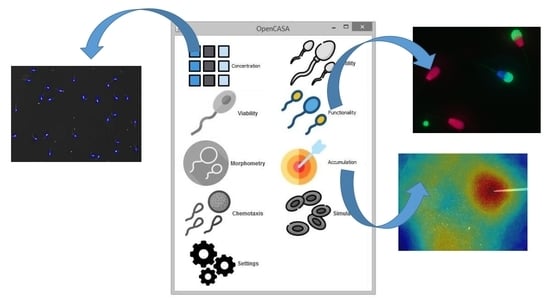
Expanding the Limits of Computer-Assisted Sperm Analysis through the Development of Open Software
Abstract
1. Introduction
2. Materials and Methods
2.1. Software Design
2.2. Experimental Design and Statistical Analyses for Validation of the Modules
2.2.1. Chemotactic Sperm Accumulation Module
2.2.2. Sperm Functionality Module
2.2.3. Sperm Concentration Module
3. Results
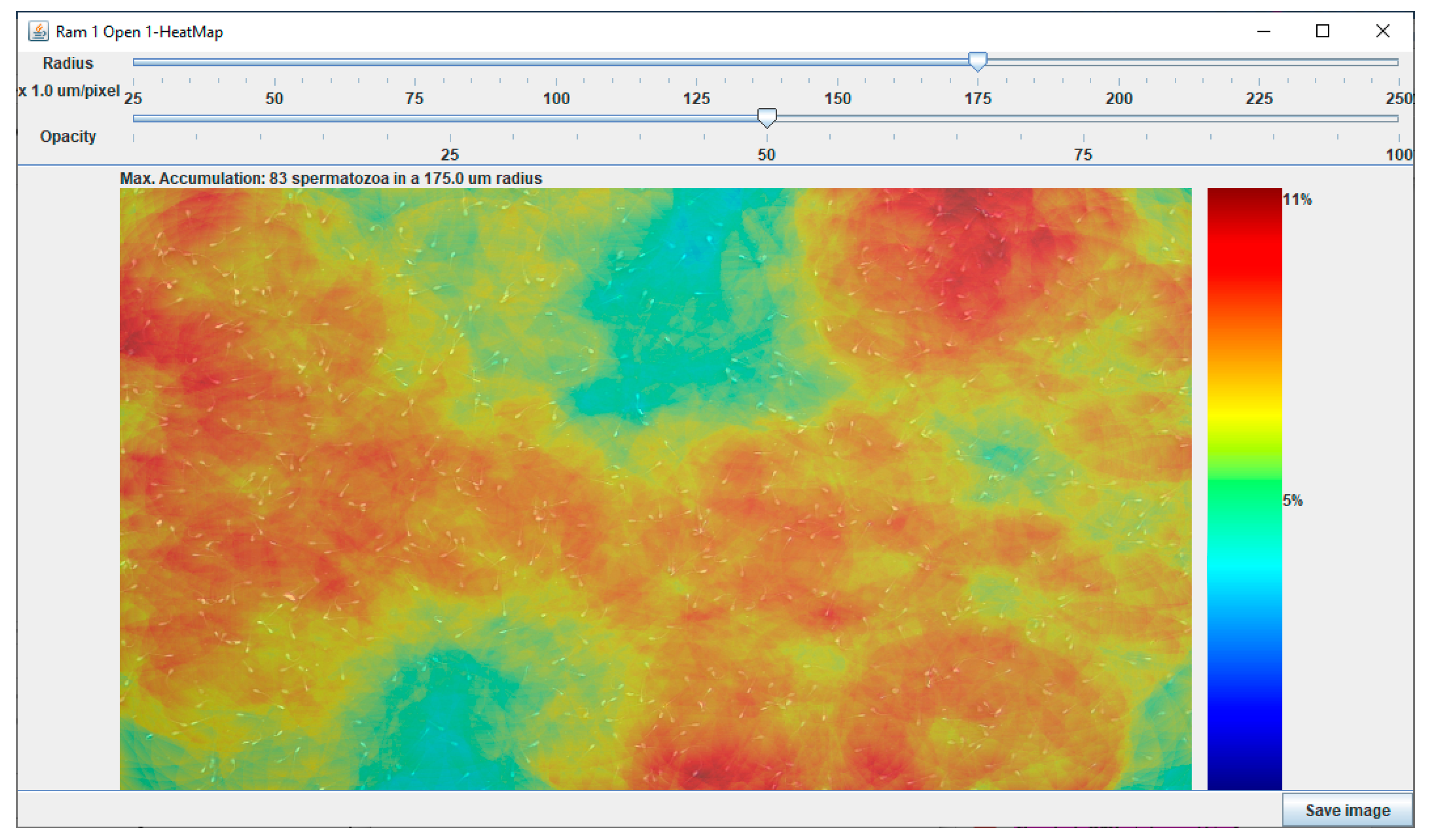
3.1. Development and Validation of the Chemotactic Sperm Accumulation Module
3.1.1. Development of the Chemotactic Sperm Accumulation Module
3.1.2. Validation of the Chemotactic Sperm Accumulation Module
3.2. Development and Validation of the Sperm Functionality Module
3.2.1. Development of the Sperm Functionality Module
3.2.2. Validation of the Sperm Functionality Module
3.3. Development and Validation of the Sperm Concentration Module
3.3.1. Development of the Sperm Concentration Module
3.3.2. Validation of the Sperm Concentration Module
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
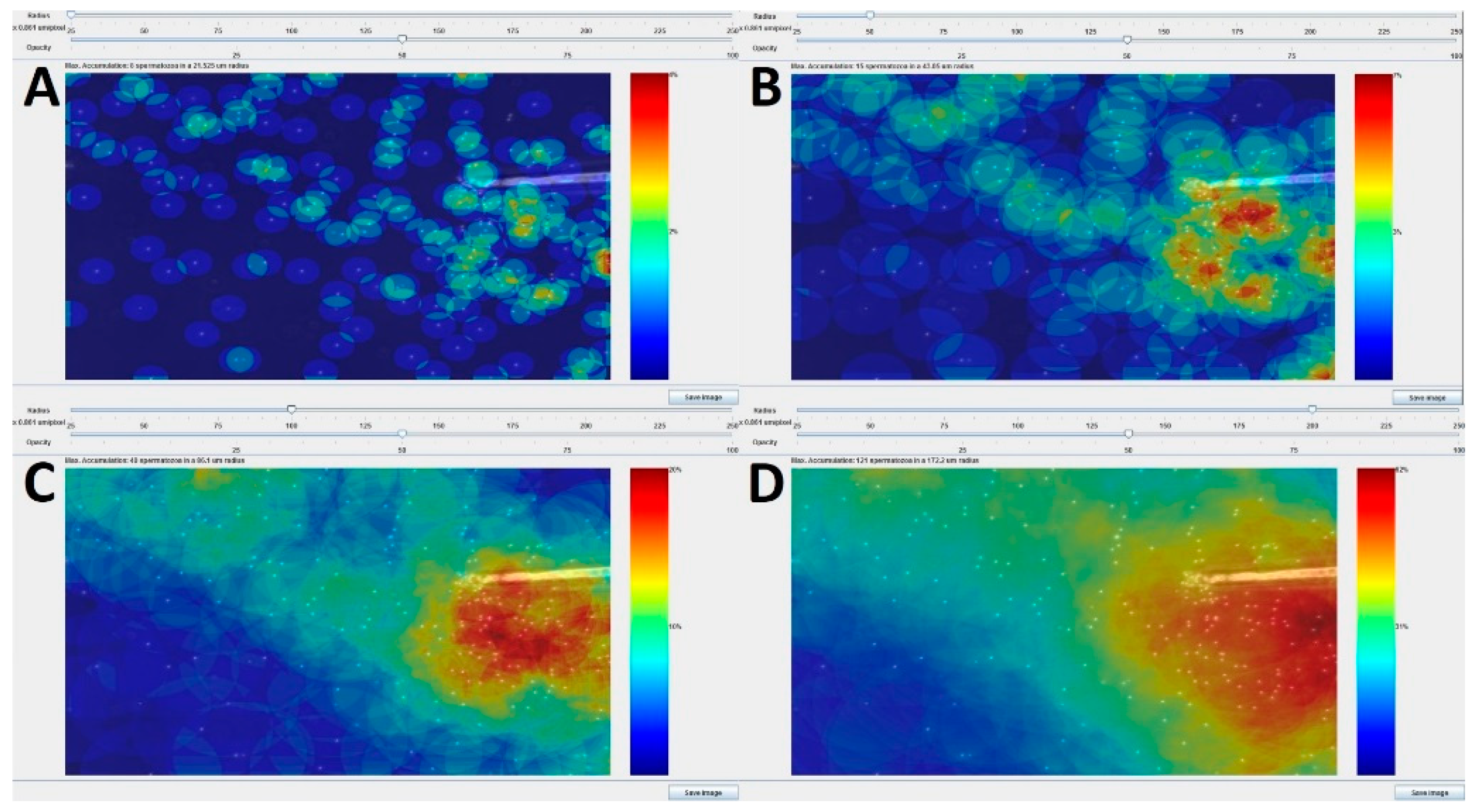
Appendix A.1. Study of the Influence of Parameter Settings on the Results of the Chemotactic Sperm Accumulation Module
Appendix A.2. Radius
Appendix A.3. Window Size
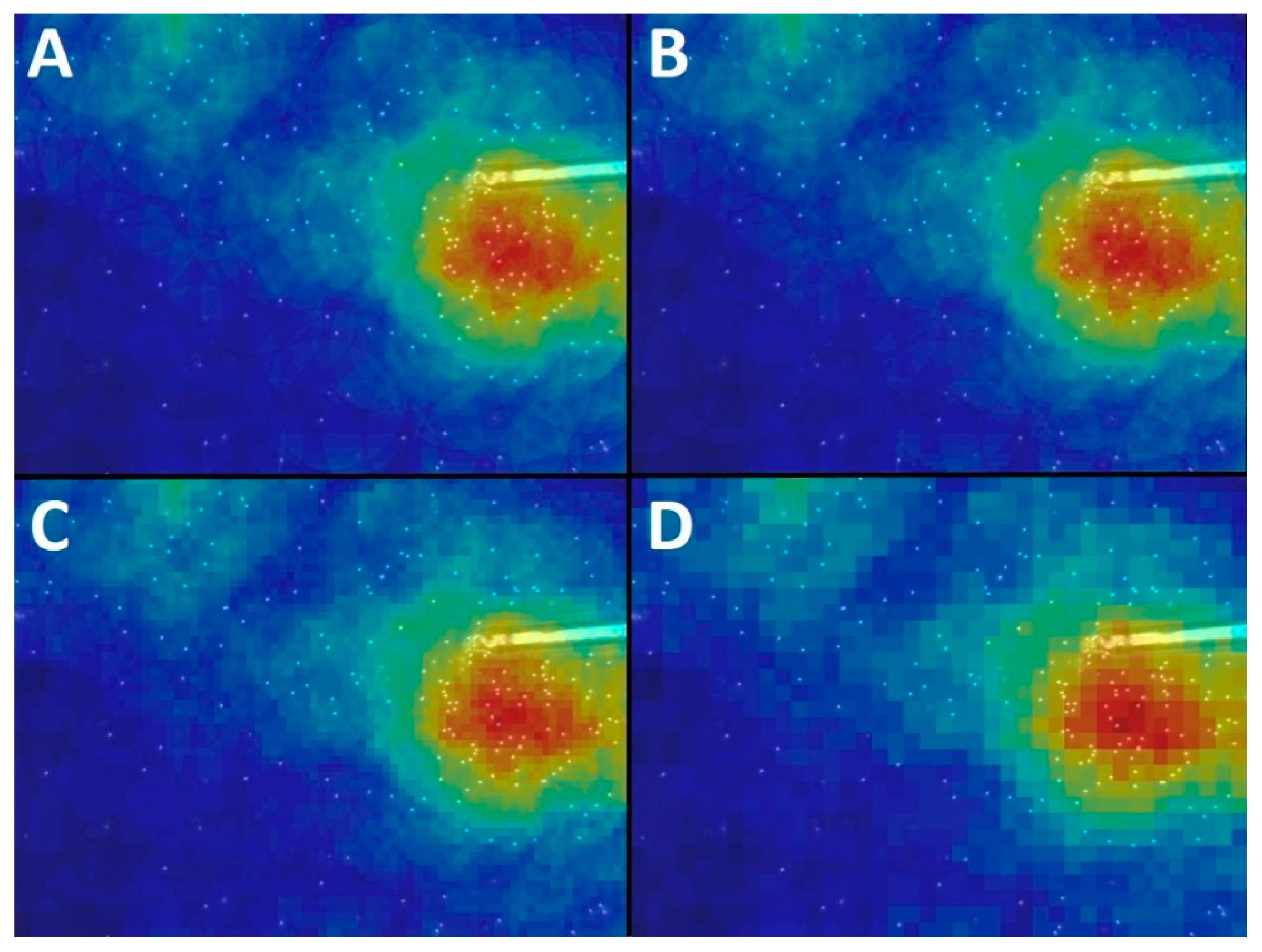
Appendix A.4. Sampling Factor
Appendix A.5. Maximal Accumulation
References
- Gallagher, M.T.; Smith, D.J.; Kirkman-Brown, J.C. CASA: Tracking the past and plotting the future. Reprod. Fertil. Dev. 2018, 30, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Baker, H.W.; Clarke, G.N. Sperm morphology: Consistency of assessment of the same sperm by different observers. Clin. Reprod. Fertil. 1987, 5, 37–43. [Google Scholar] [PubMed]
- Yaniz, J.L.; Silvestre, M.A.; Santolaria, P.; Soler, C. CASA-Mot in mammals: An update. Reprod. Fertil. Dev. 2018, 30, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.P.; Hammerstedt, R.H. Validation of a System for Computerized Measurements of Spermatozoal Velocity and Percentage of Motile Sperm. Biol. Reprod. 1980, 23, 647–656. [Google Scholar] [CrossRef]
- Katz, D.F.; Overstreet, J.W. Sperm Motility Assessment by Videomicrography. Fertil. Steril. 1981, 35, 188–193. [Google Scholar] [CrossRef]
- Katz, D.F.; Davis, R.O.; Delandmeter, B.A.; Overstreet, J.W. Real-Time Analysis of Sperm Motion Using Automatic Video Image Digitization. Comput. Methods Programs Biomed. 1985, 21, 173–182. [Google Scholar] [CrossRef]
- Gallagher, M.T.; Cupples, G.; Ooi, E.H.; Kirkman-Brown, J.C.; Smith, D.J. Rapid sperm capture: High-throughput flagellar waveform analysis. Hum. Reprod. 2019, 34, 1173–1185. [Google Scholar] [CrossRef]
- Hansen, J.N.; Rassmann, S.; Jikeli, J.F.; Wachten, D. SpermQ(-)A Simple Analysis Software to Comprehensively Study Flagellar Beating and Sperm Steering. Cells 2018, 8, 10. [Google Scholar] [CrossRef]
- Goodson, S.G.; White, S.; Stevans, A.M.; Bhat, S.; Kao, C.Y.; Jaworski, S.; Marlowe, T.R.; Kohlmeier, M.; McMillan, L.; Zeisel, S.H.; et al. CASAnova: A multiclass support vector machine model for the classification of human sperm motility patterns. Biol. Reprod. 2017, 97, 698–708. [Google Scholar] [CrossRef]
- Alquezar-Baeta, C.; Gimeno-Martos, S.; Miguel-Jimenez, S.; Santolaria, P.; Yaniz, J.; Palacin, I.; Casao, A.; Cebrian-Perez, J.A.; Muino-Blanco, T.; Perez-Pe, R. OpenCASA: A new open-source and scalable tool for sperm quality analysis. PLoS Comput. Biol. 2019, 15, e1006691. [Google Scholar] [CrossRef]
- Ortega-Ferrusola, C.; Gil, M.C.; Rodriguez-Martinez, H.; Anel, L.; Pena, F.J.; Martin-Munoz, P. Flow cytometry in Spermatology: A bright future ahead. Reprod. Domest. Anim. 2017, 52, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Peña, F.J.; Rodriguez, J.M.O.; Gil, M.C.; Ferrusola, C.O. Flow cytometry analysis of spermatozoa: Is it time for flow spermetry? Reprod. Domest. Anim. 2018, 53, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Boe-Hansen, G.B.; Satake, N. An update on boar semen assessments by flow cytometry and CASA. Theriogenology 2019, 137, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Johannisson, A.; Wallgren, M.; Nagy, S.; Siqueira, A.P.; Rodriguez-Martinez, H. Flow cytometry for the assessment of animal sperm integrity and functionality: State of the art. Asian J. Androl. 2011, 13, 406–419. [Google Scholar] [CrossRef] [PubMed]
- Gillan, L.; Evans, G.; Maxwell, W.M. Flow cytometric evaluation of sperm parameters in relation to fertility potential. Theriogenology 2005, 63, 445–457. [Google Scholar] [CrossRef]
- Holt, W.; Watson, P.; Curry, M.; Holt, C. Reproducibility of computer-aided semen analysis—Comparison of 5 different systems used in a practical workshop. Fertil. Steril. 1994, 62, 1277–1282. [Google Scholar] [CrossRef]
- Lu, J.C.; Huang, Y.F.; Lu, N.Q. Computer-aided sperm analysis: Past, present and future. Andrologia 2014, 46, 329–338. [Google Scholar] [CrossRef]
- Boryshpolets, S.; Kowalski, R.K.; Dietrich, G.J.; Dzyuba, B.; Ciereszko, A. Different computer-assisted sperm analysis (CASA) systems highly influence sperm motility parameters. Theriogenology 2013, 80, 758–765. [Google Scholar] [CrossRef]
- Ramon, M.; Martinez-Pastor, F. Implementation of novel statistical procedures and other advanced approaches to improve analysis of CASA data. Reprod. Fertil. Dev. 2018. [Google Scholar] [CrossRef]
- Wilson-Leedy, J.G.; Ingermann, R.L. Development of a novel CASA system based on open source software for characterization of zebrafish sperm motility parameters. Theriogenology 2007, 67, 661–672. [Google Scholar] [CrossRef]
- Giaretta, E.; Munerato, M.; Yeste, M.; Galeati, G.; Spinaci, M.; Tamanini, C.; Mari, G.; Bucci, D. Implementing an open-access CASA software for the assessment of stallion sperm motility: Relationship with other sperm quality parameters. Anim. Reprod. Sci. 2017, 176, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Purchase, C.F.; Earle, P.T. Modifications to the IMAGEJ computer assisted sperm analysis plugin greatly improve efficiency and fundamentally alter the scope of attainable data. J. Appl. Ichthyol. 2012, 28, 1013–1016. [Google Scholar] [CrossRef]
- Armon, L.; Eisenbach, M. Behavioral mechanism during human sperm chemotaxis: Involvement of hyperactivation. PLoS ONE 2011, 6, e28359. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Umezu, K.; Hara, K.; Hiradate, Y.; Numabe, T.; Tanemura, K. Stromal cell-derived factor 1 regulates in vitro sperm migration towards the cumulus-oocyte complex in cattle. PLoS ONE 2020, 15, e0232536. [Google Scholar] [CrossRef] [PubMed]
- Eisenbach, M. Sperm chemotaxis. Rev. Reprod. 1999, 4, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Yániz, J.L.; Palacín, I.; Vicente-Fiel, S.; Gosálvez, J.; López-Fernández, C.; Santolaria, P. Comparison of membrane-permeant fluorescent probes for sperm viability assessment in the ram. Reprod. Domest. Anim. 2013, 48, 598–603. [Google Scholar] [CrossRef]
- Yanagimachi, R. In vitro acrosome reaction and capacitation of golden hamster spermatozoa by bovine follicular fluid and its fractions. J. Exp. Zool. 1969, 170, 269–280. [Google Scholar] [CrossRef]
- Parrish, J.J. Bovine in vitro fertilization: In vitro oocyte maturation and sperm capacitation with heparin. Theriogenology 2014, 81, 67–73. [Google Scholar] [CrossRef]
- Yaniz, J.L.; Soler, C.; Alquezar-Baeta, C.; Santolaria, P. Toward an integrative and predictive sperm quality analysis in Bos taurus. Anim. Reprod. Sci. 2017, 181, 108–114. [Google Scholar] [CrossRef]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Munafo, M.R.; Nosek, B.A.; Bishop, D.V.M.; Button, K.S.; Chambers, C.D.; du Sert, N.P.; Simonsohn, U.; Wagenmakers, E.J.; Ware, J.J.; Ioannidis, J.P.A. A manifesto for reproducible science. Nat. Hum. Behav. 2017, 1, 0021. [Google Scholar] [CrossRef]
- Woelfle, M.; Olliaro, P.; Todd, M.H. Open science is a research accelerator. Nat. Chem. 2011, 3, 745–748. [Google Scholar] [CrossRef] [PubMed]
- Cosson, J.J. Flagellar Mechanics and Sperm Guidance; Bentham Science Publishers: Cenakva, Czech Republic, 2015. [Google Scholar] [CrossRef]
- Cosson, J. Frenetic activation of fish spermatozoa flagella entails short-term motility, portending their precocious decadence. J. Fish Biol. 2010, 76, 240–279. [Google Scholar] [CrossRef] [PubMed]
- Yanagimachi, R.; Harumi, T.; Matsubara, H.; Yan, W.; Yuan, S.; Hirohashi, N.; Iida, T.; Yamaha, E.; Arai, K.; Matsubara, T.; et al. Chemical and physical guidance of fish spermatozoa into the egg through the micropyle. Biol. Reprod. 2017, 96, 780–799. [Google Scholar] [CrossRef] [PubMed]
- Kholodnyy, V.; Gadelha, H.; Cosson, J.; Boryshpolets, S. How do freshwater fish sperm find the egg? The physicochemical factors guiding the gamete encounters of externally fertilizing freshwater fish. Rev. Aquacult. 2020, 12, 1165–1192. [Google Scholar] [CrossRef]
- Bahat, A.; Eisenbach, M. Sperm thermotaxis. Mol. Cell. Endocrinol. 2006, 252, 115–119. [Google Scholar] [CrossRef]
- Miki, K.; Clapham, D.E. Rheotaxis guides mammalian sperm. Curr. Biol. 2013, 23, 443–452. [Google Scholar] [CrossRef]
- Peña, A.; Johannisson, A.; Linde-Forsberg, C. Post-thaw evaluation of dog spermatozoa using new triple fluorescent staining and flow cytometry. Theriogenology 1999, 52, 965–980. [Google Scholar] [CrossRef]
- Nagy, S.; Jansen, J.; Topper, E.K.; Gadella, B.M. A triple-stain flow cytometric method to assess plasma- and acrosome-membrane integrity of cryopreserved bovine sperm immediately after thawing in presence of egg-yolk particles. Biol. Reprod. 2003, 68, 1828–1835. [Google Scholar] [CrossRef]
- Bussalleu, E.; Pinart, E.; Yeste, M.; Briz, M.; Sancho, S.; Garcia-Gil, N.; Badia, E.; Bassols, J.; Pruneda, A.; Casas, I.; et al. Development of a protocol for multiple staining with fluorochromes to assess the functional status of boar spermatozoa. Microsc. Res. Tech. 2005, 68, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.K.; Kunze, E.; Hammerstedt, R.H. Analysis of sperm cell viability, acrosomal integrity, and mitochondrial function using flow cytometry. Biol. Reprod. 1990, 43, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Gimeno-Martos, S.; Gonzalez-Arto, M.; Casao, A.; Gallego, M.; Cebrian-Perez, J.A.; Muino-Blanco, T.; Perez-Pe, R. Steroid hormone receptors and direct effects of steroid hormones on ram spermatozoa. Reproduction 2017, 154, 469–481. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fernandez, S.; Sestelo, A.; Rivolta, M.; Cordoba, M. Capacitation and Acrosome Reaction Induction on Thawed Dama dama Deer Spermatozoa: Glycine Effect as Cryopreservation Diluent Supplement. Zoolog. Sci. 2013, 30, 1110–1116. [Google Scholar] [CrossRef]
- Seify, M.; Zarabadipour, M.; Ghaleno, L.R.; Alizadeh, A.; Valojerdi, M.R. The anti-oxidant roles of Taurine and Hypotaurine on acrosome integrity, HBA and HSPA2 of the human sperm during vitrification and post warming in two different temperature. Cryobiology 2019, 90, 89–95. [Google Scholar] [CrossRef]
- Lee, Y.S.; Lee, S.; Lee, S.H.; Yang, B.K.; Park, C.K. Effect of cholesterol-loaded-cyclodextrin on sperm viability and acrosome reaction in boar semen cryopreservation. Anim. Reprod. Sci. 2015, 159, 124–130. [Google Scholar] [CrossRef]
- Pons-Rejraji, H.; Bailey, J.L.; Leclerc, P. Cryopreservation affects bovine sperm intracellular parameters associated with capacitation and acrosome exocytosis. Reprod. Fertil. Dev. 2009, 21, 525–537. [Google Scholar] [CrossRef]
- Kumar, A.; Prasad, J.K.; Srivastava, N.; Ghosh, S.K. Strategies to Minimize Various Stress-Related Freeze-Thaw Damages During Conventional Cryopreservation of Mammalian Spermatozoa. Biopreserv. Biobank. 2019, 17, 603–612. [Google Scholar] [CrossRef]
- Pini, T.; Leahy, T.; de Graaf, S.P. Sublethal sperm freezing damage: Manifestations and solutions. Theriogenology 2018, 118, 172–181. [Google Scholar] [CrossRef]
- Guidobaldi, H.A.; Hirohashi, N.; Cubilla, M.; Buffone, M.G.; Giojalas, L.C. An intact acrosome is required for the chemotactic response to progesterone in mouse spermatozoa. Mol. Reprod. Dev. 2017, 84, 310–315. [Google Scholar] [CrossRef]
- Cooper, T.G.; Noonan, E.; von Eckardstein, S.; Auger, J.; Baker, H.W.; Behre, H.M.; Haugen, T.B.; Kruger, T.; Wang, C.; Mbizvo, M.T.; et al. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update 2010, 16, 231–245. [Google Scholar] [CrossRef] [PubMed]



| Spearman’s Correlation | Bland–Altman | ||
|---|---|---|---|
| Sperm Subtype | r | p-Value | Bias (%) |
| IAIM | 0.801 | <0.0001 | 1.636 |
| IADM | 0.827 | <0.0001 | 0.491 |
| DAIM | 0.208 | n.s. | −0.932 |
| DADM | 0.851 | <0.0001 | −1.551 |
| IFI | 0.621 | <0.0001 | 1.854 |
| Pearson Correlation | Bland–Altman | |
|---|---|---|
| r | p-Value | Bias (%) |
| 0.958 | <0.0001 | −2.407 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yániz, J.; Alquézar-Baeta, C.; Yagüe-Martínez, J.; Alastruey-Benedé, J.; Palacín, I.; Boryshpolets, S.; Kholodnyy, V.; Gadêlha, H.; Pérez-Pe, R. Expanding the Limits of Computer-Assisted Sperm Analysis through the Development of Open Software. Biology 2020, 9, 207. https://doi.org/10.3390/biology9080207
Yániz J, Alquézar-Baeta C, Yagüe-Martínez J, Alastruey-Benedé J, Palacín I, Boryshpolets S, Kholodnyy V, Gadêlha H, Pérez-Pe R. Expanding the Limits of Computer-Assisted Sperm Analysis through the Development of Open Software. Biology. 2020; 9(8):207. https://doi.org/10.3390/biology9080207
Chicago/Turabian StyleYániz, Jesús, Carlos Alquézar-Baeta, Jorge Yagüe-Martínez, Jesús Alastruey-Benedé, Inmaculada Palacín, Sergii Boryshpolets, Vitaliy Kholodnyy, Hermes Gadêlha, and Rosaura Pérez-Pe. 2020. "Expanding the Limits of Computer-Assisted Sperm Analysis through the Development of Open Software" Biology 9, no. 8: 207. https://doi.org/10.3390/biology9080207
APA StyleYániz, J., Alquézar-Baeta, C., Yagüe-Martínez, J., Alastruey-Benedé, J., Palacín, I., Boryshpolets, S., Kholodnyy, V., Gadêlha, H., & Pérez-Pe, R. (2020). Expanding the Limits of Computer-Assisted Sperm Analysis through the Development of Open Software. Biology, 9(8), 207. https://doi.org/10.3390/biology9080207