, and ResNet-50. Each architecture was selected to represent different designs and computational approaches, providing complementary perspectives on the classification task while enabling robust performance comparison and ensemble learning opportunities.
All models incorporated a unified multimodal design that combines dermatoscopic image analysis with patient metadata integration. This approach mirrors clinical decision-making processes where dermatologists consider both visual lesion characteristics and patient demographics when making diagnostic assessments.
2.2.1. DenseNet-121 Architecture
The DenseNet-121 model served as the primary architecture, utilizing dense connectivity patterns that facilitate feature reuse and gradient flow throughout the network. The implementation employed ImageNet pre-trained weights as initialization, utilizing transfer learning to benefit from features learned on natural images while adapting to dermatoscopic imagery characteristics.
The DenseNet backbone was modified by replacing the original classification layer with a custom multimodal fusion system. Image features extracted from the final dense block (1024 dimensions) were concatenated with processed metadata features to create a comprehensive representation. The metadata processing pipeline consisted of a two-layer neural network that transformed the twelve-dimensional demographic and anatomical input into a thirty-two-dimensional dense representation through batch normalization, ReLU activation, and dropout regularization.
The classification head employed a three-layer architecture with progressive dimensionality reduction from the fused feature space (1056 dimensions) through 512- and 256-dimensional hidden layers to the final seven-class output. Each layer incorporated batch normalization and dropout (rates of 0.5 and 0.3, respectively) to prevent overfitting while maintaining robust feature learning. The complete DenseNet-121 implementation contained 7,632,743 trainable parameters, providing substantial model capacity while remaining computationally efficient (
Table 2).
2.2.2. EfficientNet-B3 Architecture
EfficientNet-B3 was selected as the second architecture to use compound scaling principles that systematically balance network depth, width, and resolution. This architecture represents a modern approach to efficient neural network design, achieving high accuracy while maintaining computational efficiency through carefully optimized scaling coefficients.
The EfficientNet-B3 backbone, initialized with ImageNet pre-trained weights, provided 1536-dimensional feature representations from its final global average pooling layer. The same metadata processing pipeline used in DenseNet was applied, creating a 1568-dimensional fused feature vector for classification. The identical classification head architecture ensured a fair comparison between models while maintaining consistent training dynamics.
With 11,637,263 trainable parameters, EfficientNet-B3 offered increased model capacity compared to DenseNet-121 while incorporating modern architectural innovations, such as mobile inverted bottleneck convolutions and squeeze-and-excitation blocks. Forward pass execution time averaged 0.616 s for a batch of four images, demonstrating acceptable computational efficiency for practical deployment scenarios (
Table 2).
2.2.3. ResNet-50 Architecture
ResNet-50 provided the third architecture, implementing residual learning principles that enable training of very deep networks through skip connections. This architecture served as a classical baseline, representing established deep learning approaches while contributing to ensemble diversity through its distinct feature learning characteristics.
The ResNet-50 backbone generated 2048-dimensional feature representations, requiring adaptation of the fusion architecture to accommodate the larger feature space. The metadata processing remained identical, but the fused feature vector expanded to 2080 dimensions before classification. Despite this increased dimensionality, the same classification head structure was maintained to ensure consistent comparison methodology.
ResNet-50 represented the largest model in comparison, with 24,711,207 trainable parameters, providing maximum model capacity at the cost of increased computational requirements. Forward pass execution time was optimized at 0.018 s per batch, demonstrating efficient inference despite the larger parameter count.
2.2.4. Multimodal Feature Integration
All three architectures implemented identical multimodal integration strategies to enable a fair performance comparison. Patient metadata, including age, sex, and anatomical location, underwent systematic preprocessing before neural network integration. Age values were normalized using z-score standardization, while categorical variables received appropriate encoding transformations.
The metadata processing network employed two fully connected layers with sixty-four and thirty-two neurons, respectively, incorporating batch normalization and ReLU activation functions. Dropout regularization (rate 0.3) was applied to prevent overfitting while maintaining generalization capability. This design created meaningful demographic representations that enhanced image-based classification through clinically relevant auxiliary information.
Feature fusion occurred through the simple concatenation of image and metadata representations, creating joint feature vectors that captured both visual and demographic patterns. This approach enabled the models to learn interactions between lesion appearance and patient characteristics, potentially improving diagnostic accuracy for cases where demographic factors influence lesion presentation.
2.2.5. Training Configuration and Optimization
All models employed identical training configurations to ensure a fair comparison and reproducible results. The AdamW optimizer was selected with a learning rate of 0.001 and weight decay of 1 × 10−4 to provide robust optimization with regularization. Learning rate scheduling utilized ReduceLROnPlateau with patience of three epochs and reduction factor of 0.5, enabling adaptive learning rate adjustment based on validation performance.
Focal Loss, with a gamma parameter of 2.0 and an alpha of 1.0, addressed the significant class imbalance present in the HAM10000 dataset. This loss function provided increased focus on difficult examples while maintaining stability during training. Gradient clipping with a maximum norm of 1.0 prevented gradient explosion and ensured stable training dynamics across all architectures.
Training proceeded for a maximum of twenty-five epochs with early stopping based on validation accuracy plateaus. The batch size was set to thirty-two samples to balance memory efficiency with stable gradient estimation. Each model was trained using the same 70/15/15 data split to enable direct performance comparison.
All experiments were conducted on an NVIDIA RTX 4060 GPU with 8 GB VRAM. The models were implemented in PyTorch (version 2.7.0) with CUDA acceleration, using Python 3.13 on a Windows-based workstation. Supporting libraries, such as scikit-learn and NumPy, were utilized for evaluation metrics, visualization, and statistical analysis.
2.2.6. Test-Time Augmentation Enhancement
Test-time augmentation (TTA) was implemented as a post-training enhancement technique to improve model robustness and accuracy without requiring model retraining. The TTA strategy employed eight distinct augmentation transformations applied to each test sample: original image, horizontal flip, vertical flip, fifteen-degree rotation, negative fifteen-degree rotation, top-left crop, bottom-right crop, and ninety-percent scaling.
During inference, each test image underwent all eight transformations, generating multiple predictions that were ensemble-averaged to produce the final classification result. This approach utilized the principle that consistent predictions across multiple image variations indicate robust model confidence, while averaging reduces prediction variance and improves overall accuracy.
The TTA implementation maintained metadata consistency across all augmented versions while transforming only the image component. The probability distributions from all eight predictions were averaged before final class selection, providing a more robust inference mechanism than single-image prediction. This technique required no additional training time while potentially improving test accuracy by 2–3 percentage points.
2.2.7. Model Evaluation Methodology
Comprehensive evaluation protocols were established to ensure robust and reproducible assessment of model performance across all architectures. The evaluation framework incorporated multiple complementary metrics and visualization techniques to provide a thorough analysis of classification accuracy, per-class performance, and model reliability.
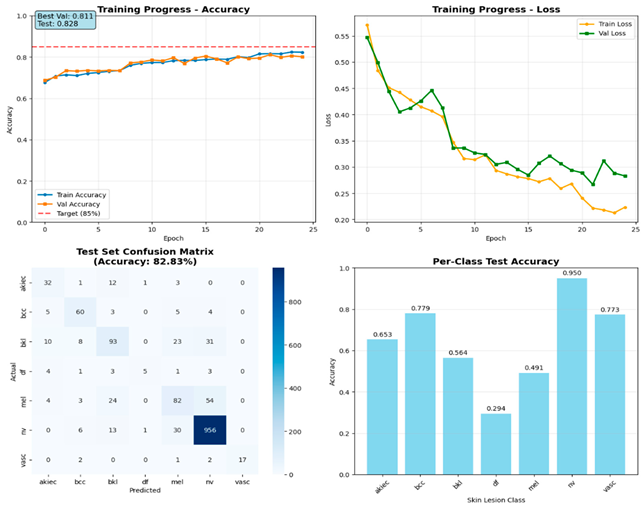
All models were evaluated using identical protocols, including overall accuracy calculation, precision, recall, and F1-score for each diagnostic category. Classification reports generated detailed per-class statistics with four-decimal precision to enable precise performance comparisons between architectures. Confusion matrices provided a visual assessment of classification patterns and common misclassification errors across different lesion types.
The stratified 70/15/15 data split ensured unbiased evaluation with sufficient sample sizes for statistical significance testing. Test set evaluation was performed only once per model using the best validation checkpoint to prevent data leakage and maintain evaluation integrity. Per-class accuracy calculations accounted for class imbalance by analyzing performance within each diagnostic category separately.
Comprehensive visualization pipelines generated standardized plots for direct model comparison. Training progression plots displayed accuracy and loss curves across epochs with target threshold lines at eighty-five percent accuracy. Confusion matrices employed consistent color schemes and annotation formats to enable visual comparison between architectures. Per-class accuracy bar charts highlighted strengths and weaknesses across different lesion types with sample size annotations.
To validate the statistical significance of observed performance differences, comprehensive statistical testing was conducted using paired t-tests, effect size analysis, and confidence interval estimation across all reported accuracy metrics (test accuracy, validation accuracy, and test-time augmentation accuracy).
The evaluation methodology ensured fair comparison between architectures while maintaining rigorous standards for medical AI research, providing the foundation for reliable performance assessment and clinical applicability analysis. The comprehensive multi-architecture approach enabled thorough evaluation of different deep learning paradigms while providing opportunities for ensemble learning and robust performance assessment across varying computational constraints.
2.3. Explainability Implementation (XRAI)
The implementation of explainable artificial intelligence (XAI) capabilities utilized the XRAI (eXplanation with Region-based Attribution for Images) algorithm, specifically adapted for medical imaging applications through the PAIRML saliency library. XRAI was selected over alternative explanation methods due to its good performance in medical image analysis, providing region-based explanations that align with clinical diagnostic reasoning patterns. Unlike pixel-level attribution methods, such as Grad-CAM or LIME, XRAI generates coherent, spatially connected explanations that correspond to anatomically meaningful structures in dermatoscopic images. The XRAI algorithm operates by partitioning the input image into hierarchical regions and computing attribution scores based on ranked area integrals. This approach ensures that explanations maintain spatial coherence while reflecting the model’s decision-making process. For skin lesion classification, this methodology proves particularly valuable as dermatologists naturally evaluate lesions by examining specific regions and structures rather than individual pixels.
2.3.1. Technical Implementation Framework
The explainability system was integrated with the best-performing EfficientNet-B3 architecture to provide interpretable predictions. The implementation required careful adaptation of the PAIRML saliency framework to accommodate the multimodal nature of the skin lesion classification model, incorporating both image features and patient metadata during explanation generation. The explainability framework utilized PyTorch hooks to capture intermediate feature representations and gradients from the final convolutional layer of EfficientNet-B3. Specifically, the system registered forward and backward hooks on the Conv2d layer with dimensions (384, 1536, kernel_size = (1,1)), enabling extraction of both feature maps and gradient information required for attribution computation. The hook implementation employed tensor dimension manipulation to ensure compatibility between PyTorch’s channel-first format and XRAI’s expected channel-last configuration. The explanation generation process maintained consistency with the training preprocessing pipeline, applying identical normalization parameters derived from the HAM10000 dataset statistics. The images underwent standardized resizing to 224 × 224 pixels, followed by tensor conversion and GPU memory allocation for efficient computation. The preprocessing function incorporated gradient requirement activation, enabling backpropagation through the network during attribution computation.
2.3.2. Multimodal Explanation Generation
The explainability implementation addressed the unique challenge of generating explanations for multimodal inputs, combining dermatoscopic images with patient demographics. The system developed a specialized model call function that integrated metadata features during explanation computation while maintaining focus on visual features most relevant to clinical interpretation. During explanation generation, patient metadata, including age, sex, and anatomical location, was processed through the same encoding pipeline used during training. For cases where specific metadata was unavailable, the system employed default values (age = 45, female encoding for unknown sex, zero-encoded anatomical location) to ensure explanation generation. In analysis modes, the system used zero-filled metadata vectors, focusing purely on image-based explanations.
The explanation framework computed attributions for both predicted classes and alternative diagnostic possibilities, providing comprehensive insight into model decision-making processes. For each input image, the system generated XRAI attributions targeting the predicted class while also computing explanations for other clinically relevant classes, enabling a comparative analysis of model focus patterns across different diagnostic hypotheses. This was achieved by iterating through all the output logits individually for each of the target classes using class-specific backpropagation to obtain gradients. Calculations of the gradients were batched together to make them execute quickly, and the attributions were calculated using Integrated Gradients, which were refined using the region-growing technique of XRAI. Computed attribution tensors were stored in dictionaries grouped by image ID, as well as class label, allowing for the retrieval of the attribution for easy interactive visualization within the user interface.
2.3.3. Explanation Visualization and Interpretation
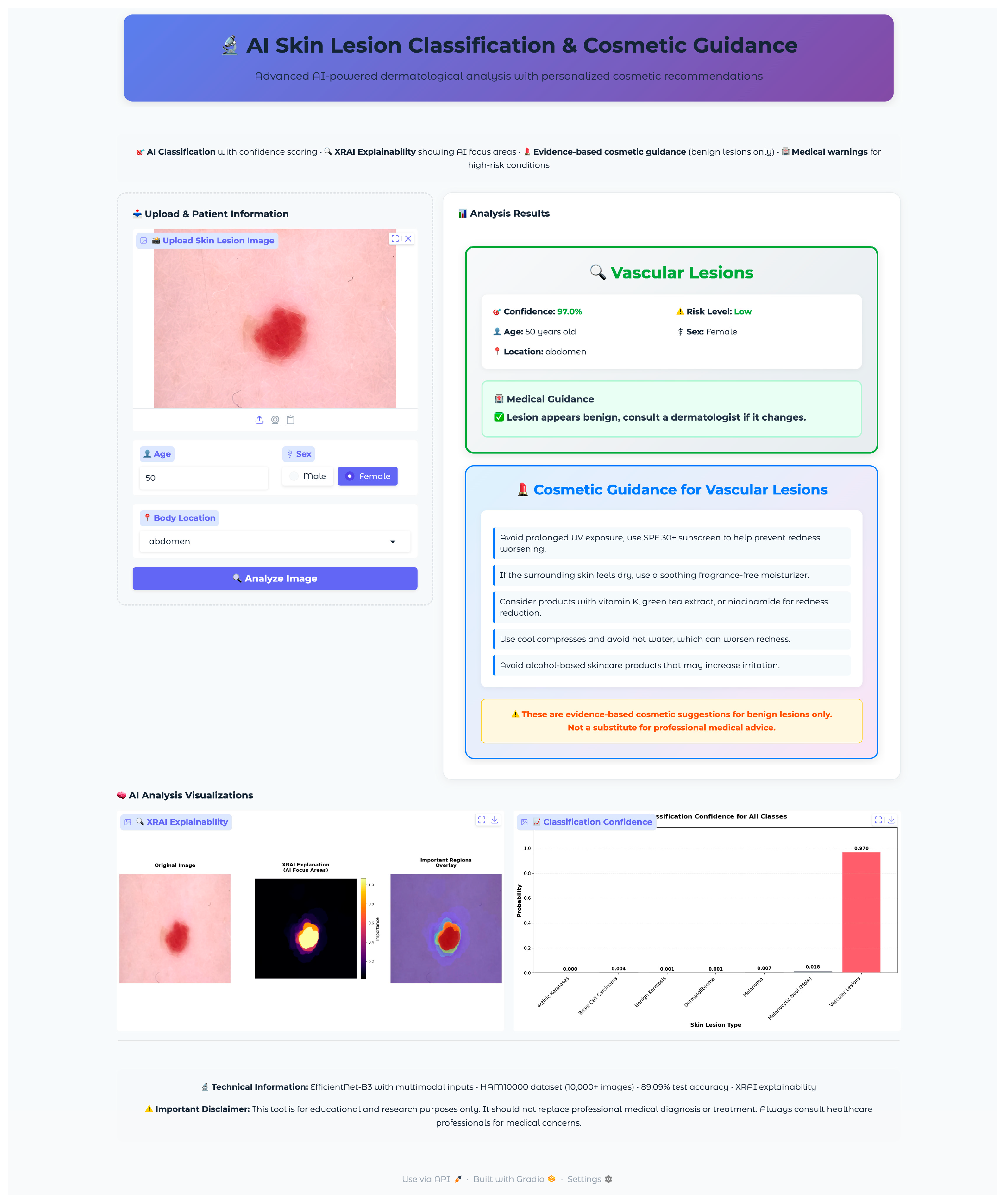
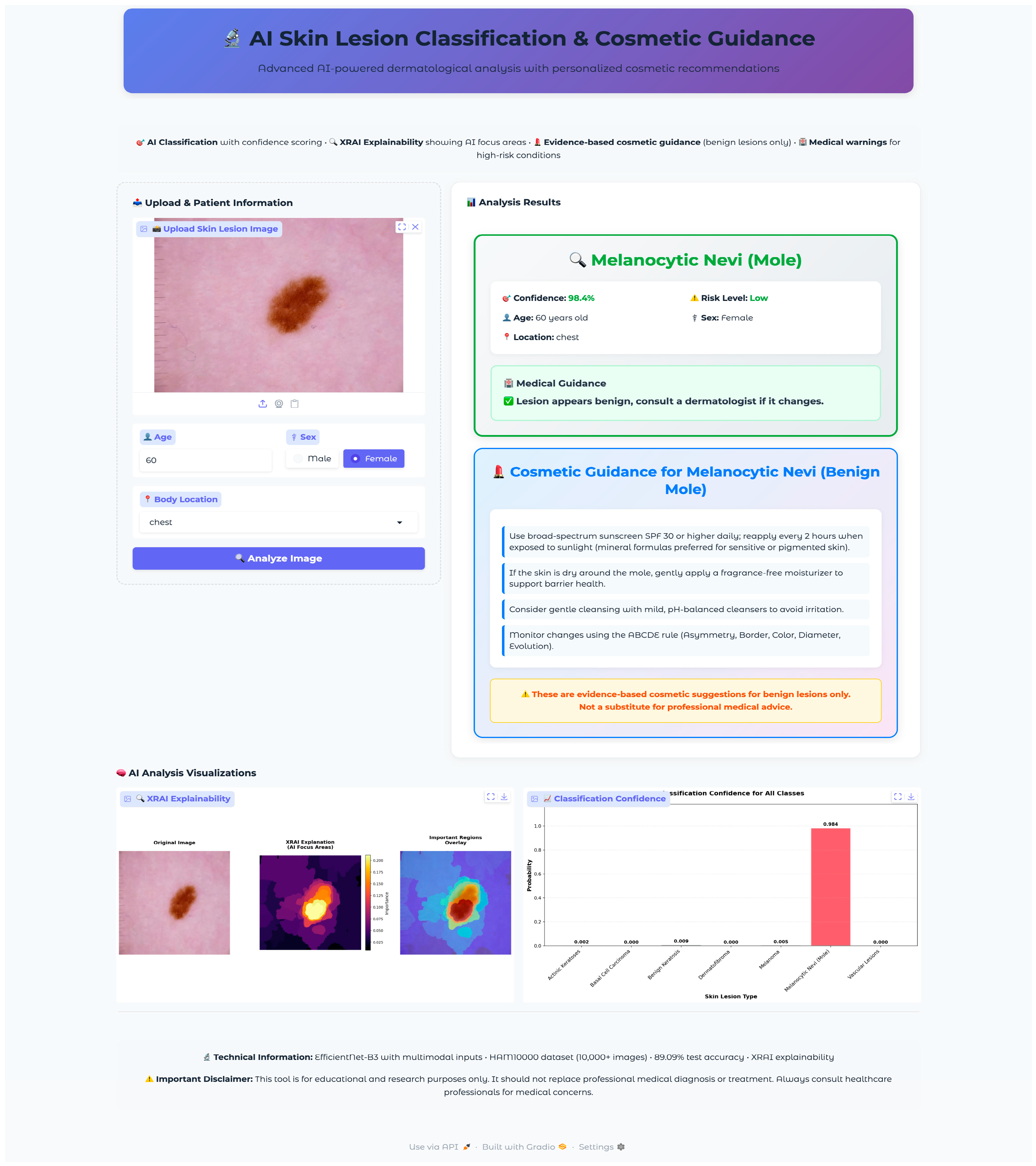
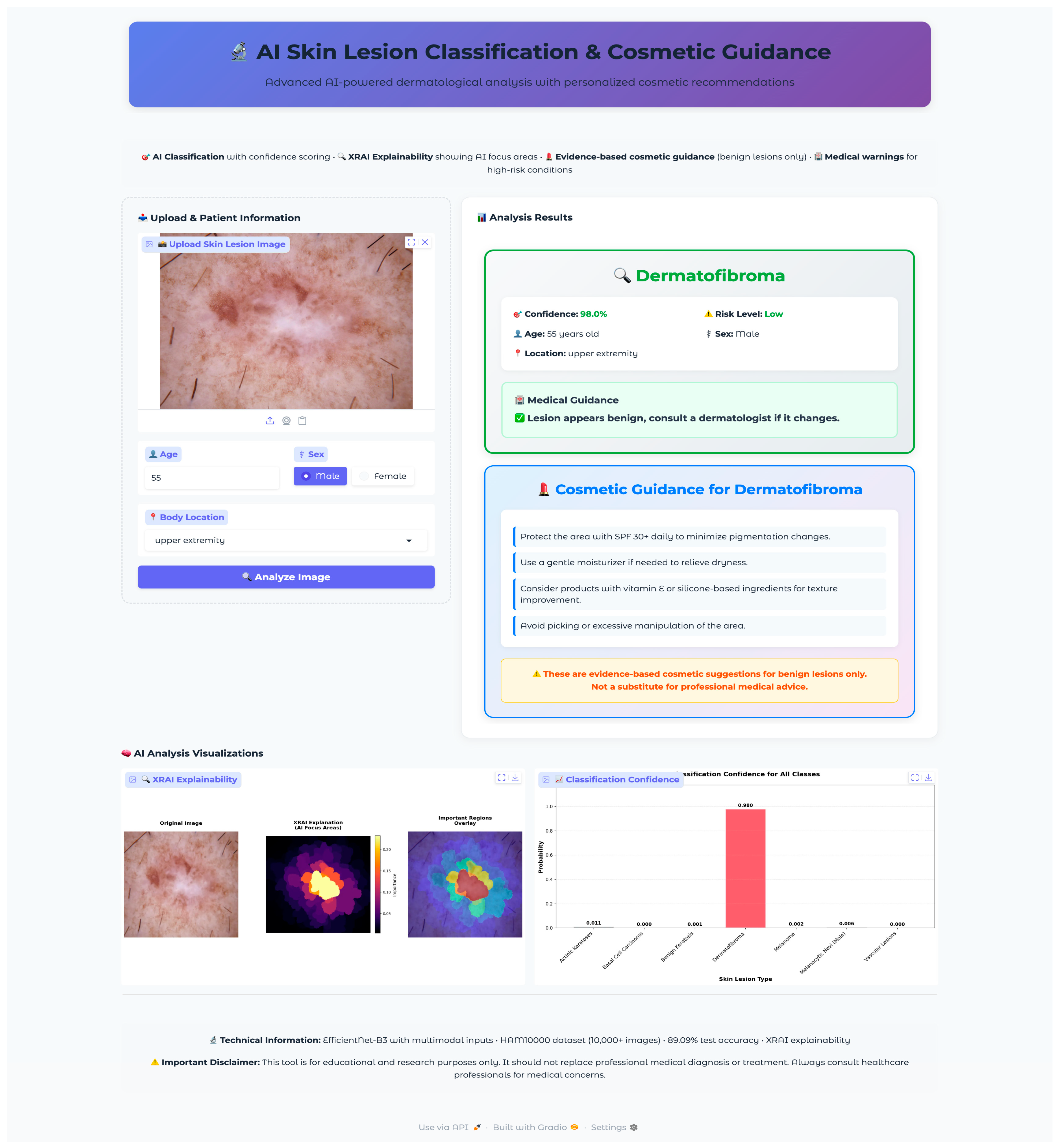
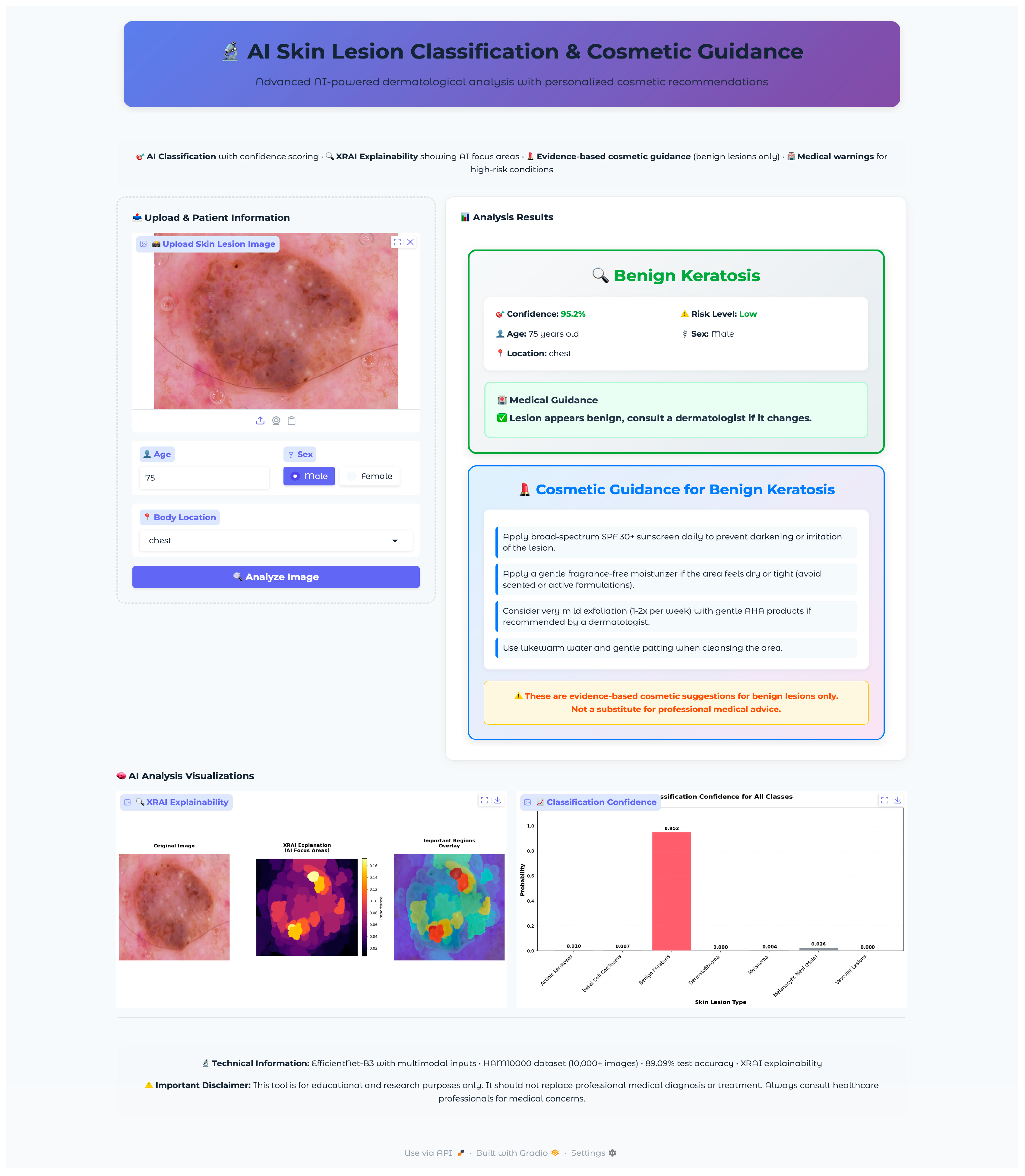
The visualization framework generated comprehensive explanation displays, incorporating multiple complementary views to enhance clinical interpretability. The system produced standardized outputs, including original images, XRAI heatmaps, importance-filtered regions, and overlay visualizations, to support different aspects of clinical reasoning. The explanation system computed and displayed regions at multiple importance thresholds, typically showing the top ten percent, twenty percent, and thirty percent most important areas. This hierarchical approach enables clinicians to understand both primary diagnostic features and secondary supporting evidence used by the model. The visualization employed the inferno colormap for heatmap generation, providing intuitive color coding, where brighter regions indicate higher diagnostic importance. Each explanation included a comprehensive statistical analysis of attribution patterns, computing maximum importance scores, mean attribution values, and percentile thresholds for quantitative assessment. The system calculated focus ratios to distinguish between highly localized attention patterns and more distributed decision-making strategies, providing insight into model confidence and reasoning patterns for different lesion types.
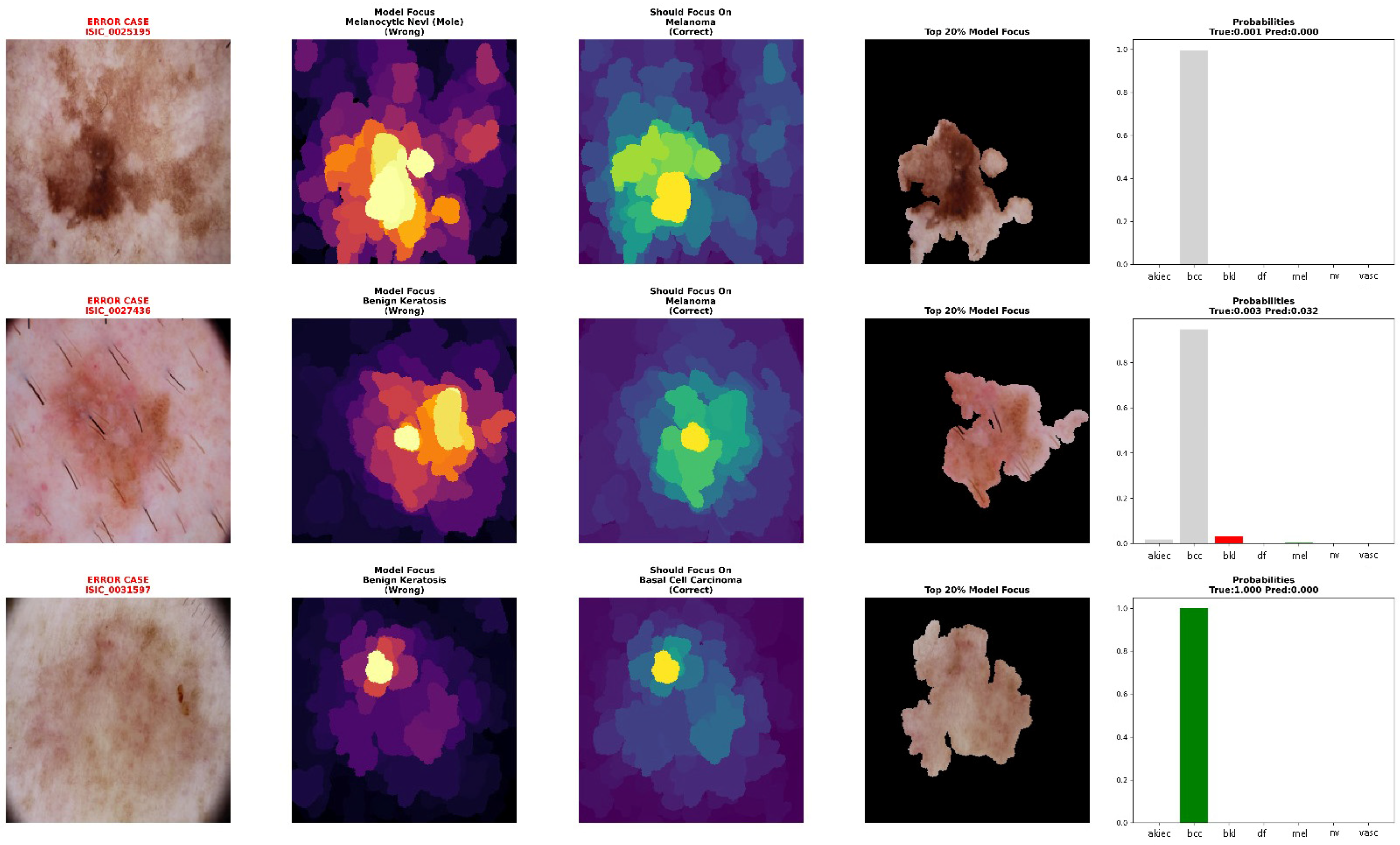
2.3.4. Error Analysis and Validation Framework
The explainability implementation incorporated error analysis capabilities to understand model limitations and validate explanation quality. The system specifically analyzed misclassified cases to identify patterns in model failures and compare attribution patterns between correct and incorrect predictions.
The framework prioritized analysis of clinically dangerous errors, particularly cases where malignant lesions (melanoma, basal cell carcinoma) were misclassified as benign conditions. For each error case, the system generated dual explanations showing both the model’s actual focus (leading to incorrect prediction) and the regions that should have received attention for correct classification. This comparative analysis revealed systematic patterns in model failures and validated the reliability of XRAI explanations.
The error analysis computed correlation coefficients between attribution patterns for predicted versus true classes, quantifying the similarity or divergence in model focus. Correlation values below 0.3 indicated completely different focus patterns that explained prediction errors, while values above 0.6 suggested close diagnostic calls where model and ground truth reasoning showed substantial overlap.
2.4. Web Application Architecture
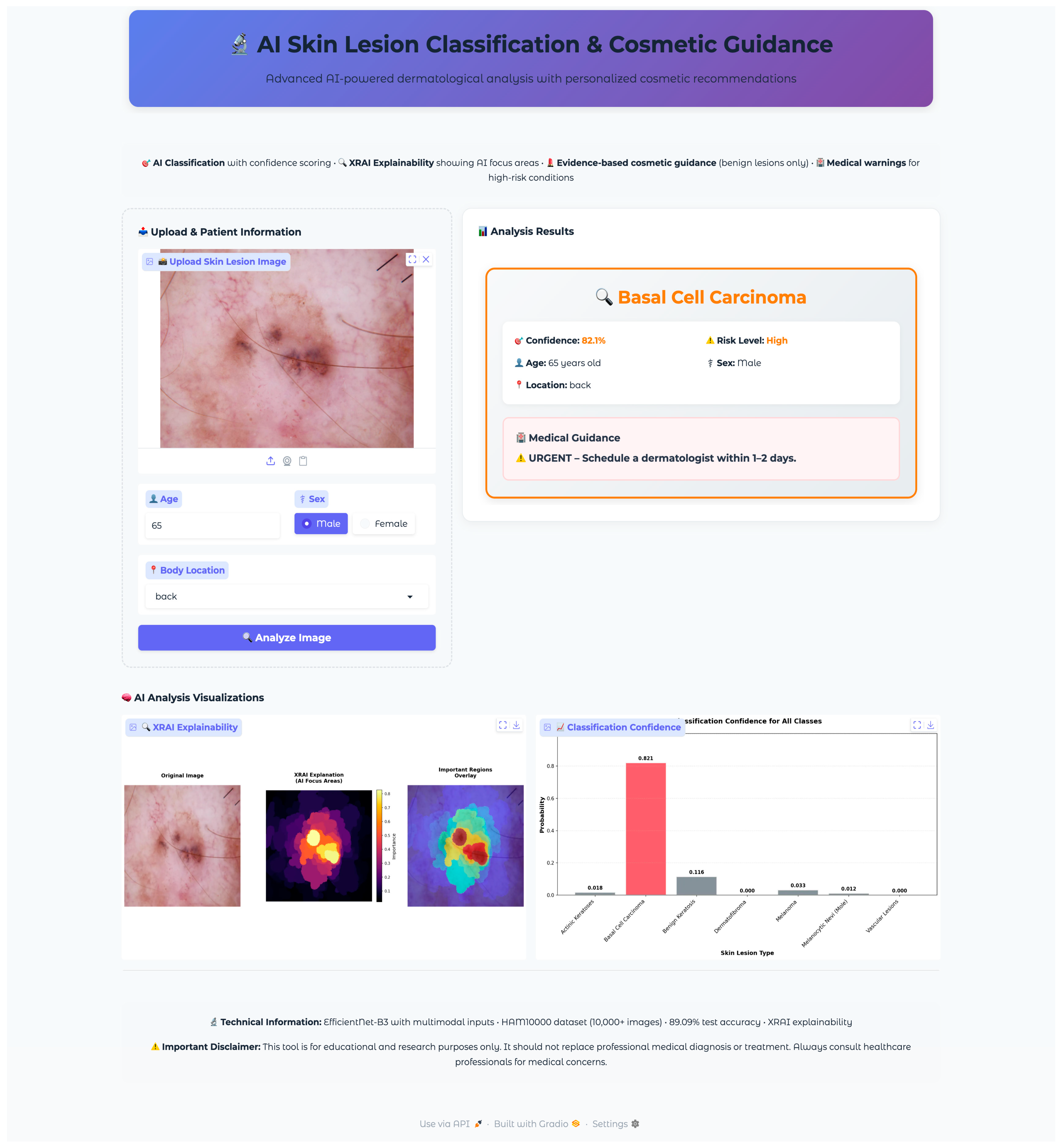
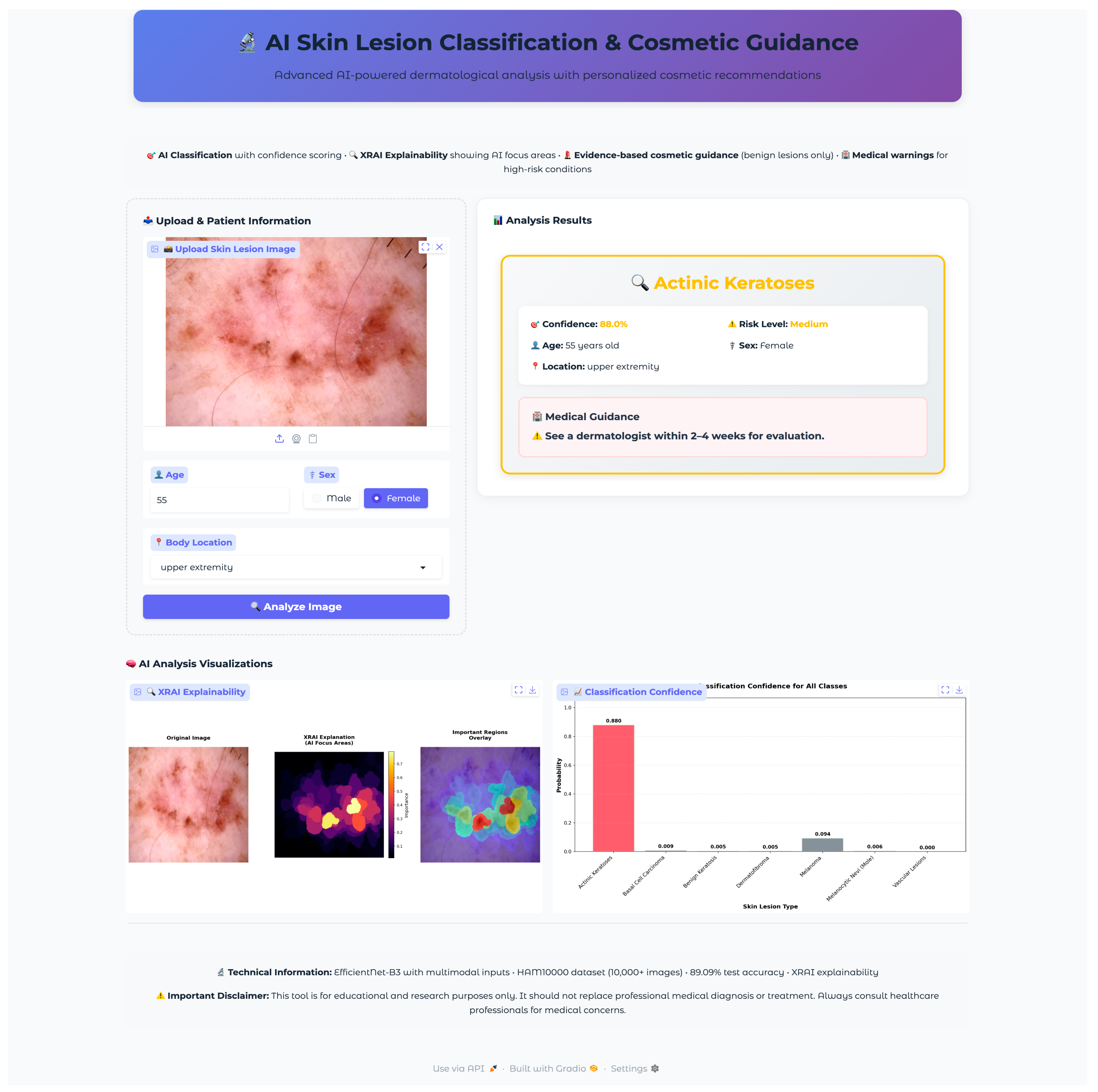
The web application was developed using the Gradio framework to provide an intuitive, clinically oriented interface for real-time skin lesion analysis. The interface design prioritized accessibility, clinical workflow integration, and clear presentation of diagnostic information while maintaining professional medical standards. The application architecture employed responsive design principles with a maximum container width of 1400 pixels to ensure optimal viewing across different device types and screen resolutions.
The application employed a two-column grid layout, with the left column dedicated to input collection (scale = 2) and the right column for displaying the results (scale = 3). This asymmetric layout prioritized result visibility while maintaining efficient space utilization for input controls. The upload section featured a distinctive dashed border and light gray background (#f8f9fa) to clearly delineate the input area, while the results section employed white backgrounds with subtle shadows for professional presentation.
2.4.1. Real-Time Inference Pipeline
The application implemented a real-time inference pipeline capable of processing dermatoscopic images with integrated XRAI explainability generation. The system was designed to handle variable input formats while maintaining consistent preprocessing standards and delivering results within acceptable latency constraints for clinical use.
The preprocessing pipeline maintained strict adherence to training data standards, applying identical normalization parameters derived from HAM10000 dataset statistics (mean = [0.763, 0.544, 0.568], std = [0.141, 0.152, 0.169]). The input images underwent automatic format detection and conversion, supporting both numpy arrays and PIL Image objects. The system implemented standardized resizing to 224 × 224 pixels using high-quality interpolation algorithms to ensure optimal model performance while preserving diagnostic features.
The application integrated patient demographic information with image analysis through a comprehensive metadata encoding system. Age values underwent normalization to a 0–1 scale (age/100), while categorical variables, including sex and anatomical location, received appropriate encoding. The location mapping system supported twelve anatomical regions (abdomen, back, chest, face, foot, hand, lower extremity, neck, scalp, trunk, upper extremity) with one-hot encoding implementation to maintain consistency with the training data structure. This structured approach ensured that both numerical and categorical patient data were combined with image-derived features for downstream model training and inference.
2.4.2. Model Integration and Deployment
The web application integrated the best-performing EfficientNet-B3 model with comprehensive error handling and degradation capabilities. The system implemented automatic model loading with state dictionary restoration from trained weights, incorporating proper device management for both CPU and GPU deployment scenarios.
The application employed robust model initialization procedures with exception handling to ensure reliable deployment across different computational environments. The system automatically detected available hardware (CUDA-enabled GPU or CPU fallback) and configured device placement accordingly. Asynchronous inference requests were also supported through a task queue mechanism using background workers, preventing UI blocking and ensuring scalability for multiple simultaneous users.
For XRAI explainability integration, the application implemented PyTorch hook registration on the final convolutional layer of the EfficientNet-B3 backbone. Forward hooks captured feature maps while backward hooks collected gradient information, storing outputs in globally accessible dictionaries for attribution computation. The hook system employed proper tensor dimension manipulation to ensure compatibility between PyTorch’s channel-first format and XRAI’s expected channel-last configuration. The XRAI attributions were computed by combining integrated gradients with region-based segmentation masks generated via a hierarchical superpixel algorithm, enabling pixel-level contribution mapping. The resulting attribution maps were normalized to a 0–1 range, converted to NumPy arrays, and overlaid onto the original input images using OpenCV-based alpha blending for seamless visualization in the web interface.
2.4.3. Safety Protocols and Risk Stratification
The application incorporated safety protocols to ensure responsible deployment in clinical contexts while maintaining clear boundaries regarding medical advice and diagnostic authority. The system implemented risk stratification with appropriate urgency messaging and safety disclaimers. The classification system employed a four-tier risk stratification framework: very high (melanoma), high (basal cell carcinoma), medium (actinic keratoses), and low (benign conditions). Each risk level triggered specific color coding (red for very high, orange for high, yellow for medium, green for low) and corresponding urgency messaging. Critical cases (melanoma, basal cell carcinoma) generated emergency alerts recommending immediate dermatological consultation, while lower-risk cases provided appropriate guidance for routine monitoring or evaluation. The application consistently emphasized its educational and research purpose through disclaimers positioned both within individual results and in the global footer information. The system explicitly stated that outputs should not replace professional medical diagnosis or treatment, directing users to consult healthcare professionals for medical concerns. This approach maintained ethical responsibility while providing valuable educational insights.
2.4.4. Evidence-Based Cosmetic Guidance System
The cosmetic recommendation system included evidence-based guidance for four benign condition types: melanocytic nevi, benign keratosis, dermatofibroma, and vascular lesions. Each recommendation set underwent validation to ensure safety and appropriateness. The system excluded cosmetic guidance for potentially malignant conditions (melanoma, basal cell carcinoma, actinic keratoses) to prevent inappropriate self-treatment of serious conditions. Cosmetic guidance was presented through styled cards featuring gradient backgrounds with hierarchical organization. Recommendations were structured as evidence-based tips with specific product categories (broad-spectrum sunscreen SPF 30+, fragrance-free moisturizers, gentle cleansers) and application instructions.
2.4.5. Explainability Visualization Framework
The application integrated a comprehensive XRAI explainability visualization to provide clinically meaningful insights into model decision-making processes. The visualization framework generated multiple complementary views to support different aspects of clinical interpretation and model transparency. The XRAI explanation system generated three-panel visualizations, including an original image display, an attribution heatmap using the inferno colormap, and an overlay visualization combining the original image with a semi-transparent attribution overlay. This multi-panel approach enables clinicians to understand both the raw model focus and its relationship to anatomical structures within a lesion. The application generated comprehensive probability visualizations showing confidence scores across all seven diagnostic categories using horizontal bar charts. The predicted class received distinctive red highlighting, while alternative classes were displayed in neutral gray. Probability measures were rounded to three decimals for the simplicity of quantitative measures of model uncertainty as well as confidence. Grid lines, along with the value labels, were provided for the visualization for added readability as well as clinical utility.
2.4.6. Web Application Validation Protocol
A comprehensive validation protocol was established to evaluate the successful translation of the research model into a functional clinical interface. The validation methodology employed systematic stratified sampling to select representative test cases from the held-out test set, ensuring unbiased evaluation across all diagnostic categories while accounting for class imbalance inherent in the HAM10000 dataset.
Seven test images were selected, one from each diagnostic class, using a random selection algorithm that validated file accessibility and extracted corresponding metadata, including patient demographics and lesion characteristics. The selected validation cases encompassed the full spectrum of diagnostic categories with varying patient demographics and anatomical locations: vascular lesions (50-year-old female, abdomen), melanocytic nevi (60-year-old female, chest), melanoma (70-year-old male, back), dermatofibroma (55-year-old male, upper extremity), benign keratosis (75-year-old male, chest), basal cell carcinoma (65-year-old male, back), and actinic keratoses (55-year-old female, upper extremity). This diverse selection provided comprehensive coverage of age ranges, anatomical locations, and lesion morphologies representative of clinical practice.
The validation protocol evaluated three key performance areas: (1) diagnostic accuracy and confidence scoring across all lesion types, (2) safety protocol implementation, including risk stratification and emergency response systems, and (3) technical performance metrics, including inference time, probability visualization accuracy, and XRAI explainability generation consistency. Each test case underwent complete workflow validation from image upload through final diagnostic output and safety protocol activation.
4. Discussion
The comprehensive evaluation of deep learning architectures with XRAI explainability integration demonstrates significant advances in automated dermatological diagnosis, establishing a foundation for clinically deployable AI systems that bridge the gap between research innovation and practical healthcare applications. This study’s approach, combining architectural comparison, explainable AI implementation, and clinical deployment validation, addresses critical gaps in existing dermatological AI research. Similar calls for clinically relevant deployment have been emphasized by Esteva et al. [
2], Wu et al. [
3], and Aksoy et al. [
21], but few studies have combined high performance with interpretability and integration into web-based applications. The system was implemented using a modular PyTorch-based pipeline combined with Gradio, enabling integration into web applications and supporting both CPU and GPU inference through dynamic device allocation.
4.1. Model Performance and Comparative Analysis
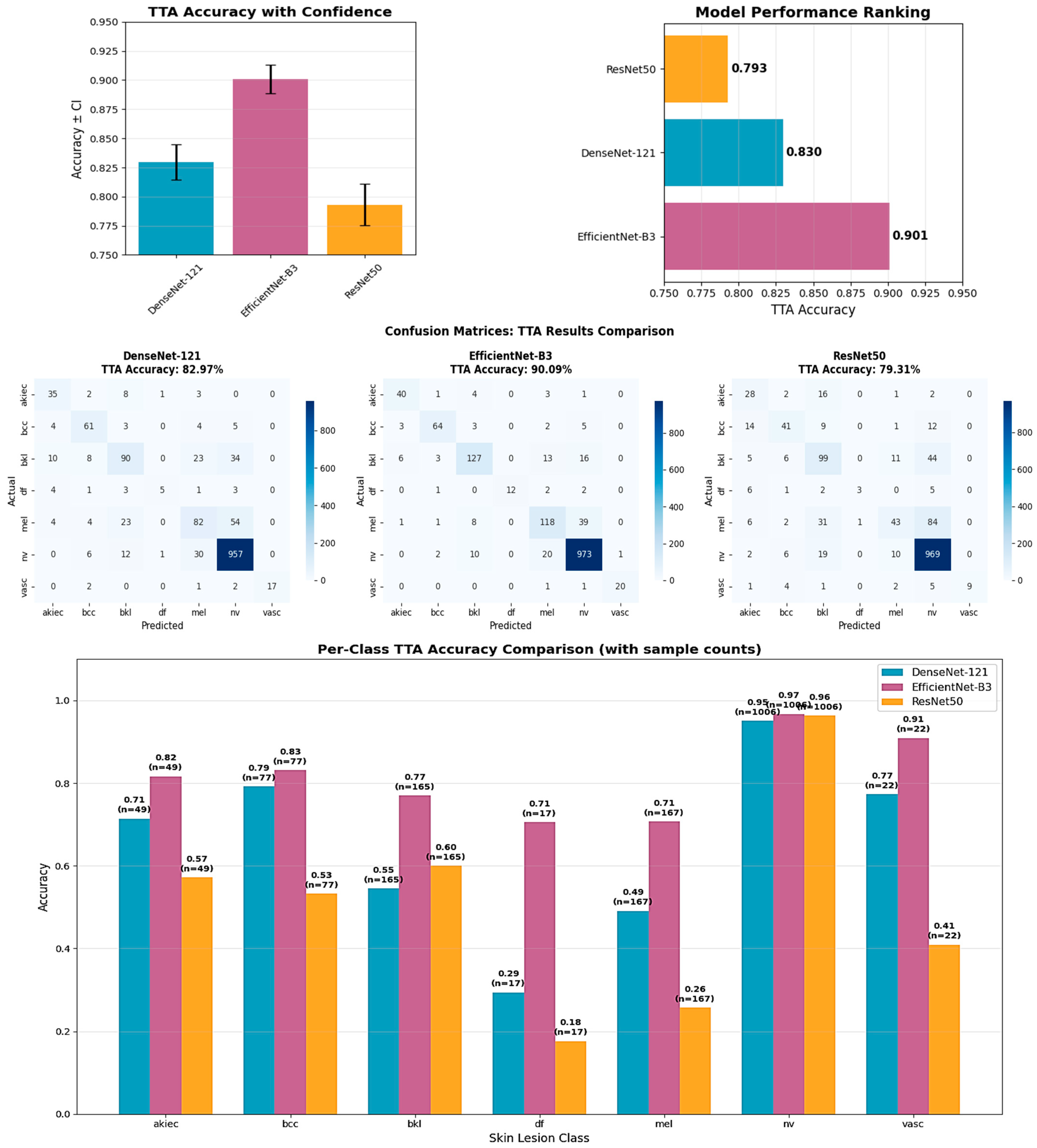
The systematic comparison of three CNN architectures revealed fundamental insights into the effectiveness of different deep learning paradigms for medical image analysis. EfficientNet-B3’s superior performance (89.09% test accuracy, 90.08% validation accuracy) demonstrates the clinical relevance of compound scaling principles that systematically balance network depth, width, and resolution. This aligns with earlier findings by Wu et al. [
3], who demonstrated that CNNs are robust baselines across architectures, and with Yao [
29], who highlighted EfficientNet’s promise for lesion classification.
The substantial performance gap between EfficientNet-B3 and ResNet-50 (89.09% vs. 78.78%), despite ResNet-50’s larger parameter count (24.7 M vs. 11.6 M), highlights the importance of architectural innovation over mere model size in medical applications. Comparable insights were reported by Arshad et al. [
10] through fusion strategies and Hussein et al. [
11] through hybrid quantum deep learning, both showing that design choices can outweigh brute parameter count.
Benchmarking against recent state-of-the-art CNN approaches reveals important performance considerations within contemporary research trends. Hussain and Toscano [
31] achieved exceptional performance using multiple CNN architectures with tailored data augmentation on HAM10000, with EfficientNetV2-B3 reaching over 98% accuracy through extensive preprocessing and class-specific augmentation strategies. While their accuracy substantially exceeds our 89.09%, their approach prioritized pure classification performance without incorporating explainability frameworks or clinical deployment considerations. Similarly, Roy et al. [
32] demonstrated competitive results with 91.17% F1-score and 90.75% accuracy using wavelet-guided attention mechanisms and gradient-based feature fusion, representing sophisticated feature engineering approaches that complement our architectural comparison findings.
Recent Vision Transformer implementations have shown remarkable promise for skin lesion analysis. Agarwal and Mahto [
33] further advanced hybrid approaches, achieving 92.81% accuracy on HAM10000 through sequential and parallel CNN–Transformer models with Convolutional Kolmogorov–Arnold Network (CKAN) fusion, showcasing the potential of architectural hybridization. Zoravar et al. [
34] explored domain adaptation challenges through Conformal Ensemble of Vision Transformers (CE-ViTs), achieving 90.38% coverage rates across multiple datasets and highlighting the importance of uncertainty quantification in clinical applications.
When positioned within existing HAM10000 research, these results reveal important methodological trade-offs. Ahmad et al.’s framework achieved 99.3% accuracy using the Butterfly Optimization Algorithm [
4], while Liu et al.’s SkinNet ensemble reached 86.7% through stacking techniques [
7]. Krishna et al. [
12] likewise pushed accuracy with Transformers and GAN-based imbalance correction. The current study’s 89.09% accuracy reflects deliberate prioritization of explainability integration and clinical deployment readiness over pure accuracy maximization. While recent state-of-the-art approaches [
31,
32,
33,
34] demonstrate higher classification accuracies, they predominantly focus on algorithmic optimization without addressing the critical gap between research performance and clinical utility. Unlike previous studies that concluded with performance metrics [
31,
32,
33,
34], this research provides a fully functional clinical prototype with comprehensive safety protocols, XRAI explainability, and evidence-based patient guidance systems.
The comparative analysis reveals distinct research philosophies within contemporary skin lesion classification. Pure accuracy-driven approaches [
31,
33] excel in controlled evaluation scenarios but lack the transparency and deployment infrastructure essential for clinical acceptance. Explainability-focused methods [
32] demonstrate sophisticated feature analysis but remain research-grade implementations without clinical interfaces. Our integrated approach bridges this gap by accepting moderate accuracy trade-offs (89.09% vs. 96–98% in pure accuracy studies) in exchange for clinical explainability, real-time deployment capability, and comprehensive patient safety protocols that are absent in higher-performing but research-only implementations.
Per-class analysis reveals critical insights for clinical deployment. EfficientNet-B3’s good performance for common conditions (95.6% for melanocytic nevi, 83.1% for basal cell carcinoma) establishes strong reliability for typical dermatological presentations. However, reduced performance for dermatofibroma (58.8% accuracy) reflects the inherent challenges of extremely rare conditions (17 test cases). Similar imbalance-related challenges were emphasized by Krishna et al. [
12], Tang and Lasser [
14], and recent studies [
31,
32,
33,
34], highlighting the importance of tailored approaches to minority-class detection in future research.
The comprehensive statistical validation provides robust evidence supporting EfficientNet-B3’s superior performance through multiple complementary analyses. All pairwise comparisons achieved statistical significance (p < 0.05) with large effect sizes (Cohen’s d > 0.8), indicating that observed differences represent meaningful practical improvements rather than statistical noise. The 95% confidence intervals that exclude zero further confirm the reliability of these performance advantages. These statistical findings validate the architectural comparison methodology and support the selection of EfficientNet-B3 for clinical deployment, addressing concerns about whether observed performance differences could be attributed to chance variation.
4.2. XRAI Explainability: Methodological Innovation
The implementation of XRAI explainability represents a significant methodological advancement over traditional attribution techniques. While previous studies employed Grad-CAM [
4,
8,
9] or LIME [
8] for visualization, these pixel-level methods often generate fragmented explanations that fail to align with clinical reasoning patterns. More advanced methods, including Patrício et al.’s concept-based explanations [
26], Ieracitano et al.’s TIxAI trustworthiness index [
27], and Metta et al.’s ABELE explainer [
28], attempted to move toward higher-level interpretability, but none have applied region-based XRAI analysis in dermatology. The XRAI approach generates coherent, spatially connected explanations corresponding to anatomically meaningful structures, addressing this gap.
The comprehensive XRAI analysis revealed clinically meaningful attention patterns that align with established dermatological diagnostic criteria. For melanoma detection, concentrated attention on irregular pigmentation patterns and asymmetric borders aligns with ABCDE criteria, while focused attention on central ulcerated areas for basal cell carcinoma reflects appropriate morphological feature recognition. This complements observations by Munjal et al. [
8] and Cino et al. [
9], who used visualizations but lacked region-level coherence. The moderately focused attention patterns across all classes (focus ratios 0.103–0.132) demonstrate appropriate balance between specific feature detection and contextual analysis required for comprehensive dermatological evaluation.
Critical error analysis provided essential insights into model failure modes, particularly for high-stakes misclassifications. The melanoma misclassified as a melanocytic nevi case showed high correlation (0.862) between model attention and ideal features, indicating “close call” scenarios reflecting inherent diagnostic challenges even for experienced clinicians. The basal cell carcinoma error with high correlation (0.968) despite 100% confidence demonstrates that sophisticated attention mechanisms can fail with atypical presentations, emphasizing the importance of maintaining clinical oversight protocols. Similar emphasis on the role of oversight was highlighted by Wu et al. [
3] and Thomas [
16], especially when integrating AI into clinical workflows.
These findings indicate that misclassifications often arise not only from the inherent ambiguity of lesion morphology but also from dataset imbalance, as the HAM10000 collection is dominated by benign nevi while rarer conditions remain underrepresented [
1]. This dual challenge underscores the need for clinical oversight and the development of balanced datasets to reduce systematic bias, consistent with observations by Tschandl et al. [
1] and Tran Van and Le [
17].
4.3. Multimodal Integration and Clinical Deployment Innovation
The successful integration of patient metadata with dermatoscopic images demonstrates significant advancement over purely image-based approaches that dominated previous HAM10000 research. This multimodal approach aligns with clinical reality, where patient characteristics significantly influence lesion presentation and diagnostic interpretation. The metadata processing pipeline enhanced classification accuracy while maintaining computational efficiency, reflecting the natural diagnostic process used by dermatologists.
The development of the first deployable web-based clinical interface with explainability and recommendations tailored to benign skin conditions represents a unique contribution that addresses the critical gap between research-grade AI models and clinical utility. Unlike previous studies focused on accuracy optimization, this research provides a fully functional clinical prototype with real-time inference, comprehensive safety protocols, and patient education components. The four-tier risk stratification system with appropriate urgency messaging and evidence-based cosmetic guidance for benign conditions demonstrates responsible medical AI deployment that prioritizes patient safety while maximizing diagnostic utility.
Validation across all diagnostic categories establishes robust performance suitable for controlled clinical trials, contrasting with previous studies that typically focused on algorithmic development without deployment validation. The integration of XRAI explainability within a real-time clinical interface enables healthcare professionals to validate AI reasoning processes during clinical decision-making, addressing transparency requirements essential for clinical acceptance.
4.4. Limitations and Future Research Directions
Several limitations require consideration for future research and clinical implementation. The persistent class imbalance challenges, particularly evident in dermatofibroma classification, highlight the need for specialized approaches to rare condition detection. Oversampling strategies, such as SMOTE or heavy synthetic augmentation, were not applied in this study, as such methods may introduce artificial dermoscopic patterns that fail to reflect true clinical presentations. While this approach preserved dataset authenticity and interpretability, it inevitably limited sensitivity in underrepresented categories, such as dermatofibroma and melanoma. Addressing this imbalance in future work will require integration of larger, multi-institutional datasets and collaborations with clinical partners to ensure more representative coverage of rare but clinically important lesions. In addition, future research should explore advanced data augmentation techniques, synthetic data generation, or federated learning approaches to address minority class limitations while maintaining diagnostic accuracy for common conditions.
Additionally, we acknowledge that the HAM10000 dataset contains multiple images per lesion, and while this study employed stratified random splitting to preserve class balance, this approach may allow images from the same lesion to appear across training, validation, and test sets. Although such stratified strategies have been commonly used in prior HAM10000 research to ensure minority-class representation, they may introduce potential data leakage. Future studies should therefore employ grouped splitting by lesion ID and extend validation to external datasets to further ensure robust generalization.
Moreover, this study emphasized clinical applicability and system deployment rather than exhaustive statistical validation. Accordingly, formal significance testing procedures (e.g., paired t-tests, confidence intervals, bootstrap resampling) were not applied in the present analysis. Future research should incorporate repeated-seed training and formal statistical validation across a broader set of models and multimodal variants to rigorously assess whether observed differences are statistically significant, thereby strengthening the robustness and reproducibility of comparative findings. Epoch selection in this study was based on validation-driven early stopping, as both training and validation curves plateaued at approximately 24 epochs. While statistical testing does not directly determine the optimal epoch count, future work will combine repeated-seed training with paired tests and bootstrap confidence intervals to evaluate whether extended training yields statistically reliable improvements. Furthermore, this study focused primarily on accuracy, F1-scores, and per-class accuracy as key evaluation metrics, given the application-oriented emphasis on clinical deployment. Operating points at fixed high-sensitivity thresholds, AUROC and PR-AUC scores, as well as per-class sensitivity and specificity with confidence intervals via bootstrapping, were not included in the present analysis. Future research should incorporate these metrics to provide an assessment of model performance, particularly for high-stakes categories, such as melanoma, basal cell carcinoma, and actinic keratoses, where sensitivity at clinically relevant thresholds is critical.
Another limitation lies in the model’s performance on cases with indistinct lesion boundaries and atypical morphologies. As shown in the error analysis, such cases often led to misclassification despite strong alignment between model attention and clinically relevant regions. This issue is exacerbated by the significant class imbalance in the HAM10000 dataset, where benign lesions vastly outnumber malignant and rare categories, limiting the model’s exposure to difficult cases. Future research should therefore explore uncertainty quantification, ensemble predictions, and longitudinal imaging analysis, alongside balanced and diverse datasets, to provide more cautious and context-aware outputs in borderline scenarios. While this study evaluated performance on HAM10000, future work should explicitly incorporate independent test sets drawn from alternative sources, such as other ISIC challenge datasets or multi-institutional cohorts collected under different imaging conditions. Such external validation is critical for demonstrating true generalizability and for ensuring robustness across sites, devices, and populations.
An important limitation of this study is the restricted demographic diversity of the HAM10000 dataset. While the dataset is comprehensive in terms of lesion types, it primarily represents lighter Fitzpatrick skin phototypes and Central European populations. This underrepresentation of darker skin tones and broader ethnic groups may limit the generalizability of the model across diverse clinical settings. Future work should therefore focus on validating performance across multi-ethnic and international cohorts to ensure equitable diagnostic accuracy. Approaches such as federated learning and cross-dataset benchmarking may provide promising strategies to mitigate demographic bias and improve global clinical utility. While this study integrated patient metadata (age, sex, and anatomical site) alongside dermatoscopic imagery, we did not explicitly measure the incremental contribution of metadata compared to image-only models or analyze subgroup performance stratified by demographic factors such as sex, age, or lesion location. Similarly, robustness to missing or erroneous metadata was not systematically evaluated, as missing values were imputed or zero-encoded in the present work. Future research should therefore investigate metadata robustness more formally, including ablation studies (image-only vs. image + metadata), subgroup performance analysis, and sensitivity testing to incomplete or noisy metadata inputs.
A further limitation is the absence of invasive squamous cell carcinoma (SCC) in the HAM10000 dataset. Although the dataset includes actinic keratoses/intraepithelial carcinoma (akiec), which represent early-stage precursors to SCC, invasive SCC lesions are not represented. This means that while the present framework already covers melanoma, basal cell carcinoma, actinic keratoses, and benign conditions, future studies should expand training and validation datasets to include SCC cases in order to provide complete coverage of all major skin cancer types. Computational requirements for XRAI explainability generation, while acceptable for clinical deployment, may benefit from optimization techniques to reduce inference latency in high-volume clinical environments.
4.5. Clinical Impact and Healthcare Translation
The successful development of a clinically deployable skin lesion classification system addresses critical gaps in dermatological care accessibility, particularly in regions with limited specialist availability. The combination of diagnostic accuracy with transparent decision-making processes through XRAI visualization provides essential foundations for clinical acceptance and integration into existing healthcare workflows.
The web-based deployment architecture offers significant advantages for underserved regions by enabling AI-assisted diagnosis with only basic internet connectivity, eliminating the need for expensive local computational infrastructure or specialized hardware. This approach democratizes access to advanced dermatological AI by allowing healthcare providers in resource-constrained environments to leverage sophisticated diagnostic capabilities through standard web browsers, dramatically reducing implementation barriers and deployment costs compared to traditional on-premise AI solutions.
The web application’s comprehensive approach to patient education, incorporating diagnostic assessment and evidence-based guidance for benign conditions, has the potential to enhance patient engagement and self-monitoring capabilities while maintaining appropriate clinical boundaries. This educational component could contribute to improved health literacy and earlier detection of concerning lesion changes in populations with limited access to specialized dermatological care, particularly benefiting remote and rural communities where internet access may be the only available connection to advanced medical technologies.
5. Conclusions
This study successfully developed and validated a clinically deployable deep learning system for automated skin lesion classification, demonstrating significant advances in both diagnostic accuracy and practical clinical utility. Through a systematic comparison of three CNN architectures, EfficientNet-B3 emerged as the optimal model, achieving 89.09% test accuracy with superior performance across most diagnostic categories, including critical malignant conditions such as melanoma (81.6% confidence) and basal cell carcinoma (82.1% confidence).
The implementation of XRAI explainability represents a crucial methodological innovation that addresses the transparency requirements essential for clinical acceptance. Unlike traditional pixel-level attribution methods, XRAI generated coherent, spatially connected explanations that align with established dermatological diagnostic criteria, providing clinicians with interpretable insights into model decision-making processes.
The successful integration of patient metadata with dermoscopic images enhanced classification accuracy while reflecting the natural diagnostic process used by clinicians who consider both visual lesion characteristics and patient demographics. This multimodal approach contributed to the system’s robust performance across diverse patient populations and anatomical locations.
The development of the first deployable web-based clinical interface with integrated explainability and evidence-based recommendations represents a significant contribution to the field. The application successfully demonstrated real-time inference capabilities with safety protocols, including a four-tier risk stratification system and appropriate emergency response protocols for malignant conditions.
This research addresses critical gaps in dermatological care accessibility, particularly in regions with limited specialist availability. The web-based deployment architecture democratizes access to advanced diagnostic capabilities by requiring only standard internet connectivity, eliminating expensive infrastructure requirements. The comprehensive patient education components, including evidence-based cosmetic guidance for benign conditions, enhance patient engagement while maintaining appropriate clinical boundaries.
Future research should focus on addressing persistent class imbalance challenges through advanced data augmentation techniques, validating performance across international datasets to ensure global generalizability, and optimizing computational requirements for high-volume clinical environments. The successful development of this clinically deployable system establishes a foundation for responsible AI integration in dermatological practice, bridging the critical gap between research innovation and practical healthcare applications while prioritizing patient safety and clinical transparency.