Potential Involvement of lncRNAs in the Modulation of the Transcriptome Response to Nodavirus Challenge in European Sea Bass (Dicentrarchus labrax L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish and Virus
2.2. Fish Infection and Sampling
2.3. RNA Isolation and High-Throughput Transcriptome Sequencing
2.4. Sequence Assembly and LncRNA Mining
2.5. Differential Expression Analysis
2.6. Genome Mapping and Identification of LncRNA Neighbouring Coding Genes
2.7. GO Enrichment Analyses
2.8. Correlation Analyses Between LncRNAs and Coding Genes
2.9. Quantitative PCR (qPCR) Validation of LncRNA Expression
3. Results
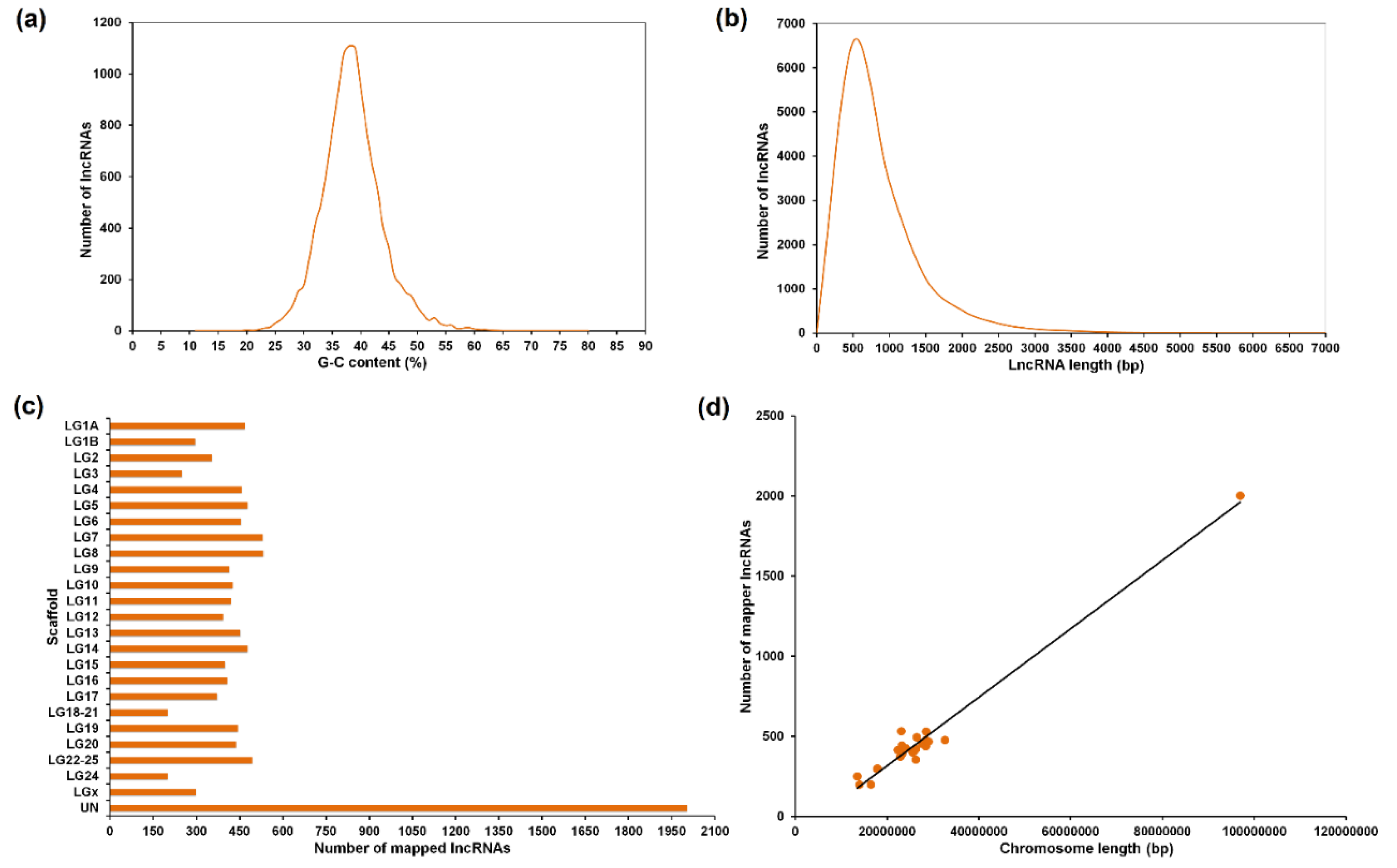
3.1. Assembly, Annotation and LncRNA Mining
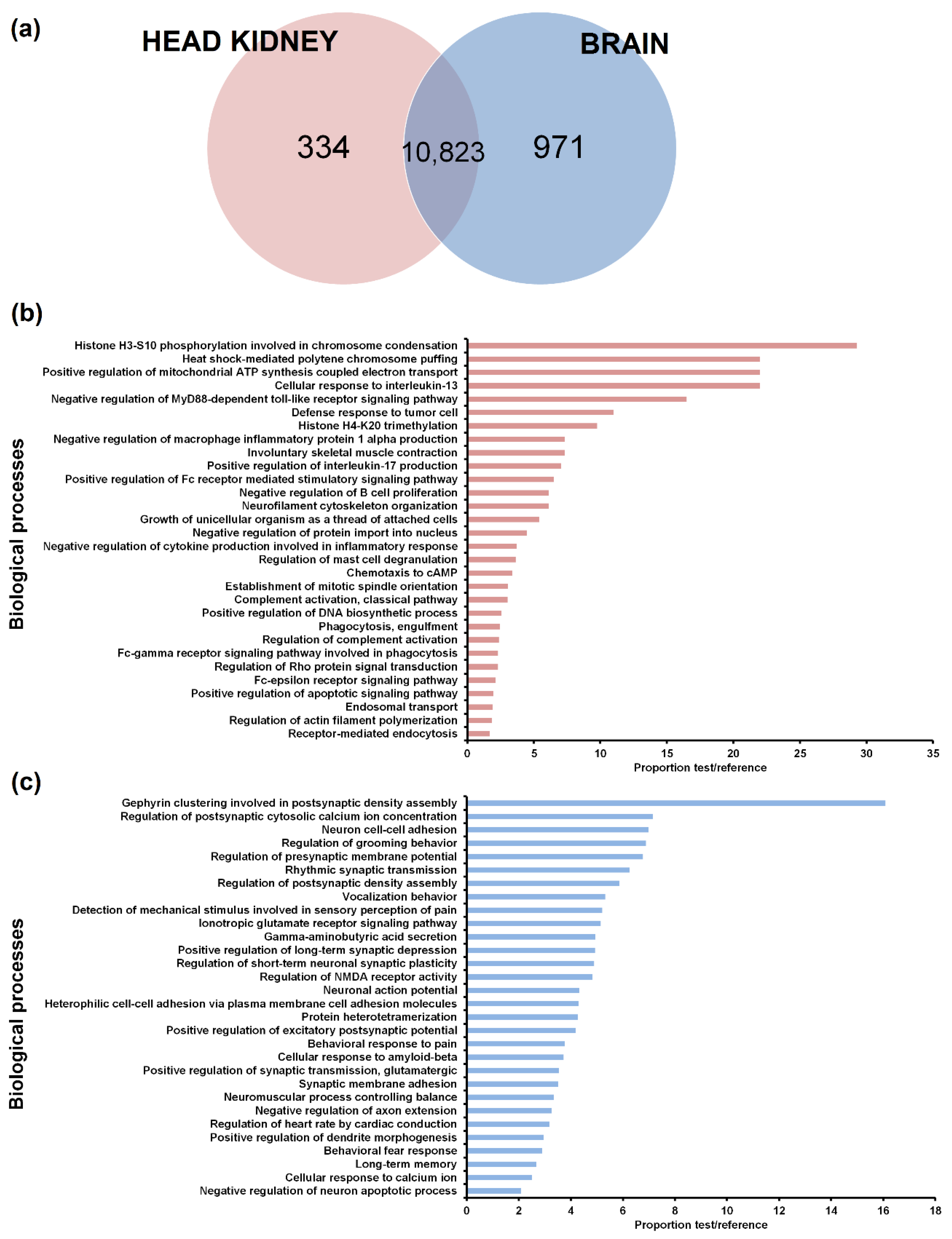
3.2. Tissue Distribution of the LncRNAs
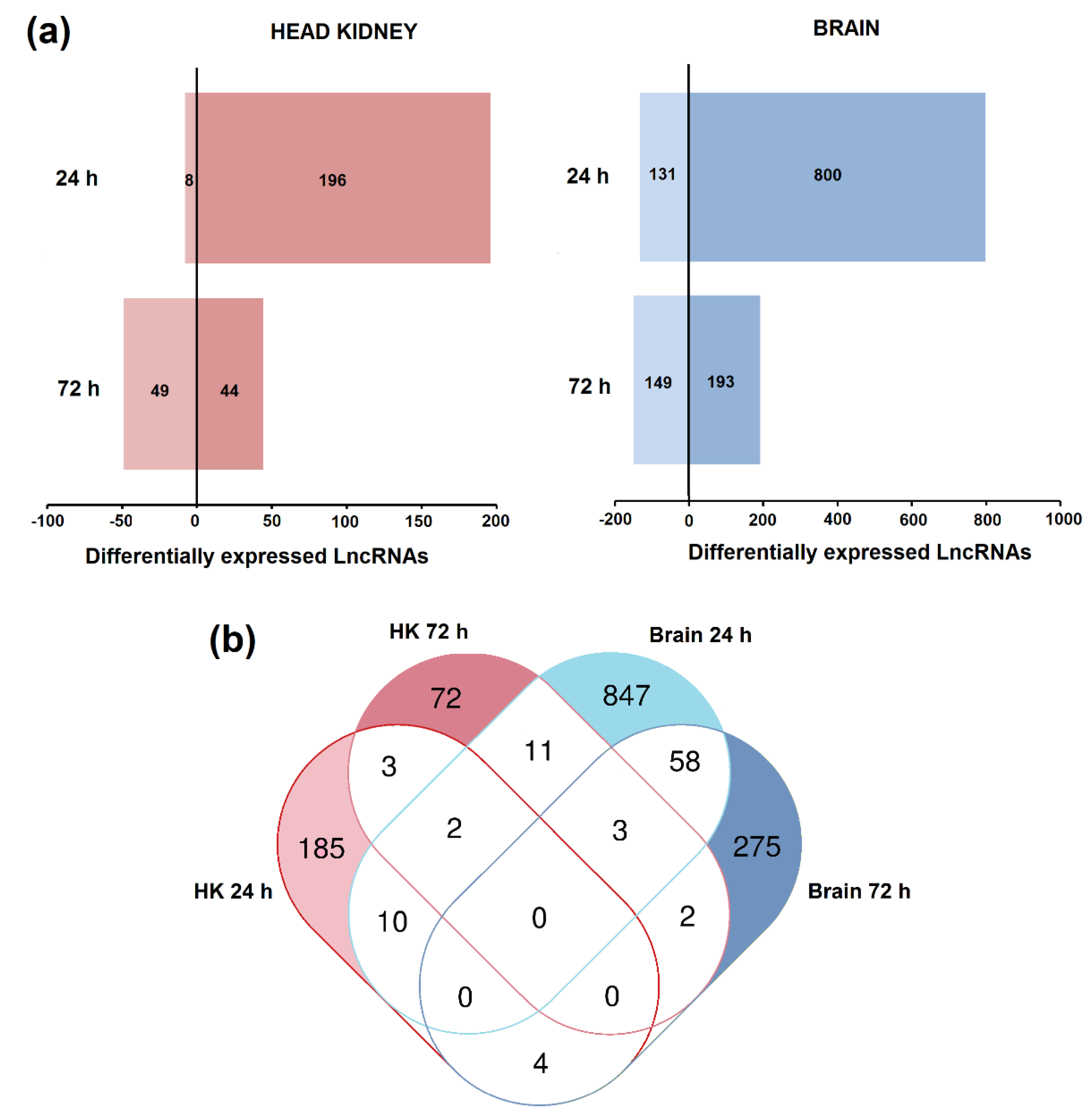
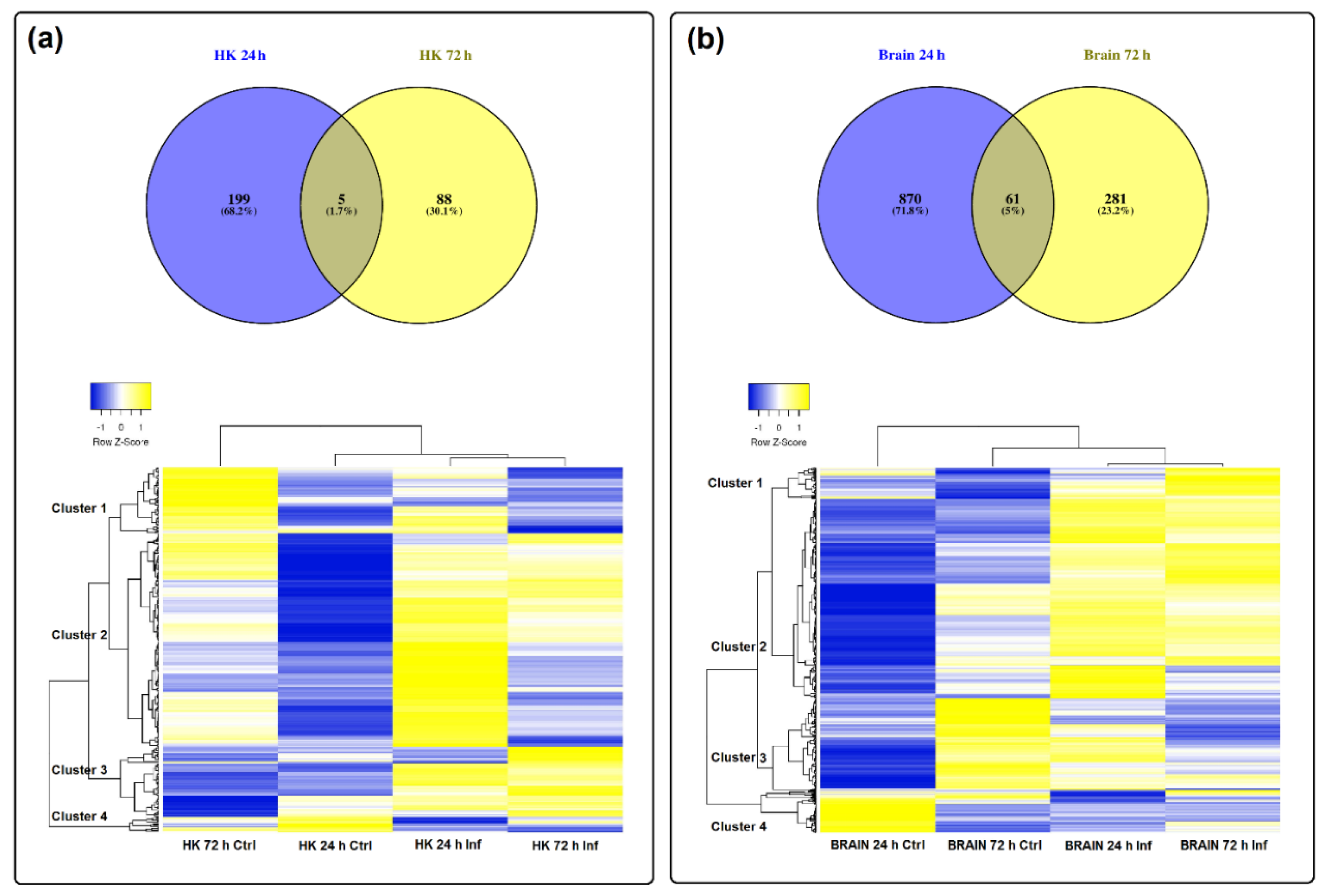
3.3. Differential Expression of LncRNAs after Nodavirus Challenge
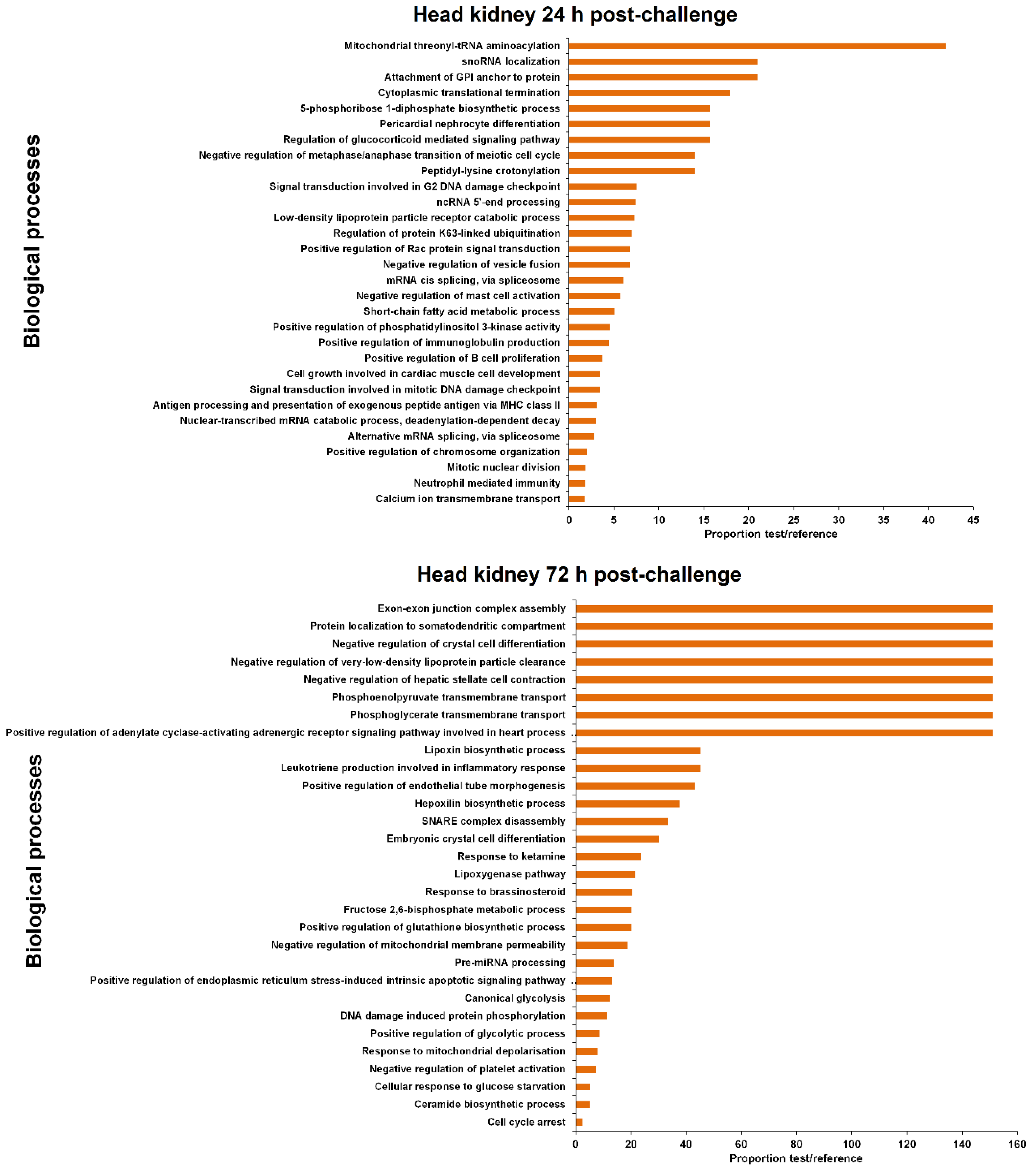
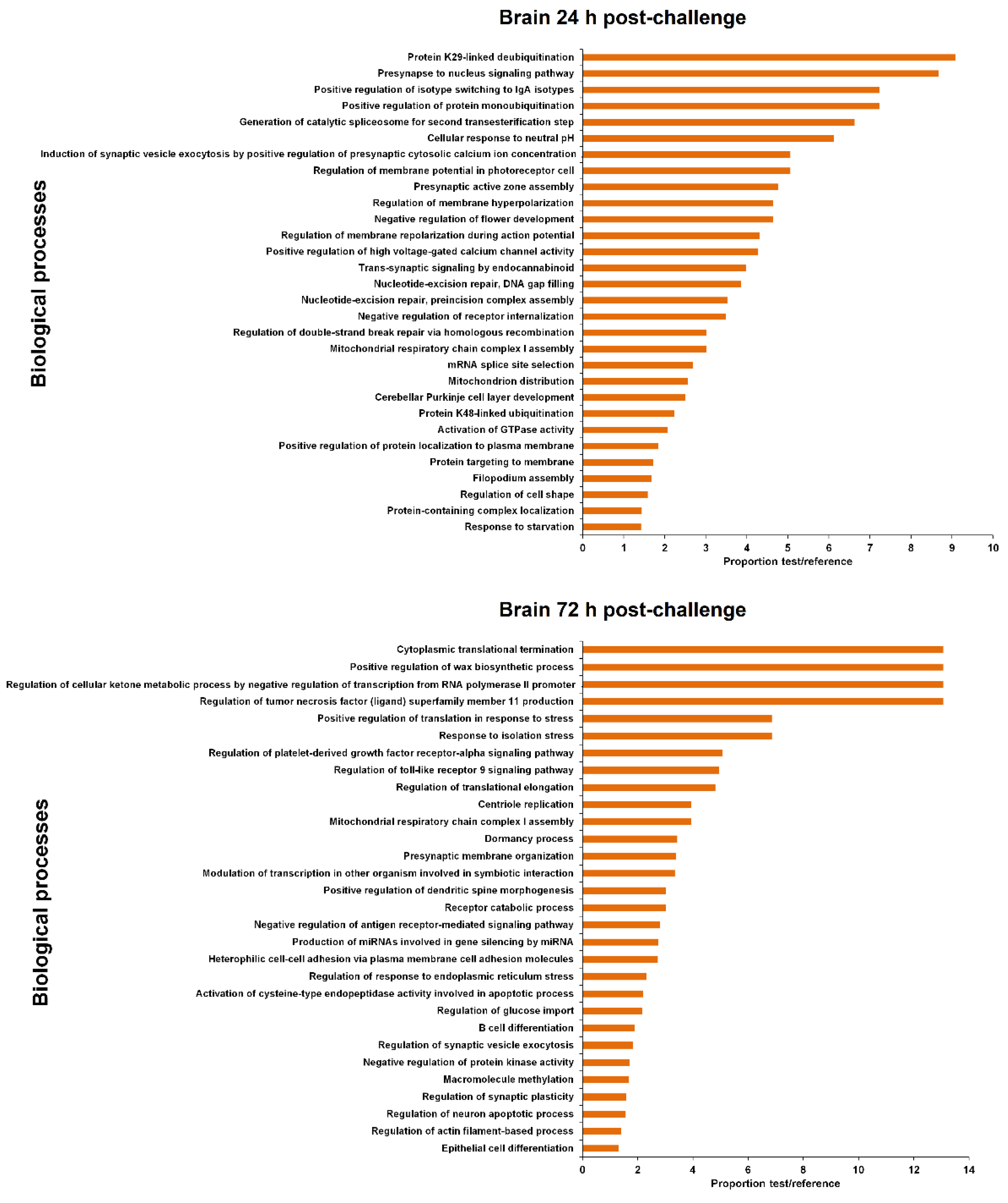
3.4. GO Enrichment of the LncRNA Neighbouring Coding Genes
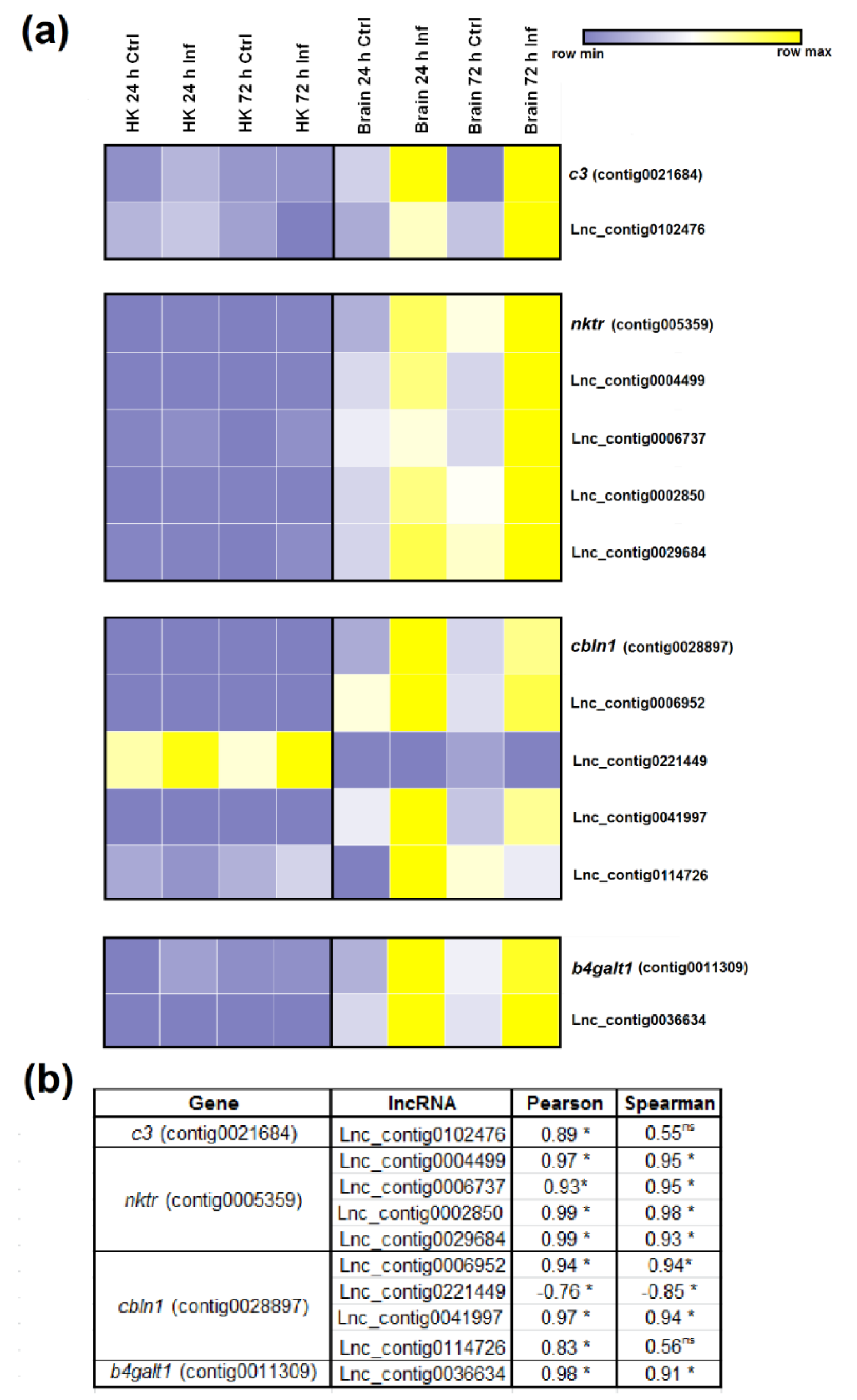
3.5. Expression Correlation of LncRNAs and Coding Genes
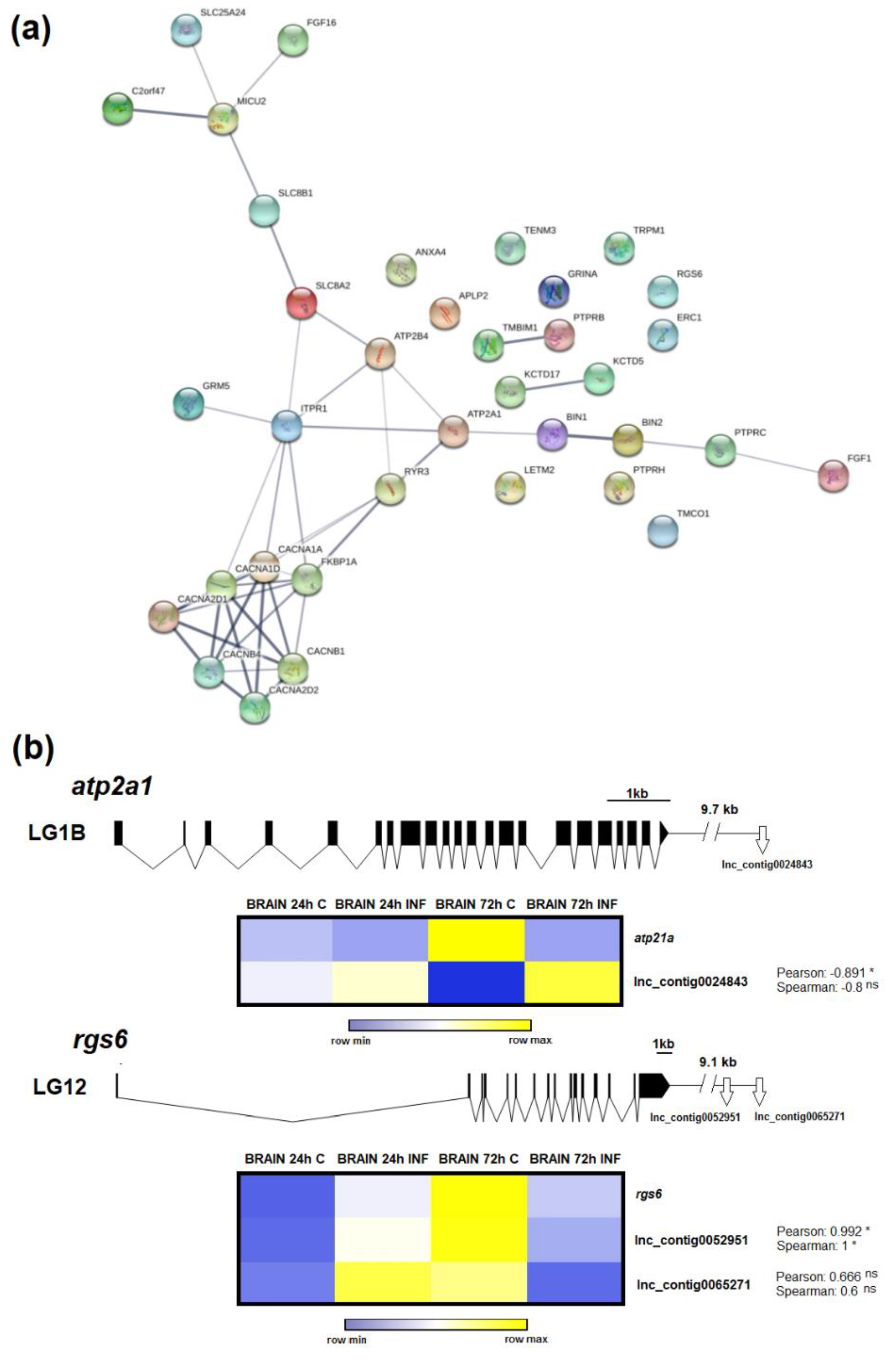
3.6. LncRNAs Could Mediate Calcium Homeostasis
3.7. qPCR Validation of LncRNA Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. Cultured Aquatic species information programme. Dicentrarchus labrax. In National Aquaculture Sector Overview; FAO: Rome, Italy, 2005. [Google Scholar]
- Doan, Q.K.; Vandeputte, M.; Chatain, B.; Morin, T.; Allal, F. Viral encephalopathy and retinopathy in aquaculture: A review. J. Fish Dis. 2017, 40, 717–742. [Google Scholar] [CrossRef]
- Munday, B.L.; Kwang, J.; Moody, N. Betanodavirus infections of teleost fish: A review. J. Fish Dis. 2002, 25, 127–142. [Google Scholar] [CrossRef]
- Poisa-Beiro, L.; Dios, S.; Montes, A.; Aranguren, R.; Figueras, A.; Novoa, B. Nodavirus increases the expression of Mx and inflammatory cytokines in fish brain. Mol. Immunol. 2008, 45, 218–225. [Google Scholar] [CrossRef]
- Poisa-Beiro, L.; Dios, S.; Ahmed, H.; Vasta, G.R.; Martínez-López, A.; Estepa, A.; Alonso-Gutiérrez, J.; Figueras, A.; Novoa, B. Nodavirus infection of sea bass (Dicentrarchus labrax) induces up-regulation of galectin-1 expression with potential anti-inflammatory activity. J. Immunol. 2009, 183, 6600–6611. [Google Scholar] [CrossRef]
- Sarropoulou, E.; Sepulcre, P.; Poisa-Beiro, L.; Mulero, V.; Meseguer, J.; Figueras, A.; Novoa, B.; Terzoglou, V.; Reinhardt, R.; Magoulas, A.; et al. Profiling of infection specific mRNA transcripts of the European seabass Dicentrarchus labrax. BMC Gen. 2009, 10, 157. [Google Scholar] [CrossRef]
- Scapigliati, G.; Buonocore, F.; Randelli, E.; Casani, D.; Meloni, S.; Zarletti, G.; Tiberi, M.; Pietretti, D.; Boschi, I.; Manchado, M.; et al. Cellular and molecular immune responses of the sea bass (Dicentrarchus labrax) experimentally infected with betanodavirus. Fish Shellfish Immunol. 2010, 28, 303–311. [Google Scholar] [CrossRef]
- Chaves-Pozo, E.; Guardiola, F.A.; Meseguer, J.; Esteban, M.A.; Cuesta, A. Nodavirus infection induces a great innate cell-mediated cytotoxic activity in resistant, gilthead seabream, and susceptible, European sea bass, teleost fish. Fish Shellfish Immunol. 2012, 33, 1159–1166. [Google Scholar] [CrossRef]
- Novel, P.; Fernandez-Trujillo, M.A.; Gallardo-Galvez, J.B.; Cano, I.; Manchado, M.; Buonocore, F.; Randelli, E.; Scapigliati, G.; Alvarez, M.C.; Bejar, J. Two Mx genes identified in European sea bass (Dicentrarchus labrax) respond differently to VNNV infection. Vet. Immunol. Immunopathol. 2013, 153, 240–248. [Google Scholar] [CrossRef]
- Buonocore, F.; Randelli, E.; Tranfa, P.; Scapigliati, G. A CD83-like molecule in sea bass (Dicentrarchus labrax): Molecular characterization and modulation by viral and bacterial infection. Fish Shellfish Immunol. 2012, 32, 1179–1184. [Google Scholar] [CrossRef]
- Buonocore, F.; Stocchi, V.; Nunez-Ortiz, N.; Randelli, E.; Gerdol, M.; Pallavicini, A.; Facchiano, A.; Bernini, C.; Guerra, L.; Scapigliati, G.; et al. Immunoglobulin T from sea bass (Dicentrarchus labrax L.): Molecular characterization, tissue localization and expression after nodavirus infection. BMC Mol. Biol. 2017, 18, 8. [Google Scholar] [CrossRef]
- Valero, Y.; Morcillo, P.; Meseguer, J.; Buonocore, F.; Esteban, M.A.; Chaves-Pozo, E.; Cuesta, A. Characterization of the interferon pathway in the teleost fish gonad against the vertically transmitted nervous necrosis virus. J. Gen. Virol. 2015, 96, 2176–2187. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Pozo, E.; Valero, Y.; Esteve-Codina, A.; Gómez-Garrido, J.; Dabad, M.; Alioto, T.; Meseguer, J.; Esteban, M.A.; Cuesta, A. Innate cell-mediated cytotoxic activity of European sea bass leucocytes against Nodavirus-infected cells: A functional and RNA-seq study. Sci. Rep. 2017, 7, 15396. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Pozo, E.; Bandín, I.; Olveira, J.G.; Esteve-Codina, A.; Gómez-Garrido, J.; Dabad, M.; Alioto, T.; Esteban, M.A.; Cuesta, A. European sea bass brain DLB-1 cell line is susceptible to nodavirus: A transcriptomic study. Fish Shellfish Immunol. 2019, 86, 14–24. [Google Scholar] [CrossRef]
- Lama, R.; Pereiro, P.; Valenzuela-Muñoz, V.; Gallardo-Escárate, C.; Tort, L.; Figueras, A.; Novoa, B. RNA-Seq analysis of European sea bass (Dicentrarchus labrax L.) infected with nodavirus reveals powerful modulation of the stress response. Vet. Res. 2020, 51, 64. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef]
- Perry, R.B.T.; Ulitsky, I. The functions of long noncoding RNAs in development and stem cells. Development 2016, 143, 3882–3894. [Google Scholar] [CrossRef]
- Guo, C.J.; Ma, X.K.; Xing, Y.H.; Zheng, C.C.; Xu, Y.F.; Shan, L.; Zhang, J.; Wang, S.; Wang, Y.; Carmichael, G.G.; et al. Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell 2020, 181, 621–636. [Google Scholar] [CrossRef]
- Aune, T.M.; Spurlock, C.F. Long non-coding RNAs in innate and adaptive immunity. Virus Res. 2016, 212, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Elling, R.; Chan, J.; Fitzgerald, K.A. Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. Eur. J. Immunol. 2016, 46, 504–512. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, S.; Wu, W.; Yu, F.; Chang, W.; Li, P.; Wang, K. Non-coding RNAs function as immune regulators in teleost fish. Front. Immunol. 2018, 9, 2801. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- Tarifeño-Saldivia, E.; Valenzuela-Miranda, D.; Gallardo-Escárate, C. In the shadow: The emerging role of long non-coding RNAs in the immune response of Atlantic salmon. Dev. Comp. Immunol. 2017, 73, 193–205. [Google Scholar] [CrossRef]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Tine, M.; Kuhl, H.; Gagnaire, P.A.; Louro, B.; Desmarais, E.; Martins, R.S.T.; Hecht, J.; Knaust, F.; Belkhir, K.; Klages, S.; et al. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat. Commun. 2014, 5, 5770. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef]
- Carpenter, S.; Aiello, D.; Atianand, M.K.; Ricci, E.P.; Gandhi, P.; Hall, L.L.; Byron, M.; Monks, B.; Henry-Bezy, M.; O’Neill, L.A.J.; et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013, 341, 789–792. [Google Scholar] [CrossRef]
- Rapicavoli, N.A.; Qu, K.; Zhang, J.; Mikhail, M.; Laberge, R.M.; Chang, H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2013, 2, e00762. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Tang, Q.; Sharma, S.; Yu, F.; Escobar, T.M.; Muljo, S.A.; Zhu, J.; Zhao, K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013, 14, 1190–1198. [Google Scholar] [CrossRef]
- Agirre, X.; Meydan, C.; Jiang, Y.; Garate, L.; Doane, A.S.; Li, Z.; Verma, A.; Paiva, B.; Martín-Subero, J.I.; Elemento, O.; et al. Long non-coding RNAs discriminate the stages and gene regulatory states of human humoral immune response. Nat. Commun. 2019, 10, 821. [Google Scholar] [CrossRef] [PubMed]
- Kambara, H.; Niazi, F.; Kostadinova, L.; Moonka, D.K.; Siegel, C.T.; Post, A.B.; Carnero, E.; Barriocanal, M.; Fortes, P.; Anthony, D.D.; et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 2014, 42, 10668–10680. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xue, Y.; Han, Y.; Lin, L.; Wu, C.; Xu, S.; Jiang, Z.; Xu, J.; Liu, Q.; Cao, X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014, 344, 310–313. [Google Scholar] [CrossRef]
- Zhang, X.; Lian, Z.; Padden, C.; Gerstein, M.B.; Rozowsky, J.; Snyder, M.; Gingeras, T.R.; Kapranov, P.; Weissman, S.M.; Newburger, P.E. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009, 113, 2526–2534. [Google Scholar] [CrossRef]
- Peng, X.; Gralinski, L.; Armour, C.D.; Ferris, M.T.; Thomas, M.J.; Proll, S.; Bradel-Tretheway, B.G.; Korth, M.J.; Castle, J.C.; Biery, M.C.; et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio 2010, 1, e00206–e00210. [Google Scholar] [CrossRef]
- Zhang, Q.; Lai, M.M.; Lou, Y.Y.; Guo, B.H.; Wang, H.Y.; Zheng, X.Q. Transcriptome altered by latent human cytomegalovirus infection on THP-1 cells using RNA-seq. Gene 2016, 594, 144–150. [Google Scholar] [CrossRef]
- Zhao, P.; Liu, S.; Zhong, Z.; Jiang, T.; Weng, R.; Xie, M.; Yang, S.; Xia, X. Analysis of expression profiles of long noncoding RNAs and mRNAs in brains of mice infected by rabies virus by RNA sequencing. Sci. Rep. 2018, 8, 11858. [Google Scholar] [CrossRef]
- Hudson, W.H.; Prokhnevska, N.; Gensheimer, J.; Akondy, R.; McGuire, D.J.; Ahmed, R.; Kissick, H.T. Expression of novel long noncoding RNAs defines virus-specific effector and memory CD8+ T cells. Nat. Commun. 2019, 10, 196. [Google Scholar] [CrossRef]
- Fortes, P.; Morris, K.V. Long noncoding RNAs in viral infections. Virus Res. 2016, 212, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, X.; Li, J.; Huang, S.; Xiang, S.; Hu, X.; Liu, C. Comprehensive analysis of coding-lncRNA gene co-expression network uncovers conserved functional lncRNAs in zebrafish. BMC Gen. 2018, 19, 112. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chen, W.; Li, J.; Huang, S.; Xu, X.; Zhang, X.; Xiang, S.; Liu, C. ZFLNC: A comprehensive and well-annotated database for zebrafish lncRNA. Database 2018, 2018, bay114. [Google Scholar] [CrossRef] [PubMed]
- Wei, N.; Pang, W.; Wang, Y.; Xiong, Y.; Xu, R.; Wu, W.; Zhao, C.; Yang, G. Knockdown of PU.1 mRNA and AS lncRNA regulates expression of immune-related genes in zebrafish Danio rerio. Dev. Comp. Immunol. 2014, 44, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Bassing, C.H.; Swat, W.; Alt, F.W. The mechanism and regulation of chromosomal V(D)J recombination. Cell 2002, 109, S45–S55. [Google Scholar] [CrossRef]
- Valenzuela-Muñoz, V.; Pereiro, P.; Álvarez-Rodríguez, M.; Gallardo-Escárate, C.; Figueras, A.; Novoa, B. Comparative modulation of lncRNAs in wild-type and rag1-heterozygous mutant zebrafish exposed to immune challenge with spring viraemia of carp virus (SVCV). Sci. Rep. 2019, 9, 14174. [Google Scholar] [CrossRef]
- Boltaña, S.; Valenzuela-Miranda, D.; Aguilar, A.; Mackenzie, S.; Gallardo-Escárate, C. Long noncoding RNAs (lncRNAs) dynamics evidence immunomodulation during ISAV-Infected Atlantic salmon (Salmo salar). Sci. Rep. 2016, 6, 22698. [Google Scholar] [CrossRef]
- Valenzuela-Miranda, D.; Gallardo-Escárate, C. Novel insights into the response of Atlantic salmon (Salmo salar) to Piscirickettsia salmonis: Interplay of coding genes and lncRNAs during bacterial infection. Fish Shellfish Immunol. 2016, 59, 427–438. [Google Scholar] [CrossRef]
- Paneru, B.; Al-Tobasei, R.; Palti, Y.; Wiens, G.D.; Salem, M. Differential expression of long non-coding RNAs in three genetic lines of rainbow trout in response to infection with Flavobacterium psychrophilum. Sci. Rep. 2016, 6, 36032. [Google Scholar] [CrossRef]
- Valenzuela-Muñoz, V.; Valenzuela-Miranda, D.; Gallardo-Escárate, C. Comparative analysis of long non-coding RNAs in Atlantic and Coho salmon reveals divergent transcriptome responses associated with immunity and tissue repair during sea lice infestation. Dev. Comp. Immunol. 2018, 87, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Kern, C.; Wang, Y.; Chitwood, J.; Korf, I.; Delany, M.; Cheng, H.; Medrano, J.F.; Van Eenennaam, A.L.; Ernst, C.; Ross, P.; et al. Genome-wide identification of tissue-specific long non-coding RNA in three farm animal species. BMC Genom. 2018, 19, 684. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.; McEwan, C.; Mills, J.D.; Janitz, M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J. Hum. Transcr. 2015, 1, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, K.; Leonard, V.E.; Kv, S.; Lalwani, M.K.; Jalali, S.; Patowary, A.; Joshi, A.; Scaria, V.; Sivasubbu, S. Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. PLoS ONE 2013, 8, e83616. [Google Scholar] [CrossRef]
- Zapata, A.; Diez, B.; Cejalvo, T.; Gutierrez-De Frias, C.; Cortes, A. Ontogeny of the immune system of fish. Fish Shellfish Immunol. 2006, 20, 126–136. [Google Scholar] [CrossRef]
- Lin, S.S.; Huang, H.P.; Yang, J.S.; Wu, J.Y.; Hsia, T.C.; Lin, C.C.; Lin, C.W.; Kuo, C.L.; Gibson-Wood, W.; Chung, J.G. DNA damage and endoplasmic reticulum stress mediated curcumin-induced cell cycle arrest and apoptosis in human lung carcinoma A-549 cells through the activation caspases cascade- and mitochondrial-dependent pathway. Cancer Lett. 2018, 272, 77–90. [Google Scholar] [CrossRef]
- Yang, J.S.; Chen, G.W.; Hsia, T.C.; Ho, H.C.; Ho, C.C.; Lin, M.W.; Lin, S.S.; Yeh, R.D.; Ip, S.W.; Lu, H.F.; et al. Diallyl disulfide induces apoptosis in human colon cancer cell line (COLO 205) through the induction of reactive oxygen species, endoplasmic reticulum stress, caspases cascade and mitochondrial-dependent pathways. Food Chem. Toxicol. 2009, 47, 171–179. [Google Scholar] [CrossRef]
- Zeng, H.; Nanayakkara, G.K.; Shao, Y.; Fu, H.; Sun, Y.; Cueto, R.; Yang, W.Y.; Yang, Q.; Sheng, H.; Wu, N.; et al. DNA checkpoint and repair factors are nuclear sensors for intracellular organelle stresses-inflammations and cancers can have high genomic risks. Front. Physiol. 2018, 9, 516. [Google Scholar] [CrossRef]
- Luftig, M.A. Viruses and the DNA damage response: Activation and antagonism. Annu. Rev. Virol. 2014, 1, 605–625. [Google Scholar] [CrossRef]
- Bagga, S.; Bouchard, M.J. Cell cycle regulation during viral infection. Methods Mol. Biol. 2014, 1170, 165–227. [Google Scholar]
- Li, S.; Kong, L.; Yu, X. The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit. Rev. Microbiol. 2015, 41, 150–164. [Google Scholar] [CrossRef]
- Zeeshan, H.M.; Lee, G.H.; Kim, H.R.; Chae, H.J. Endoplasmic reticulum stress and associated ROS. Int. J. Mol. Sci. 2016, 17, 327. [Google Scholar] [CrossRef]
- Paiva, C.N.; Bozza, M.T. Are reactive oxygen species always detrimental to pathogens? Antioxid. Redox Signal. 2014, 20, 1000–1037. [Google Scholar] [CrossRef]
- Chang, C.W.; Su, Y.C.; Her, G.M.; Ken, C.F.; Hong, J.R. Betanodavirus induces oxidative stress-mediated cell death that prevented by anti-oxidants and Zfcatalase in fish cells. PLoS ONE 2011, 6, e25853. [Google Scholar] [CrossRef] [PubMed]
- Grove, S.; Johansen, R.; Reitan, L.J.; Press, C.M.; Dannevig, B.H. Quantitative investigation of antigen and immune response in nervous and lymphoid tissues of Atlantic halibut (Hippoglossus hippoglossus) challenged with nodavirus. Fish Shellfish Immunol. 2016, 21, 525–539. [Google Scholar] [CrossRef]
- López-Muñoz, A.; Sepulcre, M.P.; García-Moreno, D.; Fuentes, I.; Béjar, J.; Manchado, M.; Álvarez, M.C.; Meseguer, J.; Mulero, V. Viral nervous necrosis virus persistently replicates in the central nervous system of asymptomatic gilthead seabream and promotes a transient inflammatory response followed by the infiltration of IgM+ B lymphocytes. Dev. Comp. Immunol. 2012, 37, 429–437. [Google Scholar] [CrossRef]
- Piazzon, M.C.; Galindo-Villegas, J.; Pereiro, P.; Estensoro, I.; Calduch-Giner, J.A.; Gómez-Casado, E.; Novoa, B.; Mulero, V.; Sitjà-Bobadilla, A.; Pérez-Sánchez, J. Differential modulation of IgT and IgM upon parasitic, bacterial, viral, and dietary challenges in a perciform fish. Front. Immunol. 2016, 7, 637. [Google Scholar] [CrossRef]
- Wu, M.S.; Chen, C.W.; Liu, Y.C.; Huang, H.H.; Lin, C.H.; Tzeng, C.S.; Chang, C.Y. Transcriptional analysis of orange-spotted grouper reacting to experimental grouper iridovirus infection. Dev. Comp. Immunol. 2012, 37, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A. Immune regulation of antibody access to neuronal tissues. Trends Mol. Med. 2017, 23, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Arundine, M.; Tymianski, M. Molecular mechanisms of calcium-dependent neurodegeneration in excitotoxicity. Cell Calcium 2003, 34, 325–337. [Google Scholar] [CrossRef]
- Dong, X.X.; Wang, Y.; Qin, Z.H. Molecular mechanisms of excitotoxicity and their relevance to pathogenesis of neurodegenerative diseases. Acta Pharmacol. Sin. 2009, 30, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Frey, T.K.; Yang, J.J. Viral calciomics: Interplays between Ca2+ and virus. Cell Calcium 2009, 46, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.M.; Hsu, C.H.; Wang, C.H.; Liu, W.; Chang, W.H.; Lin, C.S. Role of the DxxDxD motif in the assembly and stability of betanodavirus particles. Arch. Virol. 2008, 153, 1633–1642. [Google Scholar] [CrossRef]
- Chen, N.C.; Yoshimura, M.; Guan, H.H.; Wang, T.Y.; Misumi, Y.; Lin, C.C.; Chuankhayan, P.; Nakagawa, A.; Chan, S.I.; Tsukihara, T.; et al. Crystal structures of a piscine Betanodavirus: Mechanisms of capsid assembly and viral infection. PLoS Pathog. 2015, 11, e1005203. [Google Scholar] [CrossRef] [PubMed]
| READS | |
| Total reads | 679,746,504 |
| Mean reads per sample | 28,322,771 |
| ASSEMBLY | |
| Contigs | 347,317 |
| Minimum length | 200 bp |
| Maximum length | 26,142 bp |
| Average length | 723 bp |
| N50 | 1088 bp |
| Total assembled reads | 215,126,479 |
| ANNOTATION | |
| Annotated contigs | 240,274 (69%) |
| Non-annotated contigs | 107,043 |
| LncRNAs | |
| Potential lncRNAs | 12,158 |
| Minimum length | 200 |
| Maximum length | 6829 |
| Average length | 667 |
| lncRNAs mapped on genome | 11,667 (95.96%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereiro, P.; Lama, R.; Moreira, R.; Valenzuela-Muñoz, V.; Gallardo-Escárate, C.; Novoa, B.; Figueras, A. Potential Involvement of lncRNAs in the Modulation of the Transcriptome Response to Nodavirus Challenge in European Sea Bass (Dicentrarchus labrax L.). Biology 2020, 9, 165. https://doi.org/10.3390/biology9070165
Pereiro P, Lama R, Moreira R, Valenzuela-Muñoz V, Gallardo-Escárate C, Novoa B, Figueras A. Potential Involvement of lncRNAs in the Modulation of the Transcriptome Response to Nodavirus Challenge in European Sea Bass (Dicentrarchus labrax L.). Biology. 2020; 9(7):165. https://doi.org/10.3390/biology9070165
Chicago/Turabian StylePereiro, Patricia, Raquel Lama, Rebeca Moreira, Valentina Valenzuela-Muñoz, Cristian Gallardo-Escárate, Beatriz Novoa, and Antonio Figueras. 2020. "Potential Involvement of lncRNAs in the Modulation of the Transcriptome Response to Nodavirus Challenge in European Sea Bass (Dicentrarchus labrax L.)" Biology 9, no. 7: 165. https://doi.org/10.3390/biology9070165
APA StylePereiro, P., Lama, R., Moreira, R., Valenzuela-Muñoz, V., Gallardo-Escárate, C., Novoa, B., & Figueras, A. (2020). Potential Involvement of lncRNAs in the Modulation of the Transcriptome Response to Nodavirus Challenge in European Sea Bass (Dicentrarchus labrax L.). Biology, 9(7), 165. https://doi.org/10.3390/biology9070165





