Abstract
The ejaculate is heterogenous and sperm sub-populations with different kinematic patterns can be identified in various species. Nevertheless, although these sub-populations are statistically well defined, the statistical differences are not always relevant. The aim of the present study was to characterize kinematic sub-populations in sperm from two bovine species, and diluted with different commercial extenders, and to determine the statistical relevance of sub-populations through Bayesian analysis. Semen from 10 bulls was evaluated after thawing. An ISAS®v1 computer-assisted sperm analysis (CASA)-Mot system was employed with an image acquisition rate of 50 Hz and ISAS®D4C20 counting chambers. Sub-populations of motile spermatozoa were characterized using multivariate procedures such as principal components (PCs) analysis and clustering methods (k-means model). Four different sperm sub-populations were identified from three PCs that involved progressiveness, velocity, and cell undulatory movement. The proportions of the different sperm sub-populations varied with the extender used and in the two species. Despite a statistical difference (p < 0.05) between extenders, the Bayesian analysis confirmed that only one of them (Triladyl®) presented relevant differences in kinematic patterns when compared with Tris-EY and OptiXcell®. Extenders differed in the proportion of sperm cells in each of the kinematic sub-populations. Similar patterns were identified in Bos taurus and Bos indicus. Bayesian results indicate that sub-populations SP1, SP2, and SP3 were different for PC criteria and these differences were relevant. For velocity, linearity, and progressiveness, the SP4 did not show a relevant difference regarding the other sperm sub-populations. The classical approach of clustering or sperm subpopulation thus may not have a direct biological meaning. Therefore, the biological relevance of sperm sub-populations needs to be reevaluated.
1. Introduction
Fertility in cattle, as in other species, is a key determinant of productivity and, thus, understanding factors that affect fertility in beef and dairy herds is of utmost importance [1]. Most of the artificial insemination performed in dairy cattle is done with frozen-thawed semen, whereas for beef cattle it is mainly used in genetic stations for selection and in developed farms. Although it is well known that sire fertility is related to sperm motility and kinematic patterns [2] the effects of semen quality on reproductive efficiency in cattle are not yet fully understood [3]. Cryopreservation causes damages to spermatozoa resulting in lower fertilizing capacity [4]. Individual variation in semen freezability exists in most domestic species, including bulls [5] and pigs [6], and such variation may be explained, at least in part, by the patterns of motile sperm sub-populations (SPs) in the ejaculate [7].
The intrinsic variability of semen samples, as well as individual variation or differences arising as a result of treatments, can be studied by using computer-assisted sperm analysis (CASA)-Mot systems that allow for the generation of huge datasets consisting of kinematics trajectories from thousands of spermatozoa [8]. CASA systems have evolved rapidly during the last decade due to major innovations in technology [9] such as increases in computational memory [10,11], use of 3D technology [12], considerations of the effect of frame rate capture [13,14,15,16], improvements in camera acquisition [17], or new approaches for sperm cell tracking [11,18,19]. Current CASA-Mot systems can be used to analyze individual sperm kinematics more accurately and this information can be submitted to a multivariate procedure, such as cluster analysis, for an overview of distinct sperm patterns grouped into SPs or clusters [20].
Cluster analysis divides data into groups (clusters) that are meaningful, useful, or both. If the goal is to obtain meaningful groups, then the cluster should capture the natural structure of the data. In some cases, however, cluster analysis is only a useful starting point for other purposes, such as data summarization. Whether for understanding or utility, cluster analysis has long played an important role in a wide variety of fields [21]. In many areas of biology, scientists have devoted considerable effort to generate groups with hierarchical classification criteria. More recently, clustering has been employed to analyze the large amount of information that is obtained by using CASA-Mot systems. For example, clustering has been used to find groups of spermatozoa that have similar patterns. Thus, several studies in many species have identified the existence of different sperm SPs, defined by specific kinematic patterns obtained from CASA-Mot systems [13,22,23,24,25,26,27,28,29,30,31,32]
The aim of the present study was to assess whether statistical differences in kinematic patterns of bull sperm SPs were relevant according to Bayesian analyses. In addition, we examined possible differences in sperm SPs in bull semen of Bos taurus and Bos indicus diluted with different extenders.
2. Materials and Methods
This study was performed following ethical principles and with the approval of the Committee of Centro de Investigación y Desarrollo de la Agricultura Sostenible para el Trópico Húmedo at the Costa Rica Institute of Technology (CIDASTH-ITCR) according to Section 01/2019, article 1.0, DAGSC-074-2019.
2.1. Semen Collection and Processing
Bulls used in this study were of various breeds of Bos taurus and Bos indicus, with an average age of 5.7 ± 2.8 years. Straws of frozen semen were supplied by Avance Genético S.A. (Zapote, Costa Rica). Semen was collected with an artificial vagina, under a collection program of two ejaculates per week, which had no apparent changes in animal health or semen quality throughout the semen collection period. All bulls were routinely used for semen collection for commercial purposes. Data from 20 ejaculates obtained from 10 randomly selected bulls were employed in this study. The bulls had passed a standard breeding soundness evaluation, and had produced sperm with acceptable post-thaw characteristics (progressive motile sperm >60%) and fertility (non-return rate >65%).
Within 5 to 10 min of collection, the semen samples were assessed for volume, by using a conical tube graduated at 0.1 mL, gross motility, by placing 20 µL of fresh raw semen on a prewarmed slide at 37 °C, and concentration, employing a bovine photometer Accucell (IMV, L’Aigle, France) at 530 nm wavelength. The raw semen was diluted with Tris-citric acid-egg yolk extender (Tris-EY) and two commercial egg yolk extenders (OptiXcell ®-IMV, L’Aigle, France; Triladyl®, Minitube, Tiefenbach, Germany) to give a final live sperm concentration of 25 × 106 cells/straw. Diluted semen was cooled slowly to 4 °C at a linear rate of −0.3°C min−1 in a refrigerator. After cooling, equilibration took place over a period of 4–5 h at the same temperature.
Tris-EY extender was prepared by using 1.36 g citric acid (Fisher Scientific, UK), 2.42 g Tris (hydroxymethyl) aminomethane (Research Organics, USA), 1.0 g glucose (Scharlau, Spain), 14.0 mL glycerol (Merck, Germany), 20% egg yolk in 80 mL distilled water. Antibiotics, namely gentamicin sulfate (100 mg/mL; Sigma Chemical Co. St. Louis, MO, USA) was added to the Tris-EY extender. The commercial extenders were used following the manufacturer’s instructions.
Semen was packaged in 0.25 mL straws with an automatic filling and sealing machine (MRS 1, IMV Technologies, L’Aigle, France) and frozen by a programmable freezer, (Digitcool 5300, IMV, L’Aigle, France) with the following curve: 4 °C to −10 °C at 5 °C min−1, −10 °C to −100 °C at 40 °C min−1, −110 °C to −140 °C at 20 °C min−1, and then plunged into liquid nitrogen for storage. All samples were coded in such a way that the technician who performed the kinematics analysis did not know the number of the bull, the number of the ejaculate, or which ejaculate belonged to a particular bull.
2.2. Assessment of Sperm Variables
The cryopreserved semen samples were thawed in a water bath (37 °C, 60 s) and examined immediately after thawing. Disposable counting chambers for the analysis of motility and kinematics variables (ISAS®D4C20, Proiser R+D, S.L., Paterna, Spain) were used after pre-warming them to 37 °C. After thorough mixing of the diluted semen samples, 2.7 µL of diluted semen were placed in the counting chamber tracks by capillarity. Analyses were conducted with the CASA-Mot system ISAS®v1 (Integrated Semen Analysis System, Proiser R+D, Paterna, Spain) fitted with a video-camera (Proiser 782M, Proiser R+D), a frame rate of 50 frames per second (fps) and a final resolution of 768 × 576 pixels. The camera was attached to a microscope UB203 (UOP/Proiser R+D) with a 1X eyepiece and a 10× negative-phase contrast objective (AN 0.25), and an integrated heated stage maintained at 37 ± 0.5 °C.
2.3. Computerized Kinematic Analysis
CASA analyses were performed recording seven microscope fields with a total of at least 600 cells per sample. The CASA-Mot variables assessed in this study included: straight-line velocity (VSL, µm·s−1), corresponding to the straight line from the beginning to the end of the track; curvilinear velocity (VCL, µm·s−1), measured over the actual point-to-point track followed by the cell; average path velocity (VAP, µm·s−1), the average velocity over the smoothed cell path; amplitude of lateral head displacement (ALH, µm), defined as the maximum of the measured width of the head oscillation as the sperm swims; beat-cross frequency (BCF, Hz), defined as the frequency with which the actual track crosses the smoothed track in either direction; motility (%), the percentage of total motile cells and progressive motility (%), corresponding to spermatozoa swimming rapidly forward in a straight line (assessed as straightness index ≥45%; VAP ≥25 µm·s−1). Three progression ratios, expressed as percentages, were calculated from the velocity measurements described above: linearity of forward progression (LIN = VSL/VCL·100), straightness (STR = VSL/VAP·100), and wobble (WOB = VAP/VCL·100).
2.4. Statistical Analysis
The data obtained for the analysis of all sperm parameters were first assessed for normality and homoscedasticity by using Shapiro-Wilks and Levene tests. A normal probability plot was used to assess normal distribution. Multivariate procedures were performed to identify sperm SPs from the set of sperm motility data. All the values for kinematic variables were standardized to avoid any scale effect.
2.4.1. Multivariate Analysis
The first process carried out was a principal component analysis (PCA) of these data to derive a small number of linear combinations that still retained as much information as possible from the original variables. The number of principal components (PC) used in the next part of the analysis was determined from the Kaiser criterion, namely selecting only those with an eigenvalue (variance extracted of each PC) >1. Furthermore, Bartlett’s sphericity test and the KMO (Kaiser-Meyer-Olkin) were performed [33]. As a rotation method, the varimax method with Kaiser normalization was used [34].
The second process was conducted to perform a non-hierarchical analysis with the k-means model that uses Euclidean distances from the quantitative variables after standardization of these data, so the cluster centers were the means of the observations assigned to each cluster [35]. The multivariate k-means cluster analysis was made to classify the spermatozoa into a reduced number of SPs (clusters) according to their kinematic variables. In the final process, to determine the optimal number of clusters, the final centroids were clustered hierarchically using the Ward method [36]. Thus, the clustering procedure enables for the identification of sperm SPs because each cluster contributed to a final cluster formed by the spermatozoa linked to the centroids. The analysis of variance (ANOVA) and χ2-test procedures were applied to evaluate statistical differences in the distributions of observations (individual spermatozoa) within SPs and then a generalized linear model (GLM) procedure was used to determine the effects of the breed and extender type on the mean kinematic variable values defining the different sperm SPs (i.e., the cluster centers). Differences between means were analyzed by Bonferroni test. Results are presented as mean ± standard error of the mean (SEM). Statistical significance was considered at p < 0.05. All data were analyzed using IBM SPSS package, version 23.0 for Windows (SPSS Inc., Chicago, IL, USA).
2.4.2. Bayesian Analysis
Differences in sperm kinematic patterns were estimated with a model including the effect of extender and species as a permanent effect. Male number was included as a random effect. All analyses were performed using Bayesian methodology. The posterior mean of the difference between extenders (D), the highest posterior density region at 95% (HPD95%) and the probability of the difference being positive when D > 0 or negative when D < 0 (P0) were calculated. Bounded uniform priors were used for all effects. Residuals were a priori normally distributed with mean 0 and variance σ2e. We considered one third of the standard deviation (s.d.) of a trait as a relevant value (R), and we also calculated the probability of relevance (PR; i.e., the probability of the difference being greater than R when D > 0 or lower than R when D < 0). The priors for the variances were also bounded uniform. Features of the marginal posterior distributions for all unknowns were estimated using Gibbs sampling. Convergence was tested using the Z criterion of Geweke [37] and Monte Carlo sampling errors were computed using time series procedures described in [38]. The Rabbit program, developed by the Institute for Animal Science and Technology (Valencia, Spain), was used for all procedures.
3. Results
3.1. Overall Motility Parameters
The mean (± SEM) total motility (%) of samples cryopreserved in the different extenders was 51.50 ± 8.38 (OptiXcell®), 53.83 ± 3.78 (Triladyl®) and 58.38 ± 4.63 (Tris-EY) with an overall range of 18–79%. The progressive motility of sperm (%) for OptiXcell®, Triladyl® and Tris-EY was, respectively, 36.33 ± 6.81, 40.83 ± 2.44 and 37.75 ± 4.39. Average total motility (%, mean ± SEM) for Bos taurus and Bos indicus bulls was 58.33 ± 3.05 and 49.88 ± 6.45, respectively. The progressive motility (%) for the two species was 41.92 ± 2.09 (Bos taurus) and 32.75 ± 5.56 (Bos indicus).
3.2. Subpopulation Structure
After multivariate cluster analysis (k-means model), four motile sperm SPs with different kinematic patterns were identified. There was extender effect (p < 0.05) on SPs distribution of the percentages motile- and progressively motile spermatozoa (Figure 1). Summary data for kinematics of the SPs are presented in Table 1. They can be summarized as follows: subpopulation 1 (SP1) included sperm with medium velocity (intermediate values of VCL, VSL, and VAP) and linear and progressive motility (inferred from intermediate LIN and STR values). This population included 20.02% of the total motile sperm. Subpopulation 2 (SP2) contained active, fast, linear, and progressive sperm, as indicated by the greater value of VCL, VSL, and VAP, together with the lower values of LIN and STR, and intermediate value of BCF. Moreover, the ALH value was the lowest seen in all SPs, indicating movement with few undulatory characteristics. About 28.29% of the total motile sperm were assigned to this subpopulation. Subpopulation 3 (SP3) included 19.14% of the total sperm and was represented by slow motile sperm with the lowest velocity but non-undulatory as revealed by values of VAP, VCL, VSL, and BCF. This population had lesser progressive motility than the other SPs as indicated by LIN and ALH values. Subpopulation 4 (SP4) contained 32.54% of the total motile sperm population, and these cells had the highest motility and progressive motility, but movements were undulatory, as indicated by the high values of VCL, VSL, VAP, ALH, and BCF, together with the highest values of LIN, STR, and WOB.
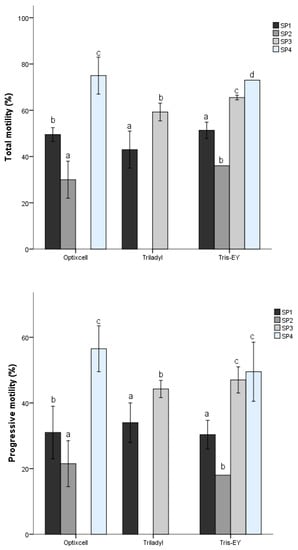
Figure 1.
Distribution of total motility and progressiveness of sperm within sub-populations (SPs) between extenders. Different letters (a–d) indicate differences within SPs between extenders. p < 0.05.

Table 1.
Kinematic variables (mean ± SEM) of the four sperm sub-populations (SPs) defined at 60 min post-thawing bull semen samples.
3.3. Sperm Kinematics within Sub-Populations for Different Extenders
Results from PCA revealed three PCs in all extenders (Table 2). PC1 in OptiXcell®, Triladyl® and Tris-EY was referred to as “progressiveness” and was represented by LIN, STR, and WOB. The larger eigenvectors corresponded to LIN (OptiXcell®: 0.931; Triladyl®: 0.977), or WOB (Tris-EY: 0.942). PC2 showed a “velocity” component, with high values for VCL and ALH. The greater effect was due to VCL in all extenders (OptiXcell®: 0.960; Triladyl®: 0.915; Tris-EY: 0.899). PC3, represented by STR and BCF, was named “undulatory movement”. It mainly related to BCF (OptiXcell®: 0.801; Triladyl®: 0.952), or STR (Tris-EY: 0.844). These results indicate that sperm progressiveness has a relatively greater effect on the total variance than the other variables (Table 2).

Table 2.
Eigenvectors of principal components (PCs) * for kinematic variables of bull sperm in three commercial extenders.
There were differences between sperm SPs within each extender (p < 0.05) with VCL and LIN exhibiting the highest differences between the sub-populations. The SP distribution was not the same for each extender. The highest VCL values were seen in SP2 for OptiXcell® (165.50 ± 1.20 µm·s−1), and SP3 for Triladyl® (168.80 ± 1.27 µm·s−1) and Tris-EY (168.04 ± 0.97 µm·s−1). For LIN, the highest values were in SP2 in OptiXcell® (87.16 ± 0.53%), SP1 in Triladyl® (85.91 ± 0.61%) and SP2 in Tris-EY (88.45 ± 0.57%). When each sperm subpopulation was compared between extenders, kinematics differences (p < 0.05) were found. The variables with the greatest differences between extenders for all SPs were VCL, LIN and WOB (Table 3).

Table 3.
Kinematic variables (means ± SEM) of motile bull sperm SPs in different commercial extenders.
3.4. Bayesian Analysis of Sperm Sub-Populations
Bayesian analysis of the data showed that VCL, VAP, LIN and BCF exhibited relevant differences between Tris-EY and Triladyl®, that VCL and VAP presented relevant differences between Tris-EY and OptiXcell®, and that only WOB showed relevant differences between Triladyl ® and OptiXcell®. In the other cases, despite differences being significant (p < 0.05) between all extenders (VSL) or at least two extenders (LIN, WOB), they were not relevant (Table 4).

Table 4.
Kinematic bull sperm variables and estimated marginal posterior distributions of differences between commercial extenders Tris-EY, Triladyl® and OptiXcell®.
Bayesian analysis of the data (posterior distribution) when comparing Bos taurus and Bos indicus showed that even when differences (p < 0.05) were identified ensuing frequentist statistics (confidence distribution), these differences between kinematic variables were considered non-relevant (Table 5).

Table 5.
Descriptive parameters and features of the estimated marginal posterior distributions of differences for kinematic bull sperm variables between Bos taurus and Bos indicus.
When the relevance of differences between sperm sub-populations was analyzed, the Bayesian analyses of data showed relevant differences between the following comparisons of SPs: SP1−SP2 (for VSL and LIN); SP1−SP3 (all kinematic variables except VCL, WOB and BCF); SP1−SP4 (for VSL); SP2−SP3 (all variables except VSL); SP2−SP4 (for STR, WOB, ALH, and BCF). Thus, the sub-populations SP1, SP2 and SP3 showed differences (p < 0.05) for most of the kinematic variables when using the classical approach, and these differences were also revealed by the Bayesian approach. On the other hand, the SP3−SP4 comparison did not show relevant differences for any kinematic variable. Even though subpopulation SP4 showed differences regarding SP2 or SP3, such differences should be regarded as non-relevant for VCL, VSL, VAP, LIN, WOB, or BCF (Table 6).

Table 6.
Kinematic bull sperm variables and estimated marginal posterior distribution of differences between motile sperm SPs.
3.5. Distribution of the Sperm Subpopulation
Analysis of the proportion of sperm cells in each subpopulation for the three extenders revealed differences between extenders and SPs for each extender. The sperm SPs were unevenly distributed for each extender. The sperm sub-populations with highest number of cells were associated with a specific extender [(OptiXcell®: SP2, 36.06%; Triladyl®: SP4, 35.62%; Tris-EY: SP4, 34.66%). On the other hand, sub-populations with lower percentages of cells in Tris-EY and OptiXcell® were associated with SP1 (12.00%) and SP3 (13.60%), respectively (Table 7).

Table 7.
Percentage of sperm cells from bulls Bos taurus and Bos indicus in each kinematic subpopulation characterized in semen diluted in three commercial extenders.
4. Discussion
Cryopreservation is frequently used in animal production and there are several sources of variation in the success of semen survival during this procedure [4]. During freezing and thawing variation relates, for instance, to the effect of extender, which can influence the kinematic patterns of spermatozoa. Breed and inter-individual variation post-thawing can relate to differences in plasma membrane properties in response to freezing and thawing. Cryopreservation can result in alterations in the semen quality due to increases of oxidative stress which in turn can generate changes in sperm motility and kinematic patterns, DNA fragmentation, alterations in timing of capacitation or the acrosome reaction [39]. The evaluation of these changes is very difficult, if not impossible, when employing a classical analysis of semen quality and, thus, the use of CASA technology is important for the improvement of sperm quality assessments [2,40,41,42]. CASA-Mot systems have demonstrated greater accuracy in motility evaluation compared with subjective methods [10]. Moreover, these systems provide abundant information on many kinematic variables for each spermatozoon [8,43] that can be used as a basis for the analysis of the heterogeneity of sperm populations. However, a strategy used by artificial insemination centers is to compensate damage to spermatozoa by increasing the number of motile sperm cells in the insemination doses, and this can mask the relevance of sperm SPs.
In bulls, it has been suggested that sperm SPs with greater percentages of fast yet nonlinear spermatozoa seem to have greater fertilization capacity [25]. Other work indicates that fast and nonlinear sperm movements are in fact specific of hyperactivated spermatozoa [44]. Results of other studies suggest that the sperm SP containing fast and linear spermatozoa is the one with a higher likelihood of sperm-oocyte interaction [28]. Despite these attempts at linking SP structure and function, including sperm fertility, the abundant data provided by CASA-Mot systems for multivariate procedures, and the resulting SP, are still insufficient to categorize an ejaculate in relation to its quality. Moreover, the classification of SPs through classical clustering procedures remains uncertain and even controversial. This relates to the observation that the multivariate procedures of clustering will find several kinematics patterns in the data set, even if there are no natural clusters in the data [45].
The multivariate approach of PCs and cluster analysis conducted for several species underscores that ejaculates are nevertheless heterogeneous in that they contain spermatozoa with different motility and kinematic patterns [17,22,31,46,47,48,49,50,51,52,53]. In the present study, we identified four different sperm SPs described from three PCs that represented velocity, progressiveness, and cell undulatory movement. The proportions of the different SPs varied with the extender used and among species. The cluster of fast and linear sperm did not follow a consistent pattern between SPs; in any case, the samples diluted with Triladyl® showed the greatest kinematic activity. In our study, despite differences (p < 0.05) between extenders, the Bayesian analysis revealed that only Triladyl® presented relevant differences in comparison to Tris-EY and OptiXcell® for these kinematic patterns. Our results indicate that subpopulation distribution was also different between and within bovine species. In contrast, other studies have indicated that the sperm subpopulation distribution was not different within species [5]. On the other hand, for the two bull species (Bos taurus, Bos indicus), the overall distribution of sperm subpopulation structure showed little variation among individuals.
The application of biostatistical analysis tools to the study of sperm kinematic variables revealed the existence of sperm SPs in the ejaculate [9,13,23,54,55,56,57,58,59], but their significance and biological relevance does not appear to be entirely clear [8,20,27,31,60]. Our findings of varying biological relevance of sperm SPs in the ejaculate strengthen the idea that a specific sub-populational structure, depicted by spermatozoa with different kinematic patterns, must be revised. Other authors have suggested some sperm SPs can be affected by a treatment [26] or by specific processes (e.g., freezing-thawing) indicating that some sperm SPs may have biological relevance [25] whereas others, co-existing in the same sample, may not [8,26,61,62]. Our Bayesian results indicated that SP1, SP2, and SP3 were different and that these differences were relevant. The biological relevance of these SPs may thus lead to an invalidation of the classical definition of the sperm cluster or subpopulation. Relevance may relate to fertility. The variability of spermatozoa in an ejaculate can correlate with the variability of fertility among males [63], and lesser motility can also associate with lower fertility rates [2]. If velocity is a principal component that explains the kinematic variance of sperm SPs, and velocity can relate to fertility, we thus need to further clarify the biological relevance of these SPs.
Sperm SPs should then be assessed using a criterion of utility in relation to biological importance. To our knowledge, this is the first study in which an assessment of statistically relevance of ejaculate SPs has been carried out. Distinction between species, and between breeds, should be taken into account and implemented in this type of analysis. Previous studies have considered these distinction and comparisons carried out between species with regards to kinematics [16] and morphology have identified differences [64]. The fact that there are differences between species suggest that new approaches may be required when managing the ejaculates. In any case, these differences must be relevant from the biological point of view and statistical differences may not be sufficient as criterion. There is thus a need to further examine the issue of biological relevance in the analysis of sperm SPs and to develop means to explore the biological meaning of data sets of clustered cells deriving from automated semen analyses.
5. Conclusions
Current approaches for the analysis of sperm kinematic SPs may not have a direct biological meaning and therefore, the biological relevance of sperm SPs needs to be reevaluated.
Author Contributions
Conceptualization, A.V. E.R.S.R., and C.S.; methodology, L.V., A.V.; software, L.V.; validation, L.V., V.B.; formal analysis, A.V.; investigation, L.V., V.B., A.V.; resources, A.V.; data curation, A.V.; writing—original draft preparation, A.V.; writing—review and editing, E.R.S.R.; A.V.; visualization, E.R.S.R., C.S., A.V.; supervision, A.V.; project administration, A.V.; funding acquisition, A.V. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Fundación para el Fomento y Promoción de la Investigación y Transferencia de Tecnología Agropecuaria de Costa Rica (FITTACORI) and Costa Rica Institute of Technology (Vice-Chancellor’s office of Research and Extension; VIE (Vicerrectoría de investigación y Extensión); Project 5402-2151-1014-). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
The authors thank the Costa Rica Institute of Technology (ITCR) and the Fundación para el Fomento y Promoción de la Investigación y Transferencia de Tecnología Agropecuaria de Costa Rica (FITTACORI) for financing this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ferraz, M.A.M.M.; Morató, R.; Yeste, M.; Arcarons, N.; Pena, A.I.; Tamargo, C.; Hidalgo, C.O.; Muiño, R.; Mogas, T. Evaluation of sperm subpopulation structure in relation to invitro sperm-oocyte interaction of frozen-thawed semen from Holstein bulls. Theriogenology 2014, 81, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Yániz, J.; Silvestre, M.; Santolaria, P.; Soler, C. CASA-Mot in mammals: An update. Reprod. Fertil. Dev. 2018, 30, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, H. Laboratory semen assessment and prediction of fertility: Still Utopia? Reprod. Domest. Anim. 2003, 38, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.F. The causes of reduced fertility with cryopreserved semen. Anim. Reprod. Sci. 2000, 60–61, 481–492. [Google Scholar] [CrossRef]
- Valverde, A.; Arenán, H.; Sancho, M.; Contell, J.; Yániz, J.; Fernández, A.; Soler, C. Morphometry and subpopulation structure of Holstein bull spermatozoa: Variations in ejaculates and cryopreservation straws. Asian J. Androl. 2016, 18, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Alborcia, M.J.; Valverde, A.; Parrilla, I.; Vazquez, J.M.; Martinez, E.A.; Roca, J. Detrimental effects of non-functional spermatozoa on the freezability of functional spermatozoa from Boar Ejaculate. PLoS ONE 2012, 7, e36550. [Google Scholar] [CrossRef]
- Davis, R.O.; Drobnis, E.Z.; Overstreet, J.W. Application of multivariate cluster, discriminate function, and stepwise regression analyses to variable selection and predictive modeling of sperm cryosurvival. Fertil. Steril. 1995, 63, 1051–1057. [Google Scholar] [CrossRef]
- Martínez-Pastor, F.; Tizado, E.; Garde, J.; Anel, L.; de Paz, P. Statistical Series: Opportunities and challenges of sperm motility subpopulation analysis. Theriogenology 2011, 75, 783–795. [Google Scholar] [CrossRef] [PubMed]
- van der Horst, G.; Maree, L.; du Plessis, S. Current perspectives of CASA applications in diverse mammalian spermatozoa. Reprod. Fertil. Dev. 2018, 30, 875–888. [Google Scholar] [CrossRef]
- Amann, R.; Waberski, D. Computer-assisted sperm analysis (CASA): Capabilities and potential developments. Theriogenology 2014, 81, 5–17.e3. [Google Scholar] [CrossRef]
- Gallagher, M.T.; Smith, D.J.; Kirkman-Brown, J.C. CASA: Tracking the past and plotting the future. Reprod. Fertil. Dev. 2018, 30, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Soler, C.; Picazo-Bueno, J.; Micó, V.; Valverde, A.; Bompart, D.; Blasco, F.J.; Lvarez, J.G.; García-Molina, A. Effect of counting chamber depth on the accuracy of lensless microscopy for the assessment of boar sperm motility. Reprod. Fertil. Dev. 2018, 30, 924–934. [Google Scholar] [CrossRef] [PubMed]
- Valverde, A.; Madrigal-Valverde, M.; Caldeira, C.; Bompart, D.; Núñez de Murga, J.; Arnau, S.; Soler, C. Effect of frame rate capture frequency on sperm kinematic parameters and subpopulation structure definition in boars, analyzed with a CASA-Mot system. Reprod. Domest. Anim. 2019, 54, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Castellini, C.; Dal Bosco, A.; Ruggeri, S.; Collodel, G. What is the best frame rate for evaluation of sperm motility in different species by computer-assisted sperm analysis? Fertil. Steril. 2011, 96, 24–27. [Google Scholar] [CrossRef]
- Bompart, D.; García-Molina, A.; Valverde, A.; Caldeira, C.; Yániz, J.; Núñez de Murga, M.; Soler, C. CASA-Mot technology: How results are affected by the frame rate and counting chamber. Reprod. Fertil. Dev. 2018, 30, 810–819. [Google Scholar] [CrossRef]
- Bompart, D.; Vázquez, R.; Gómez, R.; Valverde, A.; Roldán, E.; García-Molina, A.; Soler, C. Combined effects of type and depth of counting chamber, and rate of image frame capture, on bull sperm motility and kinematics. Anim. Reprod. Sci. 2019, 209, 106169. [Google Scholar] [CrossRef]
- Caldeira, C.; Hernández-Ibáñez, S.; Valverde, A.; Martin, P.; Herranz-Jusdado, J.G.; Gallego, V.; Asturiano, J.F.; Dzyuba, B.; Pšenička, M.; Soler, C. Standardization of sperm motility analysis by using CASA-Mot for Atlantic salmon (Salmo salar), European eel (Anguilla anguilla) and Siberian sturgeon (Acipenser baerii). Aquaculture 2019, 502, 223–231. [Google Scholar] [CrossRef]
- Shi, L.; Nascimento, J.; Berns, M.; Botvinick, E. Computer-based tracking of single sperm. J. Biomed. Opt. 2006, 11, 054009. [Google Scholar] [CrossRef]
- Urbano, L.; Masson, P.; VerMilyea, M.; Kam, M. Automatic Tracking and Motility Analysis of Human Sperm in Time-Lapse Images. IEEE Trans. Med. Imaging 2017, 36, 792–801. [Google Scholar] [CrossRef]
- Quintero-Moreno, A.; Rigau, T.; Rodríguez-Gil, J. Multivariate Cluster Analysis Regression Procedures as Tools to Identify Motile Sperm Subpopulations in Rabbit Semen and to Predict Semen Fertility and Litter Size. Reprod. Domest. Anim. 2007, 42, 312–319. [Google Scholar] [CrossRef]
- Tan, P.; Steinbach, M.; Karpatne, A.; Kumar, V. Cluster Analysis: Basic Concepts and Algorithms. In Introduction to Data Mining (2nd Edition) (What’s New in Computer Science); Pearson Education India: New, Delhi, India, 2018; pp. 125–146. ISBN 9780133128901. [Google Scholar]
- Abaigar, T.; Holt, W.; Harrison, R.; del Barrio, G. Sperm subpopulations in boar (Sus scrofa) and gazelle (Gazella dama mhorr) semen as revealed by pattern analysis of computer-assisted motility assessments. Biol. Reprod. 1999, 60, 32–41. [Google Scholar] [CrossRef]
- Dorado, J.; Molina, I.; Muñoz-Serrano, A.; Hidalgo, M. Identification of sperm subpopulations with defined motility characteristics in ejaculates from Florida goats. Theriogenology 2010, 74, 795–804. [Google Scholar] [CrossRef]
- García-Herreros, M. Sperm subpopulations in avian species: A comparative study between the rooster (Gallus domesticus) and Guinea fowl (Numida meleagris). Asian J. Androl. 2016, 18, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Ibanescu, I.; Siuda, M.; Bollwein, H. Motile sperm subpopulations in bull semen using different clustering approaches—Associations with flow cytometric sperm characteristics and fertility. Anim. Reprod. Sci. 2020, 215, 106329. [Google Scholar] [CrossRef] [PubMed]
- Ledesma, A.; Zalazar, L.; Fernández-Alegre, E.; Hozbor, F.; Cesari, A.; Martínez-Pastor, F. Seminal plasma proteins modify the distribution of sperm subpopulations in cryopreserved semen of rams with lesser fertility. Anim. Reprod. Sci. 2017, 184, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Muiño, R.; Tamargo, C.; Hidalgo, C.O.; Peña, A. Identification of sperm subpopulations with defined motility characteristics in ejaculates from Holstein bulls: Effects of cryopreservation and between-bull variation. Anim. Reprod. Sci. 2008, 109, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Peña, A.; Adán, S.; Quintela, L.; Becerra, J.; Herradón, P. Relationship between motile sperm subpopulations identified in frozen-thawed dog semen samples and their ability to bind to the zona pellucida of canine oocytes. Reprod. Domest. Anim. 2018, 53, 14–22. [Google Scholar] [CrossRef]
- Quintero-Moreno, A.; Miró, J.; Teresa Rigau, A.; Rodríguez-Gil, J.E. Identification of sperm subpopulations with specific motility characteristics in stallion ejaculates. Theriogenology 2003, 59, 1973–1990. [Google Scholar] [CrossRef]
- Ramió, L.; Rivera, M.M.; Ramírez, A.; Concha, I.I.; Peña, A.; Rigau, T.; Rodríguez-Gil, J.E. Dynamics of motile-sperm subpopulation structure in boar ejaculates subjected to “in vitro” capacitation and further “in vitro” acrosome reaction. Theriogenology 2008, 69, 501–512. [Google Scholar] [CrossRef]
- Rubio-Guillén, J.; González, D.; Garde, J.; Esteso, M.; Fernández-Santos, M.; Rodríguez-Gíl, J.; Madrid-Bury, N.; Quintero-Moreno, A. Effects of Cryopreservation on Bull Spermatozoa Distribution in Morphometrically Distinct Subpopulations. Reprod. Domest. Anim. 2007, 42, 354–357. [Google Scholar] [CrossRef]
- Yániz, J.; Palacín, I.; Vicente-Fiel, S.; Sánchez-Nadal, J.; Santolaria, P. Sperm population structure in high and low field fertility rams. Anim. Reprod. Sci. 2015, 156, 128–134. [Google Scholar] [CrossRef]
- Spencer, N. Essentials of Multivariate Data Analysis; Chapman and Hall/CRC Press: New York, NY, USA, 2013; ISBN 9781466584785. [Google Scholar]
- Kaiser, H.F. The varimax criterion for analytic rotation in factor analysis. Psychometrika 1958, 23, 187–200. [Google Scholar] [CrossRef]
- Kaufman, L.; Rousseeuw, P. Finding Groups in Data: An. Introduction to Cluster Analysis; Wiley: Hoboken, NJ, USA, 2005; ISBN 9780471735786. [Google Scholar]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Sorensen, D.; Gianola, D. Likelihood, Bayesian, and MCMC Methods in Quantitative Genetics; Statistics for Biology and Health; Springer: New York, NY, USA, 2002; ISBN 978-0-387-95440-0. [Google Scholar]
- Geyer, C. Practical Markov Chain Monte Carlo. Stat. Sci. 1992, 7, 473–483. [Google Scholar] [CrossRef]
- Walczak-Jedrzejowska, R.; Wolski, J.; Slowikowska-Hilczer, J. The role of oxidative stress and antioxidants in male fertility. Cent. Eur. J. Urol. 2013, 66, 60–67. [Google Scholar] [CrossRef]
- Peña, F.; Saravia, F.; García-Herreros, M.; Núñez-Martínez, I.; Tapia, J.; Johannisson, A.; Wallgren, M.; Rodríguez-Martínez, H. Identification of sperm morphometric subpopulations in two different portions of the boar ejaculate and its relation to postthaw quality. J. Androl. 2005, 26, 716–723. [Google Scholar] [CrossRef]
- Núñez-Martínez, I.; Moran, J.; Peña, F. Sperm indexes obtained using computer-assisted morphometry provide a forecast of the freezability of canine sperm. Int. J. Androl. 2007, 30, 182–189. [Google Scholar] [CrossRef]
- Mortimer, D.; Mortimer, S.; Horst, G. The future of computer-aided sperm analysis. Asian J. Androl. 2015, 17, 545. [Google Scholar] [CrossRef]
- Valverde, A.; Castro-Morales, O.; Madrigal-Valverde, M.; Soler, C. Sperm kinematics and morphometric subpopulations analysis with CASA systems: A review. Rev. Biol. Trop. 2019, 67, 1473–1487. [Google Scholar] [CrossRef]
- Kathiravan, P.; Kalatharan, J.; Karthikeya, G.; Rengarajan, K.; Kadirvel, G. Objective Sperm Motion Analysis to Assess Dairy Bull Fertility Using Computer-Aided System—A Review. Reprod. Domest. Anim. 2011, 46, 165–172. [Google Scholar] [CrossRef]
- Altman, N.; Krzywinski, M. Points of Significance: Clustering. Nat. Methods 2017, 14, 545–546. [Google Scholar] [CrossRef]
- Chantler, E.; Abraham-Peskir, J.; Roberts, C. Consistent presence of two normally distributed sperm subpopulations within normozoospermic human semen: A kinematic study. Int. J. Androl. 2004, 27, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Flores, E.; Taberner, E.; Rivera, M.M.; Peña, A.; Rigau, T.; Miró, J.; Rodríguez-Gil, J.E. Effects of freezing/thawing on motile sperm subpopulations of boar and donkey ejaculates. Theriogenology 2008, 70, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.A.; Montanero, J.; Calero, R.; Roy, T.J. Identification of sperm subpopulations with defined motility characteristics in ejaculates from Ile de France rams. Anim. Reprod. Sci. 2011, 129, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Yániz, J.; Palacín, I.; Caycho, K.; Soler, C.; Silvestre, M.; Santolaria, P. Determining the relationship between bull sperm kinematic subpopulations and fluorescence groups using an integrated sperm quality analysis technique. Reprod. Fertil. Dev. 2018, 30, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Ferrusola, C.; Macías García, B.; Suárez Rama, V.; Gallardo-Bolaños, J.; González-Fernández, L.; Tapia, J.; Rodríguez-Martinez, H.; Peña, F. Identification of Sperm Subpopulations in Stallion Ejaculates: Changes after Cryopreservation and Comparison with Traditional Statistics. Reprod. Domest. Anim. 2009, 44, 419–423. [Google Scholar] [CrossRef]
- Valverde, A.; Madrigal-Valverde, M.; Castro-Morales, O.; Gadea-Rivas, A.; Johnston, S.; Soler, C. Kinematic and head morphometric characterisation of spermatozoa from the Brown Caiman (Caiman crocodilus fuscus). Anim. Reprod. Sci. 2019, 207, 9–20. [Google Scholar] [CrossRef]
- Soler, C.; Valverde, A.; Bompart, D.; Fereidounfar, S.; Sancho, M.; Yániz, J.; Garcia-Molina, A.; Korneenko-Zhilyaev, Y. New methods of semen analysis by casa. Sel’skokhozyaistvennaya Biol. (Agric. Biol.) 2017, 52, 232–241. [Google Scholar] [CrossRef]
- Soler, C.; Contell, J.; Bori, L.; Sancho, M.; García-Molina, A.; Valverde, A.; Segarvall, J. Sperm kinematic, head morphometric and kinetic-morphometric subpopulations in the blue fox (Alopex lagopus). Asian J. Androl. 2017, 19, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Vásquez, F.; Soler, C.; Camps, P.; Valverde, A.; García-Molina, A. Spermiogram and sperm head morphometry assessed by multivariate cluster analysis results during adolescence (12–18 years) and the effect of varicocele. Asian J. Androl. 2016, 18, 824–830. [Google Scholar] [CrossRef] [PubMed]
- Ibănescu, I.; Leiding, C.; Bollwein, H. Cluster analysis reveals seasonal variation of sperm subpopulations in extended boar semen. J. Reprod. Dev. 2018, 64, 33–39. [Google Scholar] [CrossRef]
- Gallego, V.; Vílchez, M.C.; Peñaranda, D.S.; Pérez, L.; Herráez, M.P.; Asturiano, J.F.; Martínez-Pastor, F. Subpopulation pattern of eel spermatozoa is affected by post-activation time, hormonal treatment and the thermal regimen. Reprod. Fertil. Dev. 2015, 27, 529–543. [Google Scholar] [CrossRef]
- Holt, C.; Holt, W.; Moore, H. Choice of operating conditions to minimize sperm subpopulation sampling bias in the assessment of boar semen by computer-assisted semen analysis. J. Androl. 1996, 17, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Pastor, F.; Garcia-Macias, V.; Alvarez, M.; Herraez, P.; Anel, L.; de Paz, P. Sperm Subpopulations in Iberian Red Deer Epididymal Sperm and Their Changes Through the Cryopreservation Process1. Biol. Reprod. 2005, 72, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Valverde, A.; Madrigal-Valverde, M.; Lotz, J.; Bompart, D.; Soler, C. Effect of video capture time on sperm kinematic parameters in breeding boars. Livest. Sci. 2019, 220, 52–56. [Google Scholar] [CrossRef]
- Soler, C.; Alambiaga, A.; Martí, M.A.; García-Molina, A.; Valverde, A.; Contell, J.; Campos, M. Dog sperm head morphometry: Its diversity and evolution. Asian J. Androl. 2017, 19, 149–153. [Google Scholar] [CrossRef]
- Holt, W.; Moore, H.; Hillier, S. Computer-assisted measurement of sperm swimming speed in human semen: Correlation of results with in vitro fertilization assays**Supported by Medical Research Council grant G8203374SB. Fertil. Steril. 1985, 44, 112–119. [Google Scholar] [CrossRef]
- Harrison, R.A.P. Capacitation mechanisms, and the role of capacitation as seen in eutherian mammals. In Proceedings of the Reproduction, Fertility and Development; CSIRO: Canberra, Australia, 1996; Volume 8, pp. 581–594. [Google Scholar]
- Holt, W.; Van Look, K. Concepts in sperm heterogeneity, sperm selection and sperm competition as biological foundations for laboratory tests of semen quality. Reproduction 2004, 127, 527–535. [Google Scholar] [CrossRef]
- Beletti, M.; Costa, L.; Viana, M. A comparison of morphometric characteristics of sperm from fertile Bos taurus and Bos indicus bulls in Brazil. Anim. Reprod. Sci. 2005, 85, 105–116. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).