STAT3 Pathway in Gastric Cancer: Signaling, Therapeutic Targeting and Future Prospects
Abstract
1. Introduction
2. STAT3 Signaling Pathway and Its Role in Pathological Events
3. Targeting STAT3 in Clinical Trials: A Focus on Cancer Therapy
4. Search Strategy
5. STAT3 and Gastric Cancer
5.1. MicroRNA-Mediated Regulation of STAT3
5.2. Drug-Mediated Regulation of STAT3
5.3. LncRNA-Mediated Regulation of STAT3
5.4. Other Molecular Signaling Pathways Regulate STAT3
6. STAT3 as an Oncogene Factor in Gastric Cancer
7. Conclusions and Remarks
Funding
Conflicts of Interest
Abbreviations
| GC | gastric cancer |
| H. pylori | Helicobacter pylori |
| EBV | Epstein–Barr virus |
| lncRNAs | long non-coding RNAs |
| miR | microRNA |
| EMT | epithelial-to-mesenchymal transition |
| STAT | signal transducer and activator of transcription |
| SH2 | Src homology-2 |
| TAD | transcription activation domain |
| JAKs | Janus kinases |
| SOCS | suppressor of cytokine signaling |
| PIAS | protein inhibitor of activated STAT |
| SLP-2 | stomatin-like protein 2 |
| CTM | Chinese traditional medicine |
| VEGF | vascular endothelial growth factor |
| MMPs | matrix metalloproteinases |
| ECM | extracellular matrix |
| ROS | reactive oxygen species |
| ER | endoplasmic reticulum |
| APG | apigetrin |
| TXN | troxerutin |
| DHA | docosahexaenoid acid |
| PPAR-γ | peroxisome proliferator-activated receptor gamma |
| SIRT | sirtuin |
| STMN | stathmin |
| S1PR1 | sphingosine-1-phosphate receptor |
| YB-1 | Y-box binding protein-1 |
| NFIB | nuclear factor I/B |
| TME | tumor microenvironment |
| CAMs | cancer-associated macrophages |
| TAMs | tumor-associated macrophages |
| CAFs | cancer-associated fibroblasts |
| MSK1 | mitogen- and stress-activated protein kinase 1 |
| EZH2 | enhancer of zeste homolog 2 |
References
- Arai, H.; Nakajima, T.E. Recent Developments of Systemic Chemotherapy for Gastric Cancer. Cancers 2020, 12, 1100. [Google Scholar] [CrossRef] [PubMed]
- Magnelli, L.; Schiavone, N.; Staderini, F.; Biagioni, A.; Papucci, L. MAP Kinases Pathways in Gastric Cancer. Int. J. Mol. Sci. 2020, 21, 2893. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Rafiei, H.; Mohammadinejad, R.; Farkhondeh, T.; Samarghandian, S. Wnt-regulating microRNAs role in gastric cancer malignancy. Life Sci. 2020, 250, 117547. [Google Scholar] [CrossRef] [PubMed]
- Colquhoun, A.; Arnold, M.; Ferlay, J.; Goodman, K.; Forman, D.; Soerjomataram, I. Global patterns of cardia and non-cardia gastric cancer incidence in 2012. Gut 2015, 64, 1881–1888. [Google Scholar] [CrossRef]
- De Martel, C.; Forman, D.; Plummer, M. Gastric cancer: Epidemiology and risk factors. Gastroenterol. Clin. 2013, 42, 219–240. [Google Scholar] [CrossRef]
- Yoo, J.Y.; Cho, H.J.; Moon, S.; Choi, J.; Lee, S.; Ahn, C.; Yoo, K.-Y.; Kim, I.; Ko, K.-P.; Lee, J.E. Pickled Vegetable and Salted Fish Intake and the Risk of Gastric Cancer: Two Prospective Cohort Studies and a Meta-Analysis. Cancers 2020, 12, 996. [Google Scholar] [CrossRef]
- Riihimäki, M.; Hemminki, A.; Sundquist, K.; Sundquist, J.; Hemminki, K. Metastatic spread in patients with gastric cancer. Oncotarget 2016, 7, 52307. [Google Scholar] [CrossRef]
- Baj, J.; Brzozowska, K.; Forma, A.; Maani, A.; Sitarz, E.; Portincasa, P. Immunological Aspects of the Tumor Microenvironment and Epithelial-Mesenchymal Transition in Gastric Carcinogenesis. Int. J. Mol. Sci. 2020, 21, 2544. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.G.; Tang, X.L.; Huang, X.S.; Zhou, L.; Hao, Y.G.; Zheng, Y.F. Long noncoding RNA LINC00511 promoted cell proliferation and invasion via regulating miR-124-3p/EZH2 pathway in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4232–4245. [Google Scholar] [CrossRef]
- Chen, P.; Qian, X.K.; Zhang, Y.F.; Sun, X.G.; Shi, X.J.; Gao, Y.S. KLF5 promotes proliferation in gastric cancer via regulating p21 and CDK4. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4224–4231. [Google Scholar] [CrossRef]
- El-Guindy, D.M.; Wasfy, R.E.; Abdel Ghafar, M.T.; Ali, D.A.; Elkady, A.M. Oct4 expression in gastric carcinoma: Association with tumor proliferation, angiogenesis and survival. J. Egypt. Natl. Cancer Inst. 2019, 31, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hu, X.; Kuang, J.; Liao, J.; Yuan, Q. LncRNA DRAIC inhibits proliferation and metastasis of gastric cancer cells through interfering with NFRKB deubiquitination mediated by UCHL5. Cell. Mol. Biol. Lett. 2020, 25, 29. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, J.; Fang, L. LncRNA WT1-AS over-expression inhibits non-small cell lung cancer cell stemness by down-regulating TGF-beta1. BMC Pulm. Med. 2020, 20, 113. [Google Scholar] [CrossRef]
- Sun, C.B.; Wang, H.Y.; Han, X.Q.; Liu, Y.N.; Wang, M.C.; Zhang, H.X.; Gu, Y.F.; Leng, X.G. LINC00511 promotes gastric cancer cell growth by acting as a ceRNA. World J. Gastrointest. Oncol. 2020, 12, 394–404. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y.; You, Q.; Fu, H.; Zhao, X.; Lu, K.; Yan, R.; Yang, D. LncRNA PTCSC3 Alleviates the Postoperative Distant Recurrence of Gastric Cancer by Suppression of lncRNA HOXA11-AS. Cancer Manag. Res. 2020, 12, 2623–2629. [Google Scholar] [CrossRef]
- Loh, C.-Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef] [PubMed]
- Mohammadinejad, R.; Biagioni, A.; Arunkumar, G.; Shapiro, R.; Chang, K.-C.; Sedeeq, M.; Taiyab, A.; Hashemabadi, M.; Pardakhty, A.; Mandegary, A.; et al. EMT signaling: Potential contribution of CRISPR/Cas gene editing. Cell. Mol. Life Sci. 2020. [Google Scholar] [CrossRef]
- Cheng, J.-T.; Wang, L.; Wang, H.; Tang, F.-R.; Cai, W.-Q.; Sethi, G.; Xin, H.-W.; Ma, Z. Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells 2019, 8, 1178. [Google Scholar] [CrossRef]
- Ma, B.; Ma, J.; Yang, Y.; He, X.; Pan, X.; Wang, Z.; Qian, Y. Effects of miR-330-3p on Invasion, Migration and EMT of Gastric Cancer Cells by Targeting PRRX1-Mediated Wnt/beta-Catenin Signaling Pathway. OncoTargets Ther. 2020, 13, 3411–3423. [Google Scholar] [CrossRef]
- Nakayama, Y.; Mimura, K.; Kua, L.F.; Okayama, H.; Min, A.K.T.; Saito, K.; Hanayama, H.; Watanabe, Y.; Saito, M.; Momma, T.; et al. Immune suppression caused by PD-L2 expression on tumor cells in gastric cancer. Gastric Cancer J. Int. Gastric Cancer Assoc. Jpn. Gastric Cancer Assoc. 2020. [Google Scholar] [CrossRef]
- Liu, H.T.; Ma, R.R.; Lv, B.B.; Zhang, H.; Shi, D.B.; Guo, X.Y.; Zhang, G.H.; Gao, P. LncRNA-HNF1A-AS1 functions as a competing endogenous RNA to activate PI3K/AKT signalling pathway by sponging miR-30b-3p in gastric cancer. Br. J. Cancer 2020, 122, 1825–1836. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Lu, F.; Zhang, L.; Wang, G.; Geng, R.; Miao, Y. HBXIP Regulates Gastric Cancer Glucose Metabolism and Malignancy Through PI3K/AKT and p53 Signaling. Oncotargets Ther. 2020, 13, 3359–3374. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, H.; Dong, P.; Xie, G.; Zhou, Y.; Ma, Y.; Yuan, X.; Yang, J.; Han, L.; Chen, L.; et al. KIF23 activated Wnt/beta-catenin signaling pathway through direct interaction with Amer1 in gastric cancer. Aging 2020, 12, 8372–8396. [Google Scholar] [CrossRef]
- Zhou, C.; Guo, Z.; Xu, L.; Jiang, H.; Sun, P.; Zhu, X.; Mu, X. PFND1 Predicts Poor Prognosis of Gastric Cancer and Promotes Cell Metastasis by Activating the Wnt/beta-Catenin Pathway. OncoTargets Ther. 2020, 13, 3177–3186. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J.; Kim, Y.J.; Sohn, S.H.; Kim, B.; Sul, H.J.; Kim, H.S.; Zang, D.Y. Tivantinib inhibits the VEGF signaling pathway and induces apoptosis in gastric cancer cells with c-MET or VEGFA amplification. Investig. New Drugs 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Liu, D.; Jiang, Z.; Li, C.; Chen, L.; Xia, Y.; Liu, D.; Yao, Q.; Wang, D. Chrysin Induced Cell Apoptosis and Inhibited Invasion Through Regulation of TET1 Expression in Gastric Cancer Cells. OncoTargets Ther. 2020, 13, 3277–3287. [Google Scholar] [CrossRef]
- Jin, L.; Ma, X.M.; Wang, T.T.; Yang, Y.; Zhang, N.; Zeng, N.; Bai, Z.G.; Yin, J.; Zhang, J.; Ding, G.Q.; et al. Psoralen Suppresses Cisplatin-Mediated Resistance and Induces Apoptosis of Gastric Adenocarcinoma by Disruption of the miR196a-HOXB7-HER2 Axis. Cancer Manag. Res. 2020, 12, 2803–2827. [Google Scholar] [CrossRef]
- Mohan, C.D.; Rangappa, S.; Preetham, H.D.; Nayak, S.C.; Gupta, V.K.; Basappa, S.; Sethi, G.; Rangappa, K.S. Targeting STAT3 signaling pathway in cancer by agents derived from Mother Nature. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Ma, J.H.; Qin, L.; Li, X. Role of STAT3 signaling pathway in breast cancer. Cell Commun. Signal. CCS 2020, 18, 33. [Google Scholar] [CrossRef]
- Qin, J.; Shen, X.; Zhang, J.; Jia, D. Allosteric inhibitors of the STAT3 signaling pathway. Eur. J. Med. Chem. 2020, 190, 112122. [Google Scholar] [CrossRef]
- Wu, C.J.; Sundararajan, V.; Sheu, B.C.; Huang, R.Y.; Wei, L.H. Activation of STAT3 and STAT5 Signaling in Epithelial Ovarian Cancer Progression: Mechanism and Therapeutic Opportunity. Cancers 2019, 12, 24. [Google Scholar] [CrossRef]
- Orlova, A.; Wagner, C.; de Araujo, E.D.; Bajusz, D.; Neubauer, H.A.; Herling, M.; Gunning, P.T.; Keseru, G.M.; Moriggl, R. Direct Targeting Options for STAT3 and STAT5 in Cancer. Cancers 2019, 11, 1930. [Google Scholar] [CrossRef] [PubMed]
- Mohrherr, J.; Uras, I.Z.; Moll, H.P.; Casanova, E. STAT3: Versatile Functions in Non-Small Cell Lung Cancer. Cancers 2020, 12, 1107. [Google Scholar] [CrossRef] [PubMed]
- Zouein, F.A.; Altara, R.; Chen, Q.; Lesnefsky, E.J.; Kurdi, M.; Booz, G.W. Pivotal importance of STAT3 in protecting the heart from acute and chronic stress: New advancement and unresolved issues. Front. Cardiovasc. Med. 2015, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Chai, E.Z.P.; Shanmugam, M.K.; Arfuso, F.; Dharmarajan, A.; Wang, C.; Kumar, A.P.; Samy, R.P.; Lim, L.H.; Wang, L.; Goh, B.C. Targeting transcription factor STAT3 for cancer prevention and therapy. Pharmacol. Ther. 2016, 162, 86–97. [Google Scholar] [CrossRef]
- Timofeeva, O.A.; Tarasova, N.I.; Zhang, X.; Chasovskikh, S.; Cheema, A.K.; Wang, H.; Brown, M.L.; Dritschilo, A. STAT3 suppresses transcription of proapoptotic genes in cancer cells with the involvement of its N-terminal domain. Proc. Natl. Acad. Sci. USA 2013, 110, 1267–1272. [Google Scholar] [CrossRef]
- Dutta, P.; Sabri, N.; Li, J.; Li, W.X. Role of STAT3 in lung cancer. Jak-Stat 2014, 3, e999503. [Google Scholar] [CrossRef]
- Yu, H.; Kortylewski, M.; Pardoll, D. Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 2007, 7, 41–51. [Google Scholar] [CrossRef]
- Levy, D.E.; Darnell, J. Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002, 3, 651–662. [Google Scholar] [CrossRef]
- Avalle, L.; Camporeale, A.; Camperi, A.; Poli, V. STAT3 in cancer: A double edged sword. Cytokine 2017, 98, 42–50. [Google Scholar] [CrossRef]
- Alexander, W.S. Suppressors of cytokine signalling (SOCS) in the immune system. Nat. Rev. Immunol. 2002, 2, 410–416. [Google Scholar] [CrossRef]
- Shuai, K.; Liu, B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 2005, 5, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Qu, C.K. Protein tyrosine phosphatases in the JAK/STAT pathway. Front. Biosci. A J. Virtual Libr. 2008, 13, 4925–4932. [Google Scholar] [CrossRef] [PubMed]
- Krebs, D.L.; Hilton, D.J. SOCS proteins: Negative regulators of cytokine signaling. Stem Cells 2001, 19, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Kadye, R.; Stoffels, M.; Fanucci, S.; Mbanxa, S.; Prinsloo, E. A STAT3 of Addiction: Adipose Tissue, Adipocytokine Signalling and STAT3 as Mediators of Metabolic Remodelling in the Tumour Microenvironment. Cells 2020, 9, 1043. [Google Scholar] [CrossRef] [PubMed]
- Brachet-Botineau, M.; Polomski, M.; Neubauer, H.A.; Juen, L.; Hédou, D.; Viaud-Massuard, M.-C.; Prié, G.; Gouilleux, F. Pharmacological Inhibition of Oncogenic STAT3 and STAT5 Signaling in Hematopoietic Cancers. Cancers 2020, 12, 240. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Chen, X.; Chen, L.; Wan, F.; Chen, K.; Sun, Y.; Zhu, X. STAT3 signaling in ovarian cancer: A potential therapeutic target. J. Cancer 2020, 11, 837. [Google Scholar] [CrossRef]
- Hu, F.; Li, G.; Huang, C.; Hou, Z.; Yang, X.; Luo, X.; Feng, Y.; Wang, G.; Hu, J.; Cao, Z. The autophagy-independent role of BECN1 in colorectal cancer metastasis through regulating STAT3 signaling pathway activation. Cell Death Dis. 2020, 11, 304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Song, M.; Kundu, J.K.; Lee, M.H.; Liu, Z.Z. PIM Kinase as an Executional Target in Cancer. J. Cancer Prev. 2018, 23, 109–116. [Google Scholar] [CrossRef]
- Sun, Z.; Zeng, L.; Zhang, M.; Zhang, Y.; Yang, N. PIM1 inhibitor synergizes the anti-tumor effect of osimertinib via STAT3 dephosphorylation in EGFR-mutant non-small cell lung cancer. Ann. Transl. Med. 2020, 8, 366. [Google Scholar] [CrossRef]
- Liu, Q.; Li, A.; Wang, L.; He, W.; Zhao, L.; Wu, C.; Lu, S.; Ye, X.; Zhao, H.; Shen, X.; et al. Stomatin-like Protein 2 Promotes Tumor Cell Survival by Activating the JAK2-STAT3-PIM1 Pathway, Suggesting a Novel Therapy in CRC. Mol. Ther. Oncolytics 2020, 17, 169–179. [Google Scholar] [CrossRef]
- Shang, R.; Wang, M.; Dai, B.; Du, J.; Wang, J.; Liu, Z.; Qu, S.; Yang, X.; Liu, J.; Xia, C.; et al. Long noncoding RNA SLC2A1-AS1 regulates aerobic glycolysis and progression in hepatocellular carcinoma via inhibiting the STAT3/FOXM1/GLUT1 pathway. Mol. Oncol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Zhang, S.; Yin, J.; Shen, Y.X.; Wang, H.; Chen, X.S.; Tang, H. LINC01535 promotes proliferation and inhibits apoptosis in esophageal squamous cell cancer by activating the JAK/STAT3 pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3694–3700. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.M.; Chan, B.D.; Wong, W.Y.; Qu, Z.; Chan, M.S.; Leung, T.W.; Lin, Y.; Mok, D.K.; Chen, S.; Tai, W.C. Anti-cancer Activity of Centipeda minima Extract in Triple Negative Breast Cancer via Inhibition of AKT, NF-kappaB, and STAT3 Signaling Pathways. Front. Oncol. 2020, 10, 491. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Maryam, A.; Saleem, M.Z.; Shakir, H.A.; Qazi, J.I.; Li, Y.; Ma, T. Brevilin A induces ROS-dependent apoptosis and suppresses STAT3 activation by direct binding in human lung cancer cells. J. Cancer 2020, 11, 3725–3735. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Ren, Y.; Guo, X.; Wang, L.; Zhang, Q.; Li, X.; Wu, X.; Meng, Z.; Xu, K. Downregulation of SETD7 promotes migration and invasion of lung cancer cells via JAK2/STAT3 pathway. Int. J. Mol. Med. 2020, 45, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Yu, W.; Zhang, Y.; Cao, X.; Han, L.; Hu, H.; Wang, C. LNK promotes the growth and metastasis of triple negative breast cancer via activating JAK/STAT3 and ERK1/2 pathway. Cancer Cell Int. 2020, 20, 124. [Google Scholar] [CrossRef]
- Feng, J.; Jiang, W.; Liu, Y.; Huang, W.; Hu, K.; Li, K.; Chen, J.; Ma, C.; Sun, Z.; Pang, X. Blocking STAT3 by pyrvinium pamoate causes metabolic lethality in KRAS-mutant lung cancer. Biochem. Pharmacol. 2020, 177, 113960. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, W.; Ma, D.; Xiong, J.; Kuang, X.; Zhang, S.; Fang, Q.; Wang, J. Heme oxygenase-1 inhibition mediates Gas6 to enhance bortezomib-sensitivity in multiple myeloma via ERK/STAT3 axis. Aging 2020, 12, 6611–6629. [Google Scholar] [CrossRef]
- DeVaux, R.S.; Ropri, A.S.; Grimm, S.L.; Hall, P.A.; Herrera, E.O.; Chittur, S.V.; Smith, W.P.; Coarfa, C.; Behbod, F.; Herschkowitz, J.I. Long noncoding RNA BHLHE40-AS1 promotes early breast cancer progression through modulating IL-6/STAT3 signaling. J. Cell. Biochem. 2020, 121, 3465–3478. [Google Scholar] [CrossRef]
- Arnold, K.M.; Opdenaker, L.M.; Flynn, N.J.; Appeah, D.K.; Sims-Mourtada, J. Radiation induces an inflammatory response that results in STAT3-dependent changes in cellular plasticity and radioresistance of breast cancer stem-like cells. Int. J. Radiat. Biol. 2020, 96, 434–447. [Google Scholar] [CrossRef]
- Song, H.; Luo, Q.; Deng, X.; Ji, C.; Li, D.; Munankarmy, A.; Jian, W.; Zhao, J.; Fang, L. VGLL4 interacts with STAT3 to function as a tumor suppressor in triple-negative breast cancer. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Choi, H.S.; Kim, S.L.; Lee, D.S. The PAK1-Stat3 Signaling Pathway Activates IL-6 Gene Transcription and Human Breast Cancer Stem Cell Formation. Cancers 2019, 11, 1527. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Li, J.; Fu, L.; Gai, J.; Guan, J.; Li, Q. SIRT4 enhances the sensitivity of ER-positive breast cancer to tamoxifen by inhibiting the IL-6/STAT3 signal pathway. Cancer Med. 2019, 8, 7086–7097. [Google Scholar] [CrossRef]
- Zheng, T.; Ma, G.; Tang, M.; Li, Z.; Xu, R. IL-8 Secreted from M2 Macrophages Promoted Prostate Tumorigenesis via STAT3/MALAT1 Pathway. Int. J. Mol. Sci. 2018, 20, 98. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Wang, C.; Yu, Z.; He, D.; Yu, K.; Liu, Y.; Wang, S. MiR-17 Regulates Prostate Cancer Cell Proliferation and Apoptosis Through Inhibiting JAK-STAT3 Signaling Pathway. Cancer Biother. Radiopharm. 2018, 33, 103–109. [Google Scholar] [CrossRef]
- Soutto, M.; Chen, Z.; Bhat, A.A.; Wang, L.; Zhu, S.; Gomaa, A.; Bates, A.; Bhat, N.S.; Peng, D.; Belkhiri, A.; et al. Activation of STAT3 signaling is mediated by TFF1 silencing in gastric neoplasia. Nat. Commun. 2019, 10, 3039. [Google Scholar] [CrossRef]
- Anand, V.; Khandelwal, M.; Appunni, S.; Gupta, N.; Seth, A.; Singh, P.; Mathur, S.; Sharma, A. CD44 splice variant (CD44v3) promotes progression of urothelial carcinoma of bladder through Akt/ERK/STAT3 pathways: Novel therapeutic approach. J. Cancer Res. Clin. Oncol. 2019, 145, 2649–2661. [Google Scholar] [CrossRef]
- Yuan, L.; Ye, J.; Fan, D. The B7-H4 gene induces immune escape partly via upregulating the PD-1/Stat3 pathway in non-small cell lung cancer. Hum. Immunol. 2020, 81, 254–261. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, H.H.; Ryu, K.D.; Kim, M.; Ko, D.; Chung, K.S.; Hassan, A.H.E.; Lee, S.H.; Lee, J.Y.; Lee, K.T. KCP10043F Represses the Proliferation of Human Non-Small Cell Lung Cancer Cells by Caspase-Mediated Apoptosis via STAT3 Inactivation. J. Clin. Med. 2020, 9, 704. [Google Scholar] [CrossRef]
- Yun, H.H.; Kim, S.; Kuh, H.J.; Lee, J.H. Downregulation of BIS sensitizes A549 cells for digoxin-mediated inhibition of invasion and migration by the STAT3-dependent pathway. Biochem. Biophys. Res. Commun. 2020, 524, 643–648. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Ichikawa, T.; Kurozumi, K.; Otani, Y.; Fujimura, A.; Fujii, K.; Tomita, Y.; Hattori, Y.; Uneda, A.; Tsuboi, N.; et al. Annexin A2-STAT3-Oncostatin M receptor axis drives phenotypic and mesenchymal changes in glioblastoma. Acta Neuropathol. Commun. 2020, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.W.; Lee, P.M.; Bamodu, O.A.; Su, Y.K.; Fong, I.H.; Yeh, C.T.; Chien, M.H.; Kan, I.H.; Lin, C.M. Enhanced Hsa-miR-181d/p-STAT3 and Hsa-miR-181d/p-STAT5A Ratios Mediate the Anticancer Effect of Garcinol in STAT3/5A-Addicted Glioblastoma. Cancers 2019, 11, 1888. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Lv, A.; Deng, Q.; Zhang, G.; Hu, X.; Cui, H. TROP2 promotes the proliferation and metastasis of glioblastoma cells by activating the JAK2/STAT3 signaling pathway. Oncol. Rep. 2019, 41, 753–764. [Google Scholar] [CrossRef]
- Liu, Y.S.; Hsu, J.W.; Lin, H.Y.; Lai, S.W.; Huang, B.R.; Tsai, C.F.; Lu, D.Y. Bradykinin B1 receptor contributes to interleukin-8 production and glioblastoma migration through interaction of STAT3 and SP-1. Neuropharmacology 2019, 144, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Kurzrock, R.; Kim, Y.; Woessner, R.; Younes, A.; Nemunaitis, J.; Fowler, N.; Zhou, T.; Schmidt, J.; Jo, M.; et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 2015, 7, 314ra185. [Google Scholar] [CrossRef]
- Sonnenblick, A.; Salgado, R.; Brohée, S.; Zahavi, T.; Peretz, T.; Van den Eynden, G.; Rouas, G.; Salmon, A.; Francis, P.A.; Di Leo, A.; et al. p-STAT3 in luminal breast cancer: Integrated RNA-protein pooled analysis and results from the BIG 2-98 phase III trial. Int. J. Oncol. 2018, 52, 424–432. [Google Scholar] [CrossRef]
- Monnien, F.; Zaki, H.; Borg, C.; Mougin, C.; Bosset, J.F.; Mercier, M.; Arbez-Gindre, F.; Kantelip, B. Prognostic value of phosphorylated STAT3 in advanced rectal cancer: A study from 104 French patients included in the EORTC 22921 trial. J. Clin. Pathol. 2010, 63, 873–878. [Google Scholar] [CrossRef]
- Bu, X.; Zhao, C.; Wang, W.; Zhang, N. GRIM-19 inhibits the STAT3 signaling pathway and sensitizes gastric cancer cells to radiation. Gene 2013, 512, 198–205. [Google Scholar] [CrossRef]
- Anglesio, M.S.; George, J.; Kulbe, H.; Friedlander, M.; Rischin, D.; Lemech, C.; Power, J.; Coward, J.; Cowin, P.A.; House, C.M.; et al. IL6-STAT3-HIF signaling and therapeutic response to the angiogenesis inhibitor sunitinib in ovarian clear cell cancer. Clin. Cancer Res. An. J. Am. Assoc. Cancer Res. 2011, 17, 2538–2548. [Google Scholar] [CrossRef]
- Torres-Roca, J.F.; DeSilvio, M.; Mora, L.B.; Khor, L.Y.; Hammond, E.; Ahmad, N.; Jove, R.; Forman, J.; Lee, R.J.; Sandler, H.; et al. Activated STAT3 as a correlate of distant metastasis in prostate cancer: A secondary analysis of Radiation Therapy Oncology Group 86-10. Urology 2007, 69, 505–509. [Google Scholar] [CrossRef]
- Delyon, J.; Chevret, S.; Jouary, T.; Dalac, S.; Dalle, S.; Guillot, B.; Arnault, J.P.; Avril, M.F.; Bedane, C.; Bens, G.; et al. STAT3 Mediates Nilotinib Response in KIT-Altered Melanoma: A Phase II Multicenter Trial of the French Skin Cancer Network. J. Investig. Dermatol. 2018, 138, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Lee, S.H.; Han, S.W.; Kim, M.J.; Kim, T.M.; Kim, T.Y.; Heo, D.S.; Yuasa, M.; Yanagihara, Y.; Bang, Y.J. Phase I Study of OPB-31121, an Oral STAT3 Inhibitor, in Patients with Advanced Solid Tumors. Cancer Res. Treat. J. Korean Cancer Assoc. 2015, 47, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Jamali, Z.; Taheri-Anganeh, M.; Shabaninejad, Z.; Keshavarzi, A.; Taghizadeh, H.; Razavi, Z.S.; Mottaghi, R.; Abolhassan, M.; Movahedpour, A.; Mirzaei, H. Autophagy regulation by microRNAs: Novel insights into osteosarcoma therapy. IUBMB Life 2020. [Google Scholar] [CrossRef] [PubMed]
- Pourhanifeh, M.H.; Mahjoubin-Tehran, M.; Shafiee, A.; Hajighadimi, S.; Moradizarmehri, S.; Mirzaei, H.; Asemi, Z. MicroRNAs and exosomes: Small molecules with big actions in multiple myeloma pathogenesis. IUBMB Life 2020, 72, 314–333. [Google Scholar] [CrossRef] [PubMed]
- Nahand, J.S.; Karimzadeh, M.R.; Nezamnia, M.; Fatemipour, M.; Khatami, A.; Jamshidi, S.; Moghoofei, M.; Taghizadieh, M.; Hajighadimi, S.; Shafiee, A.; et al. The role of miR-146a in viral infection. IUBMB Life 2020, 72, 343–360. [Google Scholar] [CrossRef]
- Javandoost, E.; Firoozi-Majd, E.; Rostamian, H.; Khakpoor-Koosheh, M.; Mirzaei, H.R. Role of microRNAs in Chronic Lymphocytic Leukemia Pathogenesis. Curr. Med. Chem. 2020, 27, 282–297. [Google Scholar] [CrossRef]
- Savardashtaki, A.; Shabaninejad, Z.; Movahedpour, A.; Sahebnasagh, R.; Mirzaei, H.; Hamblin, M.R. miRNAs derived from cancer-associated fibroblasts in colorectal cancer. Epigenomics 2019, 11, 1627–1645. [Google Scholar] [CrossRef]
- Nahand, J.S.; Taghizadeh-boroujeni, S.; Karimzadeh, M.; Borran, S.; Pourhanifeh, M.H.; Moghoofei, M.; Bokharaei-Salim, F.; Karampoor, S.; Jafari, A.; Asemi, Z.; et al. microRNAs: New prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J. Cell. Physiol. 2019, 234, 17064–17099. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z.; Li, B.; Xia, Z.; Wang, X.; Xiu, Y.; Zhang, Z.; Chen, C.; Song, H.; Li, W.; et al. The inhibition of miR-17-5p promotes cortical neuron neurite growth via STAT3/GAP-43 pathway. Mol. Biol. Rep. 2020, 47, 1795–1802. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, X.M.; Feng, Y. MiR-769-5p inhibits cancer progression in oral squamous cell carcinoma by directly targeting JAK1/STAT3 pathway. Neoplasma 2020, 67, 528–536. [Google Scholar] [CrossRef]
- Quero, L.; Tiaden, A.N.; Hanser, E.; Roux, J.; Laski, A.; Hall, J.; Kyburz, D. miR-221-3p Drives the Shift of M2-Macrophages to a Pro-Inflammatory Function by Suppressing JAK3/STAT3 Activation. Front. Immunol. 2019, 10, 3087. [Google Scholar] [CrossRef] [PubMed]
- Guoping, M.; Ran, L.; Yanru, Q. miR-143 Inhibits Cell Proliferation of Gastric Cancer Cells Through Targeting GATA6. Oncol. Res. 2018, 26, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Du, F.; Sun, L.; Li, T.; Li, T.; Min, Y.; Nie, A.; Wang, X.; Geng, L.; Lu, Y.; et al. miR-143 and miR-145 inhibit gastric cancer cell migration and metastasis by suppressing MYO6. Cell Death Dis. 2017, 8, e3101. [Google Scholar] [CrossRef]
- Zhang, Q.; Feng, Y.; Liu, P.; Yang, J. MiR-143 inhibits cell proliferation and invasion by targeting DNMT3A in gastric cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39, 1010428317711312. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wan, X.; Zhao, X.; Song, Z.; Xu, Z.; Tao, Y.; Sun, C. MicroRNA-143 suppresses the proliferation and metastasis of human gastric cancer cells via modulation of STAT3 expression. Am. J. Transl. Res. 2020, 12, 867–874. [Google Scholar] [PubMed]
- Xiong, J.; Tu, Y.; Feng, Z.; Li, D.; Yang, Z.; Huang, Q.; Li, Z.; Cao, Y.; Jie, Z. Epigenetics mechanisms mediate the miR-125a/BRMS1 axis to regulate invasion and metastasis in gastric cancer. OncoTargets Ther. 2019, 12, 7513–7525. [Google Scholar] [CrossRef]
- Li, G.; Ao, S.; Hou, J.; Lyu, G. Low expression of miR-125a-5p is associated with poor prognosis in patients with gastric cancer. Oncol. Lett. 2019, 18, 1483–1490. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, S.; Guo, K.; Huang, H.; Qi, S.; Yao, J.; Zhang, Z. miR-125a restrains cell migration and invasion by targeting STAT3 in gastric cancer cells. OncoTargets Ther. 2019, 12, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Zabaleta, J. MicroRNA: A bridge from H. pylori infection to gastritis and gastric cancer development. Front. Genet. 2012, 3, 294. [Google Scholar] [CrossRef]
- Isomoto, H.; Matsushima, K.; Inoue, N.; Hayashi, T.; Nakayama, T.; Kunizaki, M.; Hidaka, S.; Nakayama, M.; Hisatsune, J.; Nakashima, M. Interweaving microRNAs and proinflammatory cytokines in gastric mucosa with reference to H. pylori infection. J. Clin. Immunol. 2012, 32, 290–299. [Google Scholar] [CrossRef]
- Belair, C.; Darfeuille, F.; Staedel, C. Helicobacter pylori and gastric cancer: Possible role of microRNAs in this intimate relationship. Clin. Microbiol. Infect. 2009, 15, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Link, A.; Kupcinskas, J.; Wex, T.; Malfertheiner, P. Macro-role of microRNA in gastric cancer. Dig. Dis. 2012, 30, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Z.; Gao, C.; Chen, P.; Chen, J.; Liu, W.; Xiao, S.; Lu, H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab. Investig. 2008, 88, 1358–1366. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Liu, K.; Xie, M.; Xing, Y.; Xi, T. miR-375 inhibits Helicobacter pylori-induced gastric carcinogenesis by blocking JAK2-STAT3 signaling. Cancer Immunol. Immunother. 2014, 63, 699–711. [Google Scholar] [CrossRef]
- Shi, H.; Chen, X.; Jiang, H.; Wang, X.; Yu, H.; Sun, P.; Sui, X. miR-148a suppresses cell invasion and migration in gastric cancer by targeting DNA methyltransferase 1. Oncol. Lett. 2018, 15, 4944–4950. [Google Scholar] [CrossRef]
- Li, B.; Wang, W.; Li, Z.; Chen, Z.; Zhi, X.; Xu, J.; Li, Q.; Wang, L.; Huang, X.; Wang, L.; et al. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett. 2017, 410, 212–227. [Google Scholar] [CrossRef]
- Yu, B.; Lv, X.; Su, L.; Li, J.; Yu, Y.; Gu, Q.; Yan, M.; Zhu, Z.; Liu, B. MiR-148a Functions as a Tumor Suppressor by Targeting CCK-BR via Inactivating STAT3 and Akt in Human Gastric Cancer. PLoS ONE 2016, 11, e0158961. [Google Scholar] [CrossRef]
- Hashemi Goradel, N.; Ghiyami-Hour, F.; Jahangiri, S.; Negahdari, B.; Sahebkar, A.; Masoudifar, A.; Mirzaei, H. Nanoparticles as new tools for inhibition of cancer angiogenesis. J. Cell. Physiol. 2018, 233, 2902–2910. [Google Scholar] [CrossRef]
- Mashreghi, M.; Azarpara, H.; Bazaz, M.R.; Jafari, A.; Masoudifar, A.; Mirzaei, H.; Jaafari, M.R. Angiogenesis biomarkers and their targeting ligands as potential targets for tumor angiogenesis. J. Cell. Physiol. 2018, 233, 2949–2965. [Google Scholar] [CrossRef]
- Wang, M.; Zeng, Q.; Li, Y.; Imani, S.; Xie, D.; Li, Y.; Han, Y.; Fan, J. Bevacizumab combined with apatinib enhances antitumor and anti-angiogenesis effects in a lung cancer model in vitro and in vivo. J. Drug Target. 2020, 1–29. [Google Scholar] [CrossRef]
- Yadav, A.S.; Radharani, N.N.V.; Gorain, M.; Bulbule, A.; Shetti, D.; Roy, G.; Baby, T.; Kundu, G.C. RGD functionalized chitosan nanoparticle mediated targeted delivery of raloxifene selectively suppresses angiogenesis and tumor growth in breast cancer. Nanoscale 2020, 12, 10664–10684. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Ma, Y.F.; Zhang, X.R.; Li, Y.; Zhao, H.H.; Han, S.G. Circ_0056618 promoted cell proliferation, migration and angiogenesis through sponging with miR-206 and upregulating CXCR4 and VEGF-A in colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4190–4202. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Li, B.; Han, X.; Zhang, X.; Dang, M.; Wang, H.; Du, F.; Zeng, X.; Guo, C. Soluble receptor for advanced glycation end-products promotes angiogenesis through activation of STAT3 in myocardial ischemia/reperfusion injury. Apoptosis Int. J. Program. Cell Death 2020, 25, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Yao, F.; Zhou, L.; Liang, G.; Song, X.; Jiang, G.; Zhou, M.; Zhang, L. Yu Ping Feng San Exert Anti-Angiogenesis Effects through the Inhibition of TSLP-STAT3 Signaling Pathways in Hepatocellular Carcinoma. Evid. Based Complement. Altern. Med. ECAM 2019, 2019, 1947156. [Google Scholar] [CrossRef]
- Zhu, F.; Wang, K.B.; Rui, L. STAT3 Activation and Oncogenesis in Lymphoma. Cancers 2019, 12, 19. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, J.; Zhi, X.; Xie, K.; Wang, W.; Li, Z.; Zhu, Y.; Yang, L.; Xu, H.; Xu, Z. miR-874 functions as a tumor suppressor by inhibiting angiogenesis through STAT3/VEGF-A pathway in gastric cancer. Oncotarget 2015, 6, 1605–1617. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cai, K.; Li, H.X.; Li, P.P.; Guo, Z.J.; Yang, Y. MicroRNA-449b-3p inhibits epithelial-mesenchymal transition by targeting IL-6 and through the JAK2/STAT3 signaling pathway in non-small cell lung cancer. Exp. Ther. Med. 2020, 19, 2527–2534. [Google Scholar] [CrossRef]
- Sun, X.; Ma, J.; Chen, Q.; Hou, Z.; Luo, X.; Wang, G.; Wang, J.; Hu, J.; Cao, Z. SIX4 promotes metastasis through STAT3 activation in breast cancer. Am. J. Cancer Res. 2020, 10, 224–236. [Google Scholar]
- Sun, H.; Zhang, Z.; Zhang, T.; Geng, H.; Xie, D.; Wang, Y.; Ding, D.; Zhang, T.; Yu, D. Resveratrol Reverses Cigarette Smoke-Induced Urocystic Epithelial-Mesenchymal Transition via Suppression of STAT3 Phosphorylation in SV-HUC-1-Immortalized Human Urothelial Cells. Oncotargets Ther. 2019, 12, 10227–10237. [Google Scholar] [CrossRef]
- Tao, Y.; Yang, S.; Wu, Y.; Fang, X.; Wang, Y.; Song, Y.; Han, T. MicroRNA-216a inhibits the metastasis of gastric cancer cells by targeting JAK2/STAT3-mediated EMT process. Oncotarget 2017, 8, 88870–88881. [Google Scholar] [CrossRef]
- Leng, X.; Liu, G.; Wang, S.; Song, J.; Zhang, W.; Zhang, X.; Rong, L.; Ma, Y.; Song, F. LINC01272 Promotes Migration and Invasion of Gastric Cancer Cells via EMT. Oncotargets Ther. 2020, 13, 3401–3410. [Google Scholar] [CrossRef]
- Jin, G.H.; Shi, Y.; Tian, Y.; Cao, T.T.; Mao, Y.; Tang, T.Y. HMGA1 accelerates the malignant progression of gastric cancer through stimulating EMT. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3642–3647. [Google Scholar] [CrossRef]
- Zhao, L.; Niu, H.; Liu, Y.; Wang, L.; Zhang, N.; Zhang, G.; Liu, R.; Han, M. LOX inhibition downregulates MMP-2 and MMP-9 in gastric cancer tissues and cells. J. Cancer 2019, 10, 6481–6490. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Dong, Y.; Yang, R.; Wei, L. LINC00619 restricts gastric cancer progression by preventing microRNA-224-5p-mediated inhibition of OPCML. Arch. Biochem. Biophys. 2020, 108390. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wu, J.; Tang, C. Effect of isofebrifugine on the proliferation and invasion of human gastric cancer cells via MMP. Cell. Mol. Biol. (Noisy-Le-Grandfrance) 2020, 66, 27–31. [Google Scholar] [CrossRef]
- Ma, D.H.; Li, B.S.; Liu, J.J.; Xiao, Y.F.; Yong, X.; Wang, S.M.; Wu, Y.Y.; Zhu, H.B.; Wang, D.X.; Yang, S.M. miR-93-5p/IFNAR1 axis promotes gastric cancer metastasis through activating the STAT3 signaling pathway. Cancer Lett. 2017, 408, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.J.; Chang, M.S.; Kim, D.H.; Kim, W.; Koo, B.K.; Yun, S.C.; Kim, S.H.; Kim, Y.S.; Woo, J.H. Epstein-Barr virus-encoded miR-BART5-5p upregulates PD-L1 through PIAS3/pSTAT3 modulation, worsening clinical outcomes of PD-L1-positive gastric carcinomas. Gastric Cancer J. Int. Gastric Cancer Assoc. Jpn. Gastric Cancer Assoc. 2020, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Yang, Y.; Huang, X. miR-199a-5p promotes proliferation and metastasis and epithelial-mesenchymal transition through targeting PIAS3 in cervical carcinoma. J. Cell. Biochem. 2019, 120, 13562–13572. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.X.; Qiu, M.K.; Wang, S.Q.; Pan, C.; Wang, Y.; Ou, J.M. lncRNA CASC2 suppresses the growth of hemangioma cells by regulating miR-18a-5p/FBXL3 axis. J. Biol. Regul. Homeost. Agents 2020, 34, 49–56. [Google Scholar] [CrossRef]
- Wu, W.; Takanashi, M.; Borjigin, N.; Ohno, S.I.; Fujita, K.; Hoshino, S.; Osaka, Y.; Tsuchida, A.; Kuroda, M. MicroRNA-18a modulates STAT3 activity through negative regulation of PIAS3 during gastric adenocarcinogenesis. Br. J. Cancer 2013, 108, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Li, Y.; Ning, Q.; Suo, Z.M. Regulation of miR-155 affects the invasion and migration of gastric carcinoma cells by modulating the STAT3 signaling pathway. Oncol. Lett. 2018, 16, 4137–4142. [Google Scholar] [CrossRef]
- Guo, W.; Li, W.; Yuan, L.; Mei, X.; Hu, W. MicroRNA-106a-3p Induces Apatinib Resistance and Activates Janus-Activated Kinase 2 (JAK2)/Signal Transducer and Activator of Transcription 3 (STAT3) by Targeting the SOCS System in Gastric Cancer. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 10122–10128. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.S.; Joo, M.K.; Park, J.J.; Yoo, H.S.; Choi, B.I.; Lee, B.J.; Chun, H.J.; Lee, S.W. Inhibition of STAT3 in gastric cancer: Role of pantoprazole as SHP-1 inducer. Cell Biosci. 2018, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Q. Sunitinib reverse multidrug resistance in gastric cancer cells by modulating Stat3 and inhibiting P-gp function. Cell Biochem. Biophys. 2013, 67, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Tian, D.; Qiao, X.; Li, J.; Zhang, L. Modulation of Myb-induced NF-kB -STAT3 signaling and resulting cisplatin resistance in ovarian cancer by dietary factors. J. Cell. Physiol. 2019, 234, 21126–21134. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.; Green, I.R.; Saleem, M.; Khattak, K.F.; Irshad, M.; Ali, M. Cucurbitacins as Anticancer Agents: A Patent Review. Recent Pat. Anti-Cancer Drug Discov. 2019, 14, 133–143. [Google Scholar] [CrossRef]
- Jafargholizadeh, N.; Zargar, S.J.; Aftabi, Y. The cucurbitacins D, E, and I from Ecballium elaterium (L.) upregulate the LC3 gene and induce cell-cycle arrest in human gastric cancer cell line AGS. Iran. J. Basic Med. Sci. 2018, 21, 253–259. [Google Scholar] [CrossRef]
- Wang, J.-R.; Shen, G.-N.; Luo, Y.-H.; Piao, X.-J.; Zhang, Y.; Wang, H.; Li, J.-Q.; Xu, W.-T.; Zhang, Y.; Wang, S.-N. 2-(4-methoxyphenylthio)-5, 8-dimethoxy-1, 4-naphthoquinone induces apoptosis via ROS-mediated MAPK and STAT3 signaling pathway in human gastric cancer cells. J. Chemother. 2019, 31, 214–226. [Google Scholar] [CrossRef]
- Zang, Y.Q.; Feng, Y.Y.; Luo, Y.H.; Zhai, Y.Q.; Ju, X.Y.; Feng, Y.C.; Wang, J.R.; Yu, C.Q.; Jin, C.H. Glycitein induces reactive oxygen species-dependent apoptosis and G0/G1 cell cycle arrest through the MAPK/STAT3/NF-κB pathway in human gastric cancer cells. Drug Dev. Res. 2019, 80, 573–584. [Google Scholar] [CrossRef]
- Chuang, K.C.; Chang, C.R.; Chang, S.H.; Huang, S.W.; Chuang, S.M.; Li, Z.Y.; Wang, S.T.; Kao, J.K.; Chen, Y.J.; Shieh, J.J. Imiquimod-induced ROS production disrupts the balance of mitochondrial dynamics and increases mitophagy in skin cancer cells. J. Dermatol. Sci. 2020. [Google Scholar] [CrossRef]
- Li, D.; Zhao, D.; Du, J.; Dong, S.; Aldhamin, Z.; Yuan, X.; Li, W.; Du, H.; Zhao, W.; Cui, L.; et al. Heme oxygenase-1 alleviated non-alcoholic fatty liver disease via suppressing ROS-dependent endoplasmic reticulum stress. Life Sci. 2020, 253, 117678. [Google Scholar] [CrossRef]
- Saleem, M.Z.; Nisar, M.A.; Alshwmi, M.; Din, S.R.U.; Gamallat, Y.; Khan, M.; Ma, T. Brevilin A Inhibits STAT3 Signaling and Induces ROS-Dependent Apoptosis, Mitochondrial Stress and Endoplasmic Reticulum Stress in MCF-7 Breast Cancer Cells. OncoTargets Ther. 2020, 13, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Liang, Y.; Chen, Y.; Wang, L.; Li, D.; Liang, Z.; Sun, L.; Wang, Y.; Niu, H. Glutamine affects T24 bladder cancer cell proliferation by activating STAT3 through ROS and glutaminolysis. Int. J. Mol. Med. 2019, 44, 2189–2200. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Zhao, S.; Zhang, Y.; Chen, Y.; Zhao, X.; Liu, L.; Sun, S.; Zhang, L.; Ye, B.; et al. MICAL1 facilitates breast cancer cell proliferation via ROS-sensitive ERK/cyclin D pathway. J. Cell. Mol. Med. 2018, 22, 3108–3118. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, Y.Y.; Wang, X.; Shen, P.; Jia, Q.; Yu, S.; Wang, Y.; Li, X.; Chen, W.; Wang, A.; et al. Lycopene prevents carcinogen-induced cutaneous tumor by enhancing activation of the Nrf2 pathway through p62-triggered autophagic Keap1 degradation. Aging 2020, 12, 8167. [Google Scholar] [CrossRef]
- Park, B.; Lim, J.W.; Kim, H. Lycopene treatment inhibits activation of Jak1/Stat3 and Wnt/β-catenin signaling and attenuates hyperproliferation in gastric epithelial cells. Nutr. Res. 2019, 70, 70–81. [Google Scholar] [CrossRef]
- Lin, L.; Huang, K.; Guo, W.; Zhou, C.; Wang, G.; Zhao, Q. Conditioned Medium of the Osteosarcoma Cell Line U2OS Induces hBMSCs to Exhibit Characteristics of Carcinoma-Associated Fibroblasts via Activation of IL-6/STAT3 Signaling. J. Biochem. 2020. [Google Scholar] [CrossRef]
- Lim, H.S.; Kim, O.S.; Kim, B.Y.; Jeong, S.J. Apigetrin from Scutellaria baicalensis Georgi Inhibits Neuroinflammation in BV-2 Microglia and Exerts Neuroprotective Effect in HT22 Hippocampal Cells. J. Med. Food 2016, 19, 1032–1040. [Google Scholar] [CrossRef]
- Sun, Q.; Lu, N.-N.; Feng, L. Apigetrin inhibits gastric cancer progression through inducing apoptosis and regulating ROS-modulated STAT3/JAK2 pathway. Biochem. Biophys. Res. Commun. 2018, 498, 164–170. [Google Scholar] [CrossRef]
- Puar, Y.R.; Shanmugam, M.K.; Fan, L.; Arfuso, F.; Sethi, G.; Tergaonkar, V. Evidence for the Involvement of the Master Transcription Factor NF-κB in Cancer Initiation and Progression. Biomedicines 2018, 6, 82. [Google Scholar] [CrossRef]
- Fan, Y.; Mao, R.; Yang, J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013, 4, 176–185. [Google Scholar] [CrossRef]
- Hoesel, B.; Schmid, J.A. The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer 2013, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- Esparza-Lopez, J.; Alvarado-Munoz, J.F.; Escobar-Arriaga, E.; Ulloa-Aguirre, A.; de Jesus Ibarra-Sanchez, M. Metformin reverses mesenchymal phenotype of primary breast cancer cells through STAT3/NF-kappaB pathways. BMC Cancer 2019, 19, 728. [Google Scholar] [CrossRef] [PubMed]
- Malinska, H.; Huttl, M.; Oliyarnyk, O.; Markova, I.; Poruba, M.; Racova, Z.; Kazdova, L.; Vecera, R. Beneficial effects of troxerutin on metabolic disorders in non-obese model of metabolic syndrome. PLoS ONE 2019, 14, e0220377. [Google Scholar] [CrossRef] [PubMed]
- Hoseindoost, M.; Alipour, M.R.; Farajdokht, F.; Diba, R.; Bayandor, P.; Mehri, K.; Nayebi Rad, S.; Babri, S. Effects of troxerutin on inflammatory cytokines and BDNF levels in male offspring of high-fat diet fed rats. Avicenna J. Phytomed. 2019, 9, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Thomas, N.S.; George, K.; Selvam, A.A.A. Anticancer mechanism of troxerutin via targeting Nrf2 and NF-kappaB signalling pathways in hepatocarcinoma cell line. Toxicol. in vitro Int. J. Publ. Assoc. BIBRA 2019, 54, 317–329. [Google Scholar] [CrossRef]
- Thomas, N.S.; George, K.; Selvam, A.A.A. Troxerutin subdues hepatic tumorigenesis via disrupting the MDM2-p53 interaction. Food Funct. 2018, 9, 5336–5349. [Google Scholar] [CrossRef]
- Xu, G.-Y.; Tang, X.-J. Troxerutin (TXN) potentiated 5-Fluorouracil (5-Fu) treatment of human gastric cancer through suppressing STAT3/NF-κB and Bcl-2 signaling pathways. Biomed. Pharmacother. 2017, 92, 95–107. [Google Scholar] [CrossRef]
- Samidurai, A.; Roh, S.K.; Prakash, M.; Durrant, D.; Salloum, F.N.; Kukreja, R.C.; Das, A. STAT3-miR-17/20 Signaling Axis Plays a Critical Role in Attenuating Myocardial Infarction following Rapamycin Treatment in Diabetic mice. Cardiovasc. Res. 2019. [Google Scholar] [CrossRef]
- Tang, Z.; Xu, T.; Li, Y.; Fei, W.; Yang, G.; Hong, Y. Inhibition of CRY2 by STAT3/miRNA-7-5p Promotes Osteoblast Differentiation through Upregulation of CLOCK/BMAL1/P300 Expression. Mol. Therapy. Nucleic Acids 2020, 19, 865–876. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Z.; Wang, X. miRNA-373 promotes urinary bladder cancer cell proliferation, migration and invasion through upregulating epidermal growth factor receptor. Exp. Ther. Med. 2019, 17, 1190–1195. [Google Scholar] [CrossRef]
- Zhang, X.J.; Jin, Y.; Song, J.L.; Deng, F. MiR-373 promotes proliferation and metastasis of oral squamous cell carcinoma by targeting SPOP. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5270–5276. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.L.; Wen, F.Q.; Chen, X.W.; Jiang, X.P.; Liu, S.X. miR-373 promotes neuroblastoma cell proliferation, migration, and invasion by targeting SRCIN1. OncoTargets Ther. 2019, 12, 4927–4936. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Sun, X.; Geng, Z.; Shi, M.; Chen, Z.; Chen, L.; Wang, Y.; Fu, X. Isoproterenol regulates CD44 expression in gastric cancer cells through STAT3/MicroRNA373 cascade. Biomaterials 2016, 105, 89–101. [Google Scholar] [CrossRef]
- Ji, H.G.; Piao, J.Y.; Kim, S.J.; Kim, D.H.; Lee, H.N.; Na, H.K.; Surh, Y.J. Docosahexaenoic acid inhibits Helicobacter pylori-induced STAT3 phosphorylation through activation of PPARγ. Mol. Nutr. Food Res. 2016, 60, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Zhan, H.; Bian, Q.; Gu, J. Piperlongumine inhibits gastric cancer cells via suppression of the JAK1, 2/STAT3 signaling pathway. Mol. Med. Rep. 2016, 13, 4475–4480. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Vishnoi, K.; Mahata, S.; Tripathi, S.C.; Misra, S.P.; Misra, V.; Mehrotra, R.; Dwivedi, M.; Bharti, A.C. Berberine and curcumin target survivin and STAT3 in gastric cancer cells and synergize actions of standard chemotherapeutic 5-fluorouracil. Nutr. Cancer 2015, 67, 1295–1306. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Wu, K.; Yin, M.; Han, S.; Ding, Y.; Qiao, A.; Lu, G.; Deng, B.; Bo, P.; Gong, W. Wogonin inhibits tumor-derived regulatory molecules by suppressing STAT3 signaling to promote tumor immunity. J. Immunother. 2015, 38, 167–184. [Google Scholar] [CrossRef]
- Dai, C.; Liu, P.; Wang, X.; Yin, Y.; Jin, W.; Shen, L.; Chen, Y.; Chen, Z.; Wang, Y. The Antipsychotic Agent Sertindole Exhibited Antiproliferative Activities by Inhibiting the STAT3 Signaling Pathway in Human Gastric Cancer Cells. J. Cancer 2020, 11, 849–857. [Google Scholar] [CrossRef]
- Yang, Y.; Fang, E.; Luo, J.; Wu, H.; Jiang, Y.; Liu, Y.; Tong, S.; Wang, Z.; Zhou, R.; Tong, Q. The Antioxidant Alpha-Lipoic Acid Inhibits Proliferation and Invasion of Human Gastric Cancer Cells via Suppression of STAT3-Mediated MUC4 Gene Expression. Oxidative Med. Cell. Longev. 2019, 2019, 3643715. [Google Scholar] [CrossRef]
- Jo, M.J.; Jeong, S.; Yun, H.K.; Kim, D.Y.; Kim, B.R.; Kim, J.L.; Na, Y.J.; Park, S.H.; Jeong, Y.A.; Kim, B.G.; et al. Genipin induces mitochondrial dysfunction and apoptosis via downregulation of Stat3/mcl-1 pathway in gastric cancer. BMC Cancer 2019, 19, 739. [Google Scholar] [CrossRef]
- Wang, H.; Luo, Y.H.; Shen, G.N.; Piao, X.J.; Xu, W.T.; Zhang, Y.; Wang, J.R.; Feng, Y.C.; Li, J.Q.; Zhang, Y.; et al. Two novel 1,4naphthoquinone derivatives induce human gastric cancer cell apoptosis and cell cycle arrest by regulating reactive oxygen speciesmediated MAPK/Akt/STAT3 signaling pathways. Mol. Med. Rep. 2019, 20, 2571–2582. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tang, X.; Wu, X.; Yang, M.; Wang, W.; Wang, L.; Tang, D.; Wang, D. Cardamonin exerts anti-gastric cancer activity via inhibiting LncRNA-PVT1-STAT3 axis. Biosci. Rep. 2019, 39, BSR20190357. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Khoi, P.N.; Yoon, H.J.; Lian, S.; Joo, Y.E.; Chay, K.O.; Kim, K.K.; Do Jung, Y. Piperine inhibits IL-1β-induced IL-6 expression by suppressing p38 MAPK and STAT3 activation in gastric cancer cells. Mol. Cell. Biochem. 2015, 398, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, S.; Fang, J.; Peng, B.; Zhang, Y.; Cao, T. Tanshinone IIA inhibits cell proliferation and tumor growth by downregulating STAT3 in human gastric cancer. Exp. Ther. Med. 2018, 16, 2931–2937. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, J.; Wang, G.; Chen, X.; Zhang, R.; Liu, H.; Zhu, J. Oxymatrine exhibits anti-tumor activity in gastric cancer through inhibition of IL-21R-mediated JAK2/STAT3 pathway. Int. J. Immunopathol. Pharmacol. 2018, 32. [Google Scholar] [CrossRef]
- Song, S.; Su, Z.; Xu, H.; Niu, M.; Chen, X.; Min, H.; Zhang, B.; Sun, G.; Xie, S.; Wang, H.; et al. Luteolin selectively kills STAT3 highly activated gastric cancer cells through enhancing the binding of STAT3 to SHP-1. Cell Death Dis. 2017, 8, e2612. [Google Scholar] [CrossRef]
- Li, H.; Lu, H.; Lv, M.; Wang, Q.; Sun, Y. Parthenolide facilitates apoptosis and reverses drug-resistance of human gastric carcinoma cells by inhibiting the STAT3 signaling pathway. Oncol. Lett. 2018, 15, 3572–3579. [Google Scholar] [CrossRef]
- Rajamanickam, V.; Zhu, H.; Feng, C.; Chen, X.; Zheng, H.; Xu, X.; Zhang, Q.; Zou, P.; He, G.; Dai, X.; et al. Novel allylated monocarbonyl analogs of curcumin induce mitotic arrest and apoptosis by reactive oxygen species-mediated endoplasmic reticulum stress and inhibition of STAT3. Oncotarget 2017, 8, 101112–101129. [Google Scholar] [CrossRef]
- Zheng, H.; Hong, H.; Zhang, L.; Cai, X.; Hu, M.; Cai, Y.; Zhou, B.; Lin, J.; Zhao, C.; Hu, W. Nifuratel, a novel STAT3 inhibitor with potent activity against human gastric cancer cells. Cancer Manag. Res. 2017, 9, 565–572. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, G.; Dai, C.; Gao, X.; Wu, J.; Shen, L.; Chen, Z.; Liu, P. Cryptotanshinone potentiates the antitumor effects of doxorubicin on gastric cancer cells via inhibition of STAT3 activity. J. Int. Med. Res. 2017, 45, 220–230. [Google Scholar] [CrossRef]
- Wang, G.; Jing, Y.; Cao, L.; Gong, C.; Gong, Z.; Cao, X. A novel synthetic Asiatic acid derivative induces apoptosis and inhibits proliferation and mobility of gastric cancer cells by suppressing STAT3 signaling pathway. OncoTargets Ther. 2017, 10, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Dai, Y.; Liu, Q.; Ning, S.; Liu, J.; Shen, Z.; Zhu, D.; Jiang, F.; Zhang, J.; et al. Sulforaphane improves chemotherapy efficacy by targeting cancer stem cell-like properties via the miR-124/IL-6R/STAT3 axis. Sci. Rep. 2016, 6, 36796. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.Q.; Wang, J.; Guo, X.F.; Liu, Z.; Dong, W.G. Thymoquinone inhibits proliferation in gastric cancer via the STAT3 pathway in vivo and in vitro. World J. Gastroenterol. 2016, 22, 4149–4159. [Google Scholar] [CrossRef]
- Zheng, Y.B.; Xiao, G.C.; Tong, S.L.; Ding, Y.; Wang, Q.S.; Li, S.B.; Hao, Z.N. Paeoniflorin inhibits human gastric carcinoma cell proliferation through up-regulation of microRNA-124 and suppression of PI3K/Akt and STAT3 signaling. World J. Gastroenterol. 2015, 21, 7197–7207. [Google Scholar] [CrossRef] [PubMed]
- Cheong, J.H.; Hong, S.Y.; Zheng, Y.; Noh, S.H. Eupatilin Inhibits Gastric Cancer Cell Growth by Blocking STAT3-Mediated VEGF Expression. J. Gastric Cancer 2011, 11, 16–22. [Google Scholar] [CrossRef]
- Zhu, B.H.; Chen, H.Y.; Zhan, W.H.; Wang, C.Y.; Cai, S.R.; Wang, Z.; Zhang, C.H.; He, Y.L. (-)-Epigallocatechin-3-gallate inhibits VEGF expression induced by IL-6 via Stat3 in gastric cancer. World J. Gastroenterol. 2011, 17, 2315–2325. [Google Scholar] [CrossRef]
- Xie, Y.L.; Tao, W.H.; Yang, T.X.; Qiao, J.G. Anticancer effect of cucurbitacin B on MKN-45 cells via inhibition of the JAK2/STAT3 signaling pathway. Exp. Ther. Med. 2016, 12, 2709–2715. [Google Scholar] [CrossRef]
- Liu, Y.F.; Lu, Y.M.; Qu, G.Q.; Liu, Y.; Chen, W.X.; Liao, X.H.; Kong, W.M. Ponicidin induces apoptosis via JAK2 and STAT3 signaling pathways in gastric carcinoma. Int. J. Mol. Sci. 2015, 16, 1576–1589. [Google Scholar] [CrossRef]
- Hwang, S.T.; Kim, C.; Lee, J.H.; Chinnathambi, A.; Alharbi, S.A.; Shair, O.H.; Sethi, G.; Ahn, K.S. Cycloastragenol can negate constitutive STAT3 activation and promote paclitaxel-induced apoptosis in human gastric cancer cells. Phytomedicine 2019, 59, 152907. [Google Scholar] [CrossRef]
- Yu, R.X.; Yu, R.T.; Liu, Z. Inhibition of two gastric cancer cell lines induced by fucoxanthin involves downregulation of Mcl-1 and STAT3. Hum. Cell 2018, 31, 50–63. [Google Scholar] [CrossRef]
- Jiang, X.; Wu, M.; Xu, Z.; Wang, H.; Wang, H.; Yu, X.; Li, Z.; Teng, L. HJC0152, a novel STAT3 inhibitor with promising anti-tumor effect in gastric cancer. Cancer Manag. Res. 2018, 10, 6857–6867. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Wang, W.; Wang, C. IGF-1-induced MMP-11 expression promotes the proliferation and invasion of gastric cancer cells through the JAK1/STAT3 signaling pathway. Oncol. Lett. 2018, 15, 7000–7006. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Tang, J.; Wu, M.; Chen, S.; Xu, Z.; Wang, H.; Wang, H.; Yu, X.; Li, Z.; Teng, L. BP1102 exerts an antitumor effect on the AGS human gastric cancer cell line through modulating the STAT3 and MAPK signaling pathways. Mol. Med. Rep. 2019, 19, 2698–2706. [Google Scholar] [CrossRef] [PubMed]
- Vafadar, A.; Shabaninejad, Z.; Movahedpour, A.; Mohammadi, S.; Fathullahzadeh, S.; Mirzaei, H.R.; Namdar, A.; Savardashtaki, A.; Mirzaei, H. Long Non-Coding RNAs As Epigenetic Regulators in Cancer. Curr. Pharm. Des. 2019, 25, 3563–3577. [Google Scholar] [CrossRef]
- Liang, Y.; Song, X.; Li, Y.; Chen, B.; Zhao, W.; Wang, L.; Zhang, H.; Liu, Y.; Han, D.; Zhang, N.; et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol. Cancer 2020, 19, 85. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, X.; Hou, R. lncRNA PICART1 alleviates progression of cervical cancer by upregulating TCF21. Oncol. Lett. 2020, 19, 3719–3724. [Google Scholar] [CrossRef]
- Shang, A.; Wang, W.; Gu, C.; Chen, W.; Lu, W.; Sun, Z.; Li, D. Long non-coding RNA CCAT1 promotes colorectal cancer progression by regulating miR-181a-5p expression. Aging 2020, 12, 8301. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, J.; Tang, H.; Zhai, D.; Huang, D.; Ling, L.; Wang, X.; Liu, T.; Zhang, Q.; Zhang, Z.; et al. LncRNA SNORD3A specifically sensitizes breast cancer cells to 5-FU by sponging miR-185-5p to enhance UMPS expression. Cell Death Dis. 2020, 11, 329. [Google Scholar] [CrossRef]
- Wu, Y.; Bi, Q.J.; Han, R.; Zhang, Y. Long noncoding RNA KCNQ1OT1 is correlated with human breast cancer cell development through inverse regulation of hsa-miR-107. Biochem. Cell Biol. Biochim. Et Biol. Cell 2020, 999, 1–7. [Google Scholar] [CrossRef]
- Guo, J.; Ding, Y.; Yang, H.; Guo, H.; Zhou, X.; Chen, X. Aberrant expression of lncRNA MALAT1 modulates radioresistance in colorectal cancer in vitro via miR-101-3p sponging. Exp. Mol. Pathol. 2020, 115, 104448. [Google Scholar] [CrossRef]
- Li, J.; Tian, J.; Lu, J.; Wang, Z.; Ling, J.; Wu, X.; Yang, F.; Xia, Y. LncRNA GAS5 inhibits Th17 differentiation and alleviates immune thrombocytopenia via promoting the ubiquitination of STAT3. Int. Immunopharmacol. 2020, 80, 106127. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Yao, R.; Zhou, P.; Wang, C.; Xia, Y.; Xu, S. LncRNA NEAT1 reversed the hindering effects of miR-495-3p/STAT3 axis and miR-211/PI3K/AKT axis on sepsis-relevant inflammation. Mol. Immunol. 2020, 117, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Wang, H.X.; Chen, P.; Chen, C.H. STAT3-induced upregulation of lncRNA DUXAP8 functions as ceRNA for miR-577 to promote the migration and invasion in colorectal cancer through the regulation of RAB14. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6105–6118. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Liu, R.; Sun, X. STAT3-induced upregulation of lncRNA SNHG17 predicts a poor prognosis of melanoma and promotes cell proliferation and metastasis through regulating PI3K-AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8000–8010. [Google Scholar] [CrossRef]
- Zeng, X.Y.; Jiang, X.Y.; Yong, J.H.; Xie, H.; Yuan, J.; Zeng, D.; Dou, Y.Y.; Xiao, S.S. lncRNA ABHD11-AS1, regulated by the EGFR pathway, contributes to the ovarian cancer tumorigenesis by epigenetically suppressing TIMP2. Cancer Med. 2019, 8, 7074–7085. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Li, Q.; Xie, F. LncRNA-MALAT1 regulates proliferation and apoptosis of ovarian cancer cells by targeting miR-503-5p. OncoTargets Ther. 2019, 12, 6297–6307. [Google Scholar] [CrossRef]
- Luo, J.; Wang, K.; Yeh, S.; Sun, Y.; Liang, L.; Xiao, Y.; Xu, W.; Niu, Y.; Cheng, L.; Maity, S.N.; et al. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat. Commun. 2019, 10, 2571. [Google Scholar] [CrossRef]
- Su, W.; Guo, C.; Wang, L.; Wang, Z.; Yang, X.; Niu, F.; Tzou, D.; Yang, X.; Huang, X.; Wu, J.; et al. LncRNA MIR22HG abrogation inhibits proliferation and induces apoptosis in esophageal adenocarcinoma cells via activation of the STAT3/c-Myc/FAK signaling. Aging 2019, 11, 4587–4596. [Google Scholar] [CrossRef]
- Chen, J.F.; Wu, P.; Xia, R.; Yang, J.; Huo, X.Y.; Gu, D.Y.; Tang, C.J.; De, W.; Yang, F. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol. Cancer 2018, 17, 6. [Google Scholar] [CrossRef]
- Jin, Y.; Wu, P.; Zhao, W.; Wang, X.; Yang, J.; Huo, X.; Chen, J.; De, W.; Yang, F. Long noncoding RNA LINC00165-induced by STAT3 exerts oncogenic properties via interaction with Polycomb Repressive Complex 2 to promote EMT in gastric cancer. Biochem. Biophys. Res. Commun. 2018, 507, 223–230. [Google Scholar] [CrossRef]
- Guo, J.Q.; Yang, Z.J.; Wang, S.; Wu, Z.Z.; Yin, L.L.; Wang, D.C. LncRNA SNHG16 functions as an oncogene by sponging miR-200a-3p in pancreatic cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1718–1724. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Jiang, T.; Mao, H.; Gao, R.; Zhang, H.; He, Y.; Liu, C.; Chen, Q. SNHG16 regulates invasion and migration of bladder cancer through induction of epithelial-to-mesenchymal transition. Hum. Cell 2020. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Yu, Z.; Zou, J. LncRNA-SNHG16 Silencing Inhibits Prostate Carcinoma Cell Growth, Downregulate GLUT1 Expression and Reduce Glucose Uptake. Cancer Manag. Res. 2020, 12, 1751–1757. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Li, H.; Xiang, H.; Gao, M.; Yin, C.; Wang, H.; Sun, Y.; Xiong, M. Long Chain Non-Coding RNA (lncRNA) HOTAIR Knockdown Increases miR-454-3p to Suppress Gastric Cancer Growth by Targeting STAT3/Cyclin D1. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 1537–1548. [Google Scholar] [CrossRef]
- Zhan, G.; Hu, J.; Xiao, B.; Wang, X.; Yang, Z.; Yang, G.; Lu, L. Trillin prevents proliferation and induces apoptosis through inhibiting STAT3 nuclear translocation in hepatoma carcinoma cells. Med. Oncol. (Northwoodlondonengland) 2020, 37, 44. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, M.; Wang, Z.; Gao, W.; Sun, Z.G. Activated STAT3 Could Reduce Survival in Patients with Esophageal Squamous Cell Carcinoma by Up-regulating VEGF and Cyclin D1 Expression. J. Cancer 2020, 11, 1859–1868. [Google Scholar] [CrossRef]
- Zhao, J.; Du, P.; Cui, P.; Qin, Y.; Hu, C.; Wu, J.; Zhou, Z.; Zhang, W.; Qin, L.; Huang, G. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene 2018, 37, 4094–4109. [Google Scholar] [CrossRef]
- Tan, H.Y.; Wang, C.; Liu, G.; Zhou, X. Long noncoding RNA NEAT1-modulated miR-506 regulates gastric cancer development through targeting STAT3. J. Cell. Biochem. 2019, 120, 4827–4836. [Google Scholar] [CrossRef]
- Shen, W.; Yuan, Y.; Zhao, M.; Li, J.; Xu, J.; Lou, G.; Zheng, J.; Bu, S.; Guo, J.; Xi, Y. Novel long non-coding RNA GACAT3 promotes gastric cancer cell proliferation through the IL-6/STAT3 signaling pathway. Tumor Biol. 2016, 37, 14895–14902. [Google Scholar] [CrossRef]
- Wang, X.; Kan, J.; Han, J.; Zhang, W.; Bai, L.; Wu, H. LncRNA SNHG16 Functions as an Oncogene by Sponging MiR-135a and Promotes JAK2/STAT3 Signal Pathway in Gastric Cancer. J. Cancer 2019, 10, 1013–1022. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, J.; Zhou, Z.; He, Z. Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer cell growth via inactivating the JAK2/STAT3 pathway. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39, 1010428317705335. [Google Scholar] [CrossRef] [PubMed]
- Bartosch, C.; Monteiro-Reis, S.; Almeida-Rios, D.; Vieira, R.; Castro, A.; Moutinho, M.; Rodrigues, M.; Graça, I.; Lopes, J.M.; Jerónimo, C. Assessing sirtuin expression in endometrial carcinoma and non-neoplastic endometrium. Oncotarget 2016, 7, 1144. [Google Scholar] [CrossRef] [PubMed]
- Cha, E.J.; Noh, S.J.; Kwon, K.S.; Kim, C.Y.; Park, B.-H.; Park, H.S.; Lee, H.; Chung, M.J.; Kang, M.J.; Lee, D.G. Expression of DBC1 and SIRT1 is associated with poor prognosis of gastric carcinoma. Clin. Cancer Res. 2009, 15, 4453–4459. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, B.; Gao, W.; Huang, S.; Liu, Z.; Li, W.; Jia, J. SIRT1 is downregulated in gastric cancer and leads to G1-phase arrest via NF-κB/Cyclin D1 signaling. Mol. Cancer Res. 2013, 11, 1497–1507. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, Y.; Huang, S.; Deng, C.; Zhou, S.; Yang, J.; Cao, Y.; Xu, L.; Yuan, Y.; Yang, J.; et al. SIRT1 inhibits gastric cancer proliferation and metastasis via STAT3/MMP-13 signaling. J. Cell. Physiol. 2019, 234, 15395–15406. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, A.; Yu, X.; Zhu, J.; Dai, H. SIRT6 inhibits growth of gastric cancer by inhibiting JAK2/STAT3 pathway. Oncol. Rep. 2017, 38, 1059–1066. [Google Scholar] [CrossRef]
- Janacova, L.; Faktor, J.; Capkova, L.; Paralova, V.; Pospisilova, A.; Podhorec, J.; Ebhardt, H.A.; Hrstka, R.; Nenutil, R.; Aebersold, R.; et al. SWATH-MS Analysis of FFPE Tissues Identifies Stathmin as a Potential Marker of Endometrial Cancer in Patients Exposed to Tamoxifen. J. Proteome Res. 2020. [Google Scholar] [CrossRef]
- Xu, J.; Wu, W.; Tang, Y.; Lin, Y.; Xue, Y.; Hu, J.; Lin, D. PRL-3 exerts oncogenic functions in myeloid leukemia cells via aberrant dephosphorylation of stathmin and activation of STAT3 signaling. Aging 2019, 11, 7817–7829. [Google Scholar] [CrossRef]
- Dos Santos Passaia, B.; Lima, K.; Kremer, J.L.; da Conceicao, B.B.; de Paula Mariani, B.M.; da Silva, J.C.L.; Zerbini, M.C.N.; Fragoso, M.; Machado-Neto, J.A.; Lotfi, C.F.P. Stathmin 1 is highly expressed and associated with survival outcome in malignant adrenocortical tumours. Investig. New Drugs 2020, 38, 899–908. [Google Scholar] [CrossRef]
- Shu, F.; Zou, X.; Tuo, H.; She, S.; Huang, J.; Ren, H.; Hu, H.; Peng, S.; Wang, J.; Yang, Y. Stathmin gene silencing suppresses proliferation, migration and invasion of gastric cancer cells via AKT/sCLU and STAT3 signaling. Int. J. Oncol. 2019, 54, 1086–1098. [Google Scholar] [CrossRef]
- Cassim, S.; Vucetic, M.; Zdralevic, M.; Pouyssegur, J. Warburg and Beyond: The Power of Mitochondrial Metabolism to Collaborate or Replace Fermentative Glycolysis in Cancer. Cancers 2020, 12, 1119. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Zhang, Q.; Guo, F.; Guo, S.; Yang, B.; Liu, B.; Li, P.; Li, J.; Guan, S.; Liu, X. Long Noncoding RNA PCED1B-AS1 Promotes the Warburg Effect and Tumorigenesis by Upregulating HIF-1alpha in Glioblastoma. Cell Transplant. 2020, 29, 963689720906777. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Chen, M.; Wei, W.; Zhang, X.; Zhang, M.; Yao, Y.; Lv, Y.; Ling, T.; Wang, L.; Zou, X. Crosstalk of mTOR/PKM2 and STAT3/c-Myc signaling pathways regulate the energy metabolism and acidic microenvironment of gastric cancer. J. Cell. Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Xu, X.; Mao, C.-Y.; Han, K.-K.; Xu, Y.-J.; Cao, B.-Y.; Zhang, Z.-B.; Sethi, G.; Tang, X.-W.; Mao, X.-L. RNF6 promotes myeloma cell proliferation and survival by inducing glucocorticoid receptor polyubiquitination. Acta Pharmacol. Sin. 2020, 41, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, C. Potential Influences of RNF6 on Prognosis and Metastasis of Colorectal Cancer: A Clinical Analysis. OncoTargets Ther. 2020, 13, 2031–2036. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Cai, Y.; Yang, C.; Chen, Z.; Sun, H.; Xu, Y.; Chen, W.; Xu, D.; Tian, W.; Wang, H. Knockdown of RNF6 inhibits gastric cancer cell growth by suppressing STAT3 signaling. OncoTargets Ther. 2018, 11, 6579–6587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Huang, W.C.; Zheng, L.C. MiR-125b-1-3p Exerts Antitumor Functions in Lung Carcinoma Cells by Targeting S1PR1. Chin. Med. J. 2018, 131, 1909–1916. [Google Scholar] [CrossRef]
- Lankadasari, M.B.; Aparna, J.S.; Mohammed, S.; James, S.; Aoki, K.; Binu, V.S.; Nair, S.; Harikumar, K.B. Targeting S1PR1/STAT3 loop abrogates desmoplasia and chemosensitizes pancreatic cancer to gemcitabine. Theranostics 2018, 8, 3824–3840. [Google Scholar] [CrossRef]
- Song, S.; Min, H.; Niu, M.; Wang, L.; Wu, Y.; Zhang, B.; Chen, X.; Liang, Q.; Wen, Y.; Wang, Y.; et al. S1PR1 predicts patient survival and promotes chemotherapy drug resistance in gastric cancer cells through STAT3 constitutive activation. EBioMedicine 2018, 37, 168–176. [Google Scholar] [CrossRef]
- Tao, Z.; Ruan, H.; Sun, L.; Kuang, D.; Song, Y.; Wang, Q.; Wang, T.; Hao, Y.; Chen, K. Targeting the YB-1/PD-L1 Axis to Enhance Chemotherapy and Antitumor Immunity. Cancer Immunol. Res. 2019, 7, 1135–1147. [Google Scholar] [CrossRef]
- Liu, Y.L.; Yan, Z.X.; Xia, Y.; Xie, X.Y.; Zhou, K.; Xu, L.L.; Shi, Y.L.; Wang, Q.; Bi, J.W. Ligustrazine reverts anthracycline chemotherapy resistance of human breast cancer by inhibiting JAK2/STAT3 signaling and decreasing fibrinogen gamma chain (FGG) expression. Am. J. Cancer Res. 2020, 10, 939–952. [Google Scholar] [PubMed]
- Chua, P.J.; Lim, J.P.; Guo, T.T.; Khanna, P.; Hu, Q.; Bay, B.H.; Baeg, G.H. Y-box binding protein-1 and STAT3 independently regulate ATP-binding cassette transporters in the chemoresistance of gastric cancer cells. Int. J. Oncol. 2018, 53, 2579–2589. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, H.; Yao, H.; Li, C.; Fang, J.Y.; Xu, J. Regulation of PD-L1: Emerging Routes for Targeting Tumor Immune Evasion. Front. Pharmacol. 2018, 9, 536. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Mezzadra, R.; Schumacher, T.N. Regulation and Function of the PD-L1 Checkpoint. Immunity 2018, 48, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, Y.; Zhang, J.; Liu, Y.; Qi, Q. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis. 2019, 10, 731. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Dong, L.; Wang, K.; Zou, H.; Zhao, S.; Wang, Y.; Wang, G. MiR-21 Participates in the PD-1/PD-L1 Pathway-Mediated Imbalance of Th17/Treg Cells in Patients After Gastric Cancer Resection. Ann. Surg. Oncol. 2019, 26, 884–893. [Google Scholar] [CrossRef]
- Derer, A.; Frey, B.; Fietkau, R.; Gaipl, U.S. Immune-modulating properties of ionizing radiation: Rationale for the treatment of cancer by combination radiotherapy and immune checkpoint inhibitors. Cancer Immunol. Immunother. 2016, 65, 779–786. [Google Scholar] [CrossRef]
- Yan, Y.; Kumar, A.B.; Finnes, H.; Markovic, S.N.; Park, S.; Dronca, R.S.; Dong, H. Combining Immune Checkpoint Inhibitors with Conventional Cancer Therapy. Front. Immunol. 2018, 9, 1739. [Google Scholar] [CrossRef]
- Chen, M.F.; Chen, P.T.; Chen, W.C.; Lu, M.S.; Lin, P.Y.; Lee, K.D. The role of PD-L1 in the radiation response and prognosis for esophageal squamous cell carcinoma related to IL-6 and T-cell immunosuppression. Oncotarget 2016, 7, 7913–7924. [Google Scholar] [CrossRef]
- Sun, L.; Wang, Q.; Chen, B.; Zhao, Y.; Shen, B.; Wang, H.; Xu, J.; Zhu, M.; Zhao, X.; Xu, C.; et al. Gastric cancer mesenchymal stem cells derived IL-8 induces PD-L1 expression in gastric cancer cells via STAT3/mTOR-c-Myc signal axis. Cell Death Dis. 2018, 9, 928. [Google Scholar] [CrossRef]
- Gasi Tandefelt, D.; Boormans, J.L.; van der Korput, H.A.; Jenster, G.W.; Trapman, J. A 36-gene signature predicts clinical progression in a subgroup of ERG-positive prostate cancers. Eur. Urol. 2013, 64, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Wang, W.; Ji, M.; Zhu, Q.; Feng, Y.; Zhou, F.; He, Q. TMEM119 silencing inhibits cell viability and causes the apoptosis of gastric cancer SGC-7901 cells. Oncol. Lett. 2018, 15, 8281–8286. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Wang, W.; Ji, M.; Zhu, Q.; Feng, Y.; Zhou, F.; He, Q. TMEM119 promotes gastric cancer cell migration and invasion through STAT3 signaling pathway. OncoTargets Ther. 2018, 11, 5835–5844. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ma, J.; Chen, J.; Yang, Y.; Liang, J.; Liang, Y. LncRNA LINC00461 Promotes Colorectal Cancer Progression via miRNA-323b-3p/NFIB Axis. OncoTargets Ther. 2019, 12, 11119–11129. [Google Scholar] [CrossRef]
- Zhang, W.; Zhan, F.; Li, D.; Wang, T.; Huang, H. RGMB-AS1/miR-22-3p/NFIB axis contributes to the progression of gastric cancer. Neoplasma 2020, 67, 484–491. [Google Scholar] [CrossRef]
- Wu, C.; Zhu, X.; Liu, W.; Ruan, T.; Wan, W.; Tao, K. NFIB promotes cell growth, aggressiveness, metastasis and EMT of gastric cancer through the Akt/Stat3 signaling pathway. Oncol. Rep. 2018, 40, 1565–1573. [Google Scholar] [CrossRef]
- Brown, L.C.; Murphy, A.R.; Lalonde, C.S.; Subhedar, P.D.; Miller, A.H.; Stevens, J.S. Posttraumatic stress disorder and breast cancer: Risk factors and the role of inflammation and endocrine function. Cancer 2020. [Google Scholar] [CrossRef]
- Piotrowski, I.; Kulcenty, K.; Suchorska, W. Interplay between inflammation and cancer. Rep. Pract. Oncol. Radiother. J. Greatpoland Cancer Cent. Pozn. Pol. Soc. Radiat. Oncol. 2020, 25, 422–427. [Google Scholar] [CrossRef]
- Xu, X.; Yang, C.; Chen, J.; Liu, J.; Li, P.; Shi, Y.; Yu, P. Interleukin-23 promotes the migration and invasion of gastric cancer cells by inducing epithelial-to-mesenchymal transition via the STAT3 pathway. Biochem. Biophys. Res. Commun. 2018, 499, 273–278. [Google Scholar] [CrossRef]
- Brassart-Pasco, S.; Brézillon, S.; Brassart, B.; Ramont, L.; Oudart, J.-B.; Monboisse, J.C. Tumor Microenvironment: Extracellular Matrix Alterations Influence Tumor Progression. Front. Oncol. 2020, 10, 397. [Google Scholar] [CrossRef]
- Joyce, J.A.; Pollard, J.W. Microenvironmental regulation of metastasis. Nat. Reviews. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Lan, C.; Huang, X.; Lin, S.; Huang, H.; Cai, Q.; Wan, T.; Lu, J.; Liu, J. Expression of M2-polarized macrophages is associated with poor prognosis for advanced epithelial ovarian cancer. Technol. Cancer Res. Treat. 2013, 12, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.; Herrera, A.; Dominguez, G.; Silva, J.; Garcia, V.; Garcia, J.M.; Gomez, I.; Soldevilla, B.; Munoz, C.; Provencio, M.; et al. Cancer-associated fibroblast and M2 macrophage markers together predict outcome in colorectal cancer patients. Cancer Sci. 2013, 104, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Peng, C.W.; Yang, G.F.; Hu, W.Q.; Yang, X.J.; Huang, C.Q.; Xiong, B.; Li, Y. Distribution pattern of tumor associated macrophages predicts the prognosis of gastric cancer. Oncotarget 2017, 8, 92757–92769. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Huang, J.; Li, Z.; Zhang, J.; Luo, J.; Lu, C.; Xu, H.; Xu, H. The Prognostic and Clinicopathological Significance of Tumor-Associated Macrophages in Patients with Gastric Cancer: A Meta-Analysis. PLoS ONE 2017, 12, e0170042. [Google Scholar] [CrossRef]
- Chen, L.; Shi, Y.; Zhu, X.; Guo, W.; Zhang, M.; Che, Y.; Tang, L.; Yang, X.; You, Q.; Liu, Z. IL10 secreted by cancerassociated macrophages regulates proliferation and invasion in gastric cancer cells via cMet/STAT3 signaling. Oncol. Rep. 2019, 42, 595–604. [Google Scholar] [CrossRef]
- Sawa-Wejksza, K.; Kandefer-Szerszeń, M. Tumor-associated macrophages as target for antitumor therapy. Arch. Immunol. Et Ther. Exp. 2018, 66, 97–111. [Google Scholar] [CrossRef]
- Mantovani, A.; Germano, G.; Marchesi, F.; Locatelli, M.; Biswas, S.K. Cancer-promoting tumor-associated macrophages: New vistas and open questions. Eur. J. Immunol. 2011, 41, 2522–2525. [Google Scholar] [CrossRef]
- Zheng, X.; Turkowski, K.; Mora, J.; Brüne, B.; Seeger, W.; Weigert, A.; Savai, R. Redirecting tumor-associated macrophages to become tumoricidal effectors as a novel strategy for cancer therapy. Oncotarget 2017, 8, 48436. [Google Scholar] [CrossRef]
- Yang, M.; Liu, J.; Shao, J.; Qin, Y.; Ji, Q.; Zhang, X.; Du, J. Cathepsin S-mediated autophagic flux in tumor-associated macrophages accelerate tumor development by promoting M2 polarization. Mol. Cancer 2014, 13, 43. [Google Scholar] [CrossRef]
- Zhou, Y.; Xia, L.; Liu, Q.; Wang, H.; Lin, J.; Oyang, L.; Chen, X.; Luo, X.; Tan, S.; Tian, Y.; et al. Induction of Pro-Inflammatory Response via Activated Macrophage-Mediated NF-kappaB and STAT3 Pathways in Gastric Cancer Cells. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 47, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.L.; Duan, W.; Su, C.Y.; Mao, F.Y.; Lv, Y.P.; Teng, Y.S.; Yu, P.W.; Zhuang, Y.; Zhao, Y.L. Interleukin 6 induces M2 macrophage differentiation by STAT3 activation that correlates with gastric cancer progression. Cancer Immunol. Immunother. 2017, 66, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Holthof, L.C.; Mutis, T. Challenges for Immunotherapy in Multiple Myeloma: Bone Marrow Microenvironment-Mediated Immune Suppression and Immune Resistance. Cancers 2020, 12, 988. [Google Scholar] [CrossRef] [PubMed]
- Licarete, E.; Rauca, V.F.; Luput, L.; Drotar, D.; Stejerean, I.; Patras, L.; Dume, B.; Toma, V.A.; Porfire, A.; Gherman, C.; et al. Overcoming Intrinsic Doxorubicin Resistance in Melanoma by Anti-Angiogenic and Anti-Metastatic Effects of Liposomal Prednisolone Phosphate on Tumor Microenvironment. Int. J. Mol. Sci. 2020, 21, 2968. [Google Scholar] [CrossRef] [PubMed]
- Calabretta, E.; Carlo-Stella, C. The Many Facets of CD38 in Lymphoma: From Tumor-Microenvironment Cell Interactions to Acquired Resistance to Immunotherapy. Cells 2020, 9, 802. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Ma, J.; Song, X.; Xu, X.; Mou, Y. Cancer-Associated Fibroblasts Promote the Chemo-resistance in Gastric Cancer through Secreting IL-11 Targeting JAK/STAT3/Bcl2 Pathway. Cancer Res. Treat. J. Korean Cancer Assoc. 2019, 51, 194–210. [Google Scholar] [CrossRef]
- Zuo, E.; Lu, Y.; Yan, M.; Pan, X.; Cheng, X. Increased expression of hepcidin and associated upregulation of JAK/STAT3 signaling in human gastric cancer. Oncol. Lett. 2018, 15, 2236–2244. [Google Scholar] [CrossRef]
- Hill, D.G.; Yu, L.; Gao, H.; Balic, J.J.; West, A.; Oshima, H.; McLeod, L.; Oshima, M.; Gallimore, A.; D’Costa, K.; et al. Hyperactive gp130/STAT3-driven gastric tumourigenesis promotes submucosal tertiary lymphoid structure development. Int. J. Cancer 2018, 143, 167–178. [Google Scholar] [CrossRef]
- Tuo, H.; Shu, F.; She, S.; Yang, M.; Zou, X.Q.; Huang, J.; Hu, H.D.; Hu, P.; Ren, H.; Peng, S.F.; et al. Sorcin induces gastric cancer cell migration and invasion contributing to STAT3 activation. Oncotarget 2017, 8, 104258–104271. [Google Scholar] [CrossRef]
- Uen, Y.H.; Fang, C.L.; Lin, C.C.; Hseu, Y.C.; Hung, S.T.; Sun, D.P.; Lin, K.Y. Ceramide synthase 6 predicts the prognosis of human gastric cancer: It functions as an oncoprotein by dysregulating the SOCS2/JAK2/STAT3 pathway. Mol. Carcinog. 2018, 57, 1675–1689. [Google Scholar] [CrossRef] [PubMed]
- Ollila, S.; Domenech-Moreno, E.; Laajanen, K.; Wong, I.P.; Tripathi, S.; Pentinmikko, N.; Gao, Y.; Yan, Y.; Niemela, E.H.; Wang, T.C.; et al. Stromal Lkb1 deficiency leads to gastrointestinal tumorigenesis involving the IL-11-JAK/STAT3 pathway. J. Clin. Investig. 2018, 128, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xu, F.; Shi, Y.; Chen, Y.W.; Wang, H.P.; Yu, X.; Li, Y. C-X-C motif chemokine receptor 4 promotes tumor angiogenesis in gastric cancer via activation of JAK2/STAT3. Cell Biol. Int. 2017, 41, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Sun, J.; Huang, C.; Hu, X.; Jiang, N.; Lu, C. RNAi-mediated silencing of NOX4 inhibited the invasion of gastric cancer cells through JAK2/STAT3 signaling. Am. J. Transl. Res. 2017, 9, 4440–4449. [Google Scholar] [PubMed]
- Li, X.; Na, H.; Xu, L.; Zhang, X.; Feng, Z.; Zhou, X.; Cui, J.; Zhang, J.; Lin, F.; Yang, S.; et al. DC-SIGN mediates gastric cancer progression by regulating the JAK2/STAT3 signaling pathway and affecting LncRNA RP11-181G12.2 expression. Biomed. Pharmacother. Biomed. Pharmacother. 2020, 121, 109644. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Shi, M.; Ji, J.; Cai, Q.; Jiang, J.; Zhang, H.; Zhu, Z.; Zhang, J. A self-enforcing HOXA11/Stat3 feedback loop promotes stemness properties and peritoneal metastasis in gastric cancer cells. Theranostics 2019, 9, 7628–7647. [Google Scholar] [CrossRef]
- Zhu, S.; Soutto, M.; Chen, Z.; Blanca Piazuelo, M.; Kay Washington, M.; Belkhiri, A.; Zaika, A.; Peng, D.; El-Rifai, W. Activation of IGF1R by DARPP-32 promotes STAT3 signaling in gastric cancer cells. Oncogene 2019, 38, 5805–5816. [Google Scholar] [CrossRef]
- Ikeda, T.; Nishita, M.; Hoshi, K.; Honda, T.; Kakeji, Y.; Minami, Y. Mesenchymal stem cell-derived CXCL16 promotes progression of gastric cancer cells by STAT3-mediated expression of Ror1. Cancer Sci. 2020, 111, 1254–1265. [Google Scholar] [CrossRef]
- Yuan, K.; Ye, J.; Liu, Z.; Ren, Y.; He, W.; Xu, J.; He, Y.; Yuan, Y. Complement C3 overexpression activates JAK2/STAT3 pathway and correlates with gastric cancer progression. J. Exp. Clin. Cancer Res. 2020, 39, 9. [Google Scholar] [CrossRef]
- Xu, Y.Y.; Guo, M.; Yang, L.Q.; Zhou, F.; Yu, C.; Wang, A.; Pang, T.H.; Wu, H.Y.; Zou, X.P.; Zhang, W.J.; et al. Regulation of CD44v6 expression in gastric carcinoma by the IL-6/STAT3 signaling pathway and its clinical significance. Oncotarget 2017, 8, 45848–45861. [Google Scholar] [CrossRef]
- Xu, L.; Zhou, R.; Yuan, L.; Wang, S.; Li, X.; Ma, H.; Zhou, M.; Pan, C.; Zhang, J.; Huang, N.; et al. IGF1/IGF1R/STAT3 signaling-inducible IFITM2 promotes gastric cancer growth and metastasis. Cancer Lett. 2017, 393, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tao, P.; Zhou, Q.; Li, J.; Yu, Z.; Wang, X.; Li, J.; Li, C.; Yan, M.; Zhu, Z.; et al. IL-6 secreted by cancer-associated fibroblasts promotes epithelial-mesenchymal transition and metastasis of gastric cancer via JAK2/STAT3 signaling pathway. Oncotarget 2017, 8, 20741–20750. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Tang, N.; Wang, C.; Xiao, L.; Yu, M.; Zhao, L.; Cai, H.; Han, L.; Xie, C.; Zhang, Y. TNF-alpha-inducing protein of Helicobacter pylori induces epithelial-mesenchymal transition (EMT) in gastric cancer cells through activation of IL-6/STAT3 signaling pathway. Biochem. Biophys. Res. Commun. 2017, 484, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Zhao, T.; Xu, C.; Shi, W.; Geng, B.; Shen, J.; Zhang, C.; Pan, J.; Yang, J.; Hu, S.; et al. Oncometabolite succinate promotes angiogenesis by upregulating VEGF expression through GPR91-mediated STAT3 and ERK activation. Oncotarget 2017, 8, 13174–13185. [Google Scholar] [CrossRef]
- Li, T.; Guo, H.; Zhao, X.; Jin, J.; Zhang, L.; Li, H.; Lu, Y.; Nie, Y.; Wu, K.; Shi, Y.; et al. Gastric Cancer Cell Proliferation and Survival Is Enabled by a Cyclophilin B/STAT3/miR-520d-5p Signaling Feedback Loop. Cancer Res. 2017, 77, 1227–1240. [Google Scholar] [CrossRef]
- Yuan, W.; Li, T.; Mo, X.; Wang, X.; Liu, B.; Wang, W.; Su, Y.; Xu, L.; Han, W. Knockdown of CMTM3 promotes metastasis of gastric cancer via the STAT3/Twist1/EMT signaling pathway. Oncotarget 2016, 7, 29507–29519. [Google Scholar] [CrossRef]
- Zhang, J.L.; Liu, X.Z.; Wang, P.Y.; Chen, G.W.; Jiang, Y.; Qiao, S.K.; Zhu, J.; Wang, X.; Pan, Y.S.; Liu, Y.C. Targeting HCCR expression resensitizes gastric cancer cells to chemotherapy via down-regulating the activation of STAT3. Sci. Rep. 2016, 6, 24196. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, M.; Hu, H.; Zhao, X.; Bao, L.; Huang, D.; Song, L.; Li, Y. Mitochondrial GRIM-19 as a potential therapeutic target for STAT3-dependent carcinogenesis of gastric cancer. Oncotarget 2016, 7, 41404–41420. [Google Scholar] [CrossRef]
- Piao, J.Y.; Lee, H.G.; Kim, S.J.; Kim, D.H.; Han, H.J.; Ngo, H.K.; Park, S.A.; Woo, J.H.; Lee, J.S.; Na, H.K.; et al. Helicobacter pylori Activates IL-6-STAT3 Signaling in Human Gastric Cancer Cells: Potential Roles for Reactive Oxygen Species. Helicobacter 2016, 21, 405–416. [Google Scholar] [CrossRef]
- Zhao, G.; Zhu, G.; Huang, Y.; Zheng, W.; Hua, J.; Yang, S.; Zhuang, J.; Ye, J. IL-6 mediates the signal pathway of JAK-STAT3-VEGF-C promoting growth, invasion and lymphangiogenesis in gastric cancer. Oncol. Rep. 2016, 35, 1787–1795. [Google Scholar] [CrossRef]
- Wu, X.; Yang, T.; Liu, X.; Guo, J.N.; Xie, T.; Ding, Y.; Lin, M.; Yang, H. IL-17 promotes tumor angiogenesis through Stat3 pathway mediated upregulation of VEGF in gastric cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 5493–5501. [Google Scholar] [CrossRef] [PubMed]
- Fukui, H.; Watari, J.; Zhang, X.; Ran, Y.; Tomita, T.; Oshima, T.; Hirota, S.; Miwa, H. Phosphorylated STAT3 expression linked to SOCS3 methylation is associated with proliferative ability of gastric mucosa in patients with early gastric cancer. Oncol. Lett. 2020, 19, 3542–3550. [Google Scholar] [CrossRef] [PubMed]
- Balic, J.J.; Saad, M.I.; Dawson, R.; West, A.J.; McLeod, L.; West, A.C.; D’Costa, K.; Deswaerte, V.; Dev, A.; Sievert, W.; et al. Constitutive STAT3 Serine Phosphorylation Promotes Helicobacter-Mediated Gastric Disease. Am. J. Pathol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, N.; Ushijima, T.; Tan, P. How to stomach an epigenetic insult: The gastric cancer epigenome. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 467–478. [Google Scholar] [CrossRef]
- Gordon, K.; Clouaire, T.; Bao, X.X.; Kemp, S.E.; Xenophontos, M.; de Las Heras, J.I.; Stancheva, I. Immortality, but not oncogenic transformation, of primary human cells leads to epigenetic reprogramming of DNA methylation and gene expression. Nucleic Acids Res. 2014, 42, 3529–3541. [Google Scholar] [CrossRef]
- Cai, W.; Wang, C.; Li, Y.; Yao, C.; Shen, L.; Liu, S.; Bao, X.; Schnable, P.S.; Girton, J.; Johansen, J.; et al. Genome-wide analysis of regulation of gene expression and H3K9me2 distribution by JIL-1 kinase mediated histone H3S10 phosphorylation in Drosophila. Nucleic Acids Res. 2014, 42, 5456–5467. [Google Scholar] [CrossRef]
- Qi, H.; Yang, Z.; Dai, C.; Wang, R.; Ke, X.; Zhang, S.; Xiang, X.; Chen, K.; Li, C.; Luo, J.; et al. STAT3 activates MSK1-mediated histone H3 phosphorylation to promote NFAT signaling in gastric carcinogenesis. Oncogenesis 2020, 9, 15. [Google Scholar] [CrossRef]
- Krstic, M.; Stojanovic, N.M.; Stojnev, S.; Radenkovic, G.; Cukuranovic Kokoris, J.; Mladenovic, B.; Jankovic Velickovic, L. Interplay between STAT3, Cell Adhesion Molecules and Angiogenesis-Related Parameters in Gastric Carcinoma. Does STAT3 Really Have a Prognostic Value? Medicina (Kaunaslithuania) 2019, 55, 300. [Google Scholar] [CrossRef]
- Wei, K.L.; Chou, J.L.; Chen, Y.C.; Jin, H.; Chuang, Y.M.; Wu, C.S.; Chan, M.W.Y. Methylomics analysis identifies a putative STAT3 target, SPG20, as a noninvasive epigenetic biomarker for early detection of gastric cancer. PLoS ONE 2019, 14, e0218338. [Google Scholar] [CrossRef]
- Mirnoori, S.M.; Shahangian, S.S.; Salehi, Z.; Mashayekhi, F.; Talesh Sasani, S.; Saedi, H.S. Influence of single nucleotide polymorphisms in pri-miR-124-1 and STAT3 genes on gastric cancer susceptibility. Br. J. Biomed. Sci. 2018, 75, 182–186. [Google Scholar] [CrossRef]
- Yu, L.; Wu, D.; Gao, H.; Balic, J.J.; Tsykin, A.; Han, T.S.; Liu, Y.D.; Kennedy, C.L.; Li, J.K.; Mao, J.Q.; et al. Clinical Utility of a STAT3-Regulated miRNA-200 Family Signature with Prognostic Potential in Early Gastric Cancer. Clin. Cancer Res. J. Am. Assoc. Cancer Res. 2018, 24, 1459–1472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Huang, S.; Deng, C.; Cao, Y.; Yang, J.; Chen, G.; Zhang, B.; Duan, C.; Shi, J.; Kong, B.; et al. Co-ordinated overexpression of SIRT1 and STAT3 is associated with poor survival outcome in gastric cancer patients. Oncotarget 2017, 8, 18848–18860. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Cheng, L.; Qiu, L.X.; Wang, M.Y.; Li, J.; Sun, M.H.; Yang, Y.J.; Wang, J.C.; Jin, L.; Wang, Y.N.; et al. Associations of potentially functional variants in IL-6, JAKs and STAT3 with gastric cancer risk in an eastern Chinese population. Oncotarget 2016, 7, 28112–28123. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Yan, R.; He, X.; He, J. Constitutive activation of STAT3 and cyclin D1 overexpression contribute to proliferation, migration and invasion in gastric cancer cells. Am. J. Transl. Res. 2017, 9, 5671–5677. [Google Scholar] [PubMed]
- Law, S.K.; Micklem, K.J.; Shaw, J.M.; Zhang, X.P.; Dong, Y.; Willis, A.C.; Mason, D.Y. A new macrophage differentiation antigen which is a member of the scavenger receptor superfamily. Eur. J. Immunol. 1993, 23, 2320–2325. [Google Scholar] [CrossRef]
- Schaer, D.J.; Schaer, C.A.; Buehler, P.W.; Boykins, R.A.; Schoedon, G.; Alayash, A.I.; Schaffner, A. CD163 is the macrophage scavenger receptor for native and chemically modified hemoglobins in the absence of haptoglobin. Blood 2006, 107, 373–380. [Google Scholar] [CrossRef]
- Tarin, C.; Carril, M.; Martin-Ventura, J.L.; Markuerkiaga, I.; Padro, D.; Llamas-Granda, P.; Moreno, J.A.; Garcia, I.; Genicio, N.; Plaza-Garcia, S.; et al. Targeted gold-coated iron oxide nanoparticles for CD163 detection in atherosclerosis by MRI. Sci. Rep. 2015, 5, 17135. [Google Scholar] [CrossRef]
- Karrasch, T.; Brunnler, T.; Hamer, O.W.; Schmid, K.; Voelk, M.; Herfarth, H.; Buechler, C. Soluble CD163 is increased in patients with acute pancreatitis independent of disease severity. Exp. Mol. Pathol. 2015, 99, 236–239. [Google Scholar] [CrossRef]
- Rojo-Martinez, G.; Maymo-Masip, E.; Rodriguez, M.M.; Solano, E.; Goday, A.; Soriguer, F.; Valdes, S.; Chaves, F.J.; Delgado, E.; Colomo, N.; et al. Serum sCD163 levels are associated with type 2 diabetes mellitus and are influenced by coffee and wine consumption: Results of the Di@bet.es study. PLoS ONE 2014, 9, e101250. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, D.; Gong, B.; Wang, P.; Liu, F. CD163 as a novel target gene of STAT3 is a potential therapeutic target for gastric cancer. Oncotarget 2017, 8, 87244–87262. [Google Scholar] [CrossRef]
- Nagano, O.; Saya, H. Mechanism and biological significance of CD44 cleavage. Cancer Sci. 2004, 95, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Orian-Rousseau, V. CD44, a therapeutic target for metastasising tumours. Eur. J. Cancer (Oxf. Engl. 1990) 2010, 46, 1271–1277. [Google Scholar] [CrossRef] [PubMed]
- Zoller, M. CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 2011, 11, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, T.; Nagano, O.; Yae, T.; Tamada, M.; Motohara, T.; Oshima, H.; Oshima, M.; Ikeda, T.; Asaba, R.; Yagi, H.; et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell 2011, 19, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Nagano, O.; Okazaki, S.; Saya, H. Redox regulation in stem-like cancer cells by CD44 variant isoforms. Oncogene 2013, 32, 5191–5198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Balch, C.; Chan, M.W.; Lai, H.C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.; Nephew, K.P. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef]
- Yae, T.; Tsuchihashi, K.; Ishimoto, T.; Motohara, T.; Yoshikawa, M.; Yoshida, G.J.; Wada, T.; Masuko, T.; Mogushi, K.; Tanaka, H.; et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat. Commun. 2012, 3, 883. [Google Scholar] [CrossRef]
- Fujii, S.; Ito, K.; Ito, Y.; Ochiai, A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J. Biol. Chem. 2008, 283, 17324–17332. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.Q.; Liang, J.J.; Du, J.; Ye, Z.X.; Gao, P.; Liang, Y.L. Long Noncoding RNA DLX6-AS1 Regulates the Growth and Aggressiveness of Colorectal Cancer Cells Via Mediating miR-26a/EZH2 Axis. Cancer Biother. Radiopharm. 2020. [Google Scholar] [CrossRef]
- Chen, H.; Hou, G.; Yang, J.; Chen, W.; Guo, L.; Mao, Q.; Ge, J.; Zhang, X. SOX9-activated PXN-AS1 promotes the tumorigenesis of glioblastoma by EZH2-mediated methylation of DKK1. J. Cell. Mol. Med. 2020. [Google Scholar] [CrossRef]
- Pan, Y.M.; Wang, C.G.; Zhu, M.; Xing, R.; Cui, J.T.; Li, W.M.; Yu, D.D.; Wang, S.B.; Zhu, W.; Ye, Y.J.; et al. STAT3 signaling drives EZH2 transcriptional activation and mediates poor prognosis in gastric cancer. Mol. Cancer 2016, 15, 79. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Wang, L.L.; Zheng, X.; Meng, L.N.; Lyu, B.; Jin, H.F. Expression of p-STAT3 and vascular endothelial growth factor in MNNG-induced precancerous lesions and gastric tumors in rats. World J. Gastrointest. Oncol. 2016, 8, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Wu, D.; Zhao, L.; Huang, L.; Shen, G.; Huang, J.; Chai, Y. Prognostic role of STAT3 in solid tumors: A systematic review and meta-analysis. Oncotarget 2016, 7, 19863–19883. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Liao, G.; Liu, Y.; Huang, L.; Kang, M.; Chen, L. Overexpression of STAT3/pSTAT3 was associated with poor prognosis in gastric cancer: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 20014–20023. [Google Scholar] [PubMed]
- Hajimoradi, M.; Mohammad Hassan, Z.; Ebrahimi, M.; Soleimani, M.; Bakhshi, M.; Firouzi, J.; Samani, F.S. STAT3 is Overactivated in Gastric Cancer Stem-Like Cells. Cell J. 2016, 17, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Thiem, S.; Eissmann, M.F.; Elzer, J.; Jonas, A.; Putoczki, T.L.; Poh, A.; Nguyen, P.; Preaudet, A.; Flanagan, D.; Vincan, E.; et al. Stomach-Specific Activation of Oncogenic KRAS and STAT3-Dependent Inflammation Cooperatively Promote Gastric Tumorigenesis in a Preclinical Model. Cancer Res. 2016, 76, 2277–2287. [Google Scholar] [CrossRef]
- Balic, J.J.; Garama, D.J.; Saad, M.I.; Yu, L.; West, A.C.; West, A.J.; Livis, T.; Bhathal, P.S.; Gough, D.J.; Jenkins, B.J. Serine-Phosphorylated STAT3 Promotes Tumorigenesis via Modulation of RNA Polymerase Transcriptional Activity. Cancer Res. 2019, 79, 5272–5287. [Google Scholar] [CrossRef]
- Ji, K.; Zhang, L.; Zhang, M.; Chu, Q.; Li, X.; Wang, W. Prognostic Value and Clinicopathological Significance of p-stat3 Among Gastric Carcinoma Patients: A Systematic Review and Meta-Analysis. Medicine 2016, 95, e2641. [Google Scholar] [CrossRef]
- Sun, Y.; Guo, B.F.; Xu, L.B.; Zhong, J.T.; Liu, Z.W.; Liang, H.; Wen, N.Y.; Yun, W.J.; Zhang, L.; Zhao, X.J. Stat3-siRNA inhibits the growth of gastric cancer in vitro and in vivo. Cell Biochem. Funct. 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Kashyap, D.; Tuli, H.S.; Yerer, M.B.; Sharma, A.; Sak, K.; Srivastava, S.; Pandey, A.; Garg, V.K.; Sethi, G.; Bishayee, A. Natural product-based nanoformulations for cancer therapy: Opportunities and challenges. Semin. Cancer Biol. 2019. [Google Scholar] [CrossRef]
- Hosseini, S.H.; Zohreh, N.; Karimi, N.; Gaeini, N.; Alipour, S.; Seidi, F.; Gholipour, N. Magnetic nanoparticles double wrapped into cross-linked salep/PEGylated carboxymethyl cellulose; a biocompatible nanocarrier for pH-triggered release of doxorubicin. Int. J. Biol. Macromol. 2020, 158, 994–1006. [Google Scholar] [CrossRef] [PubMed]
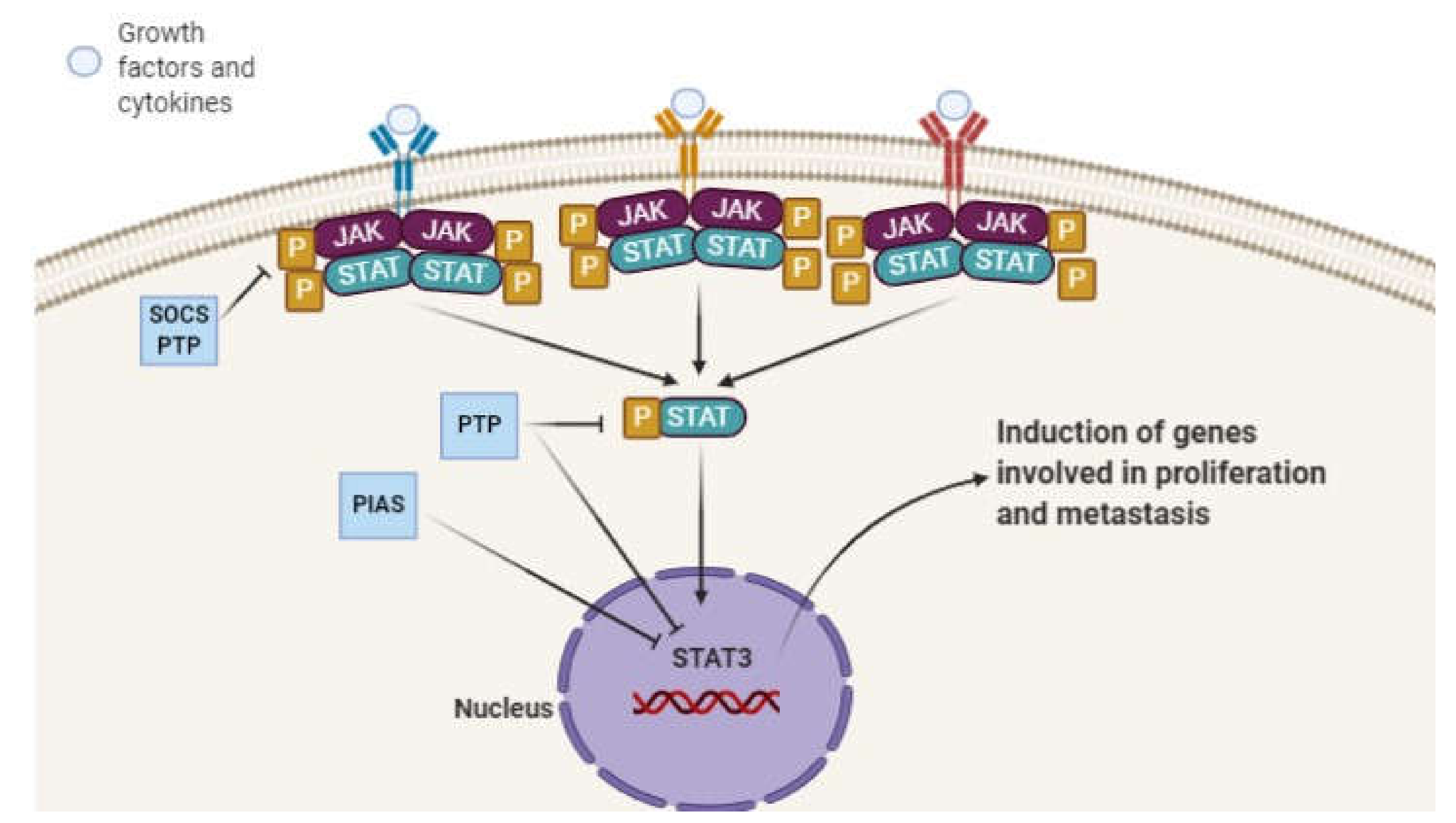
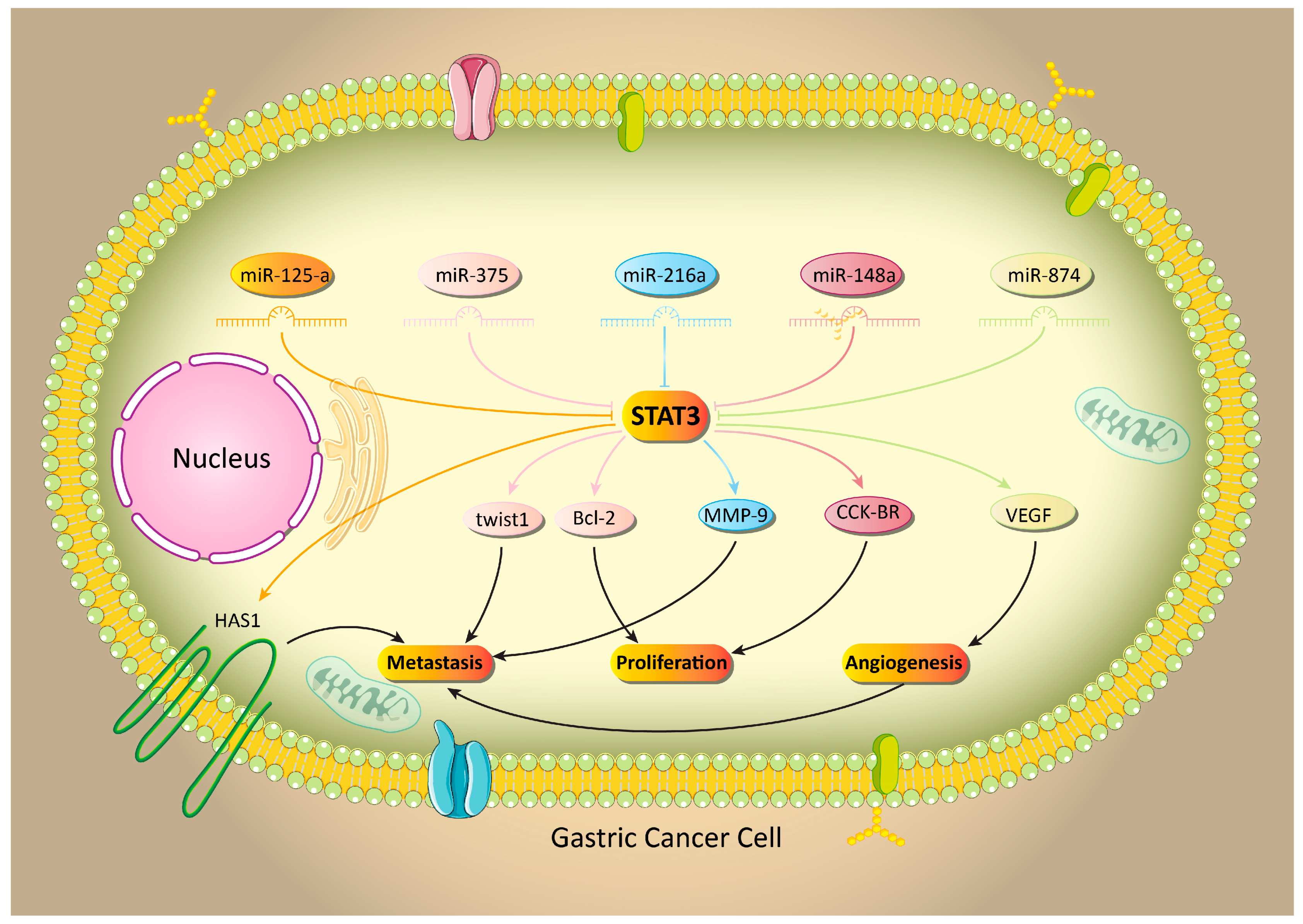

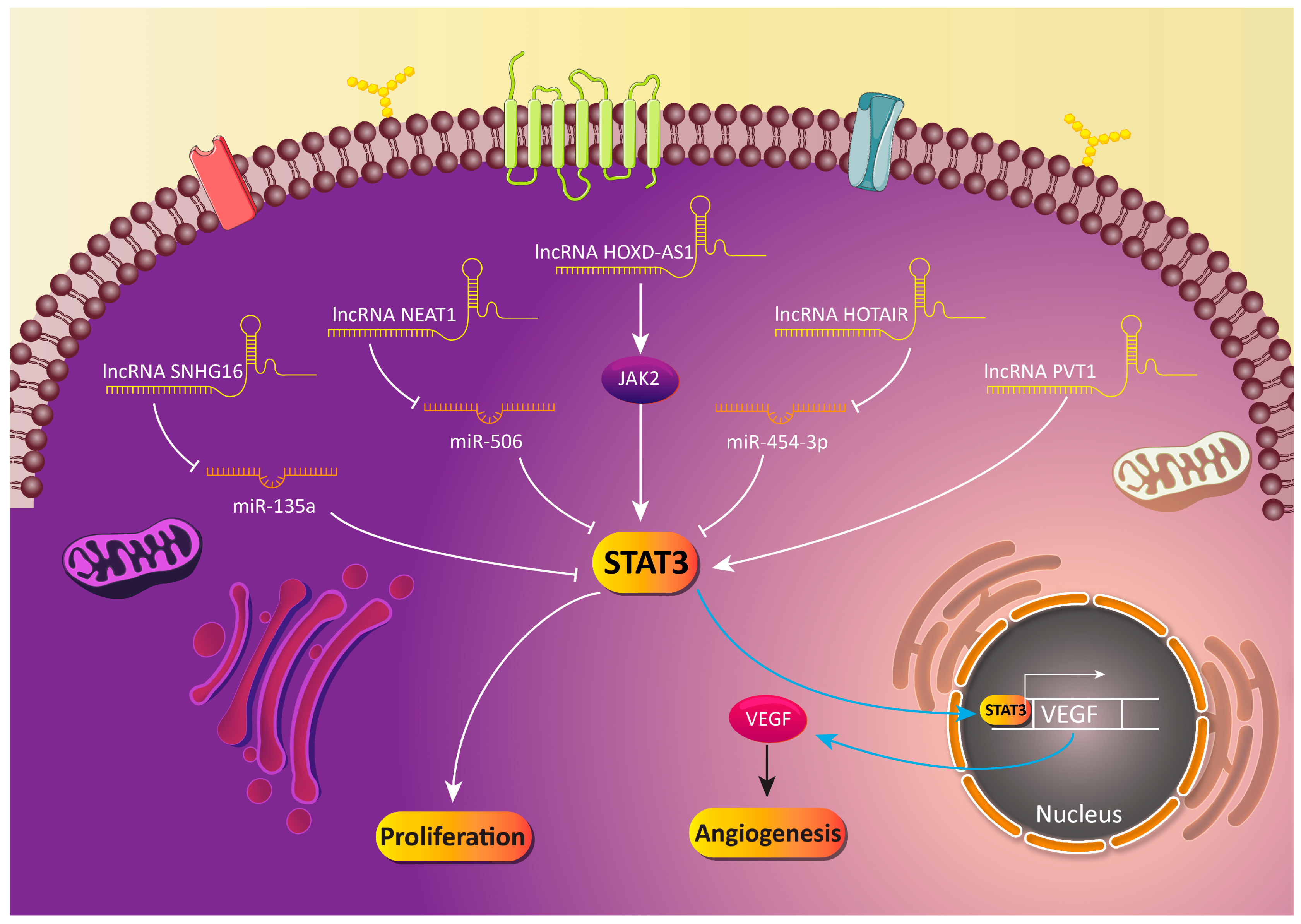
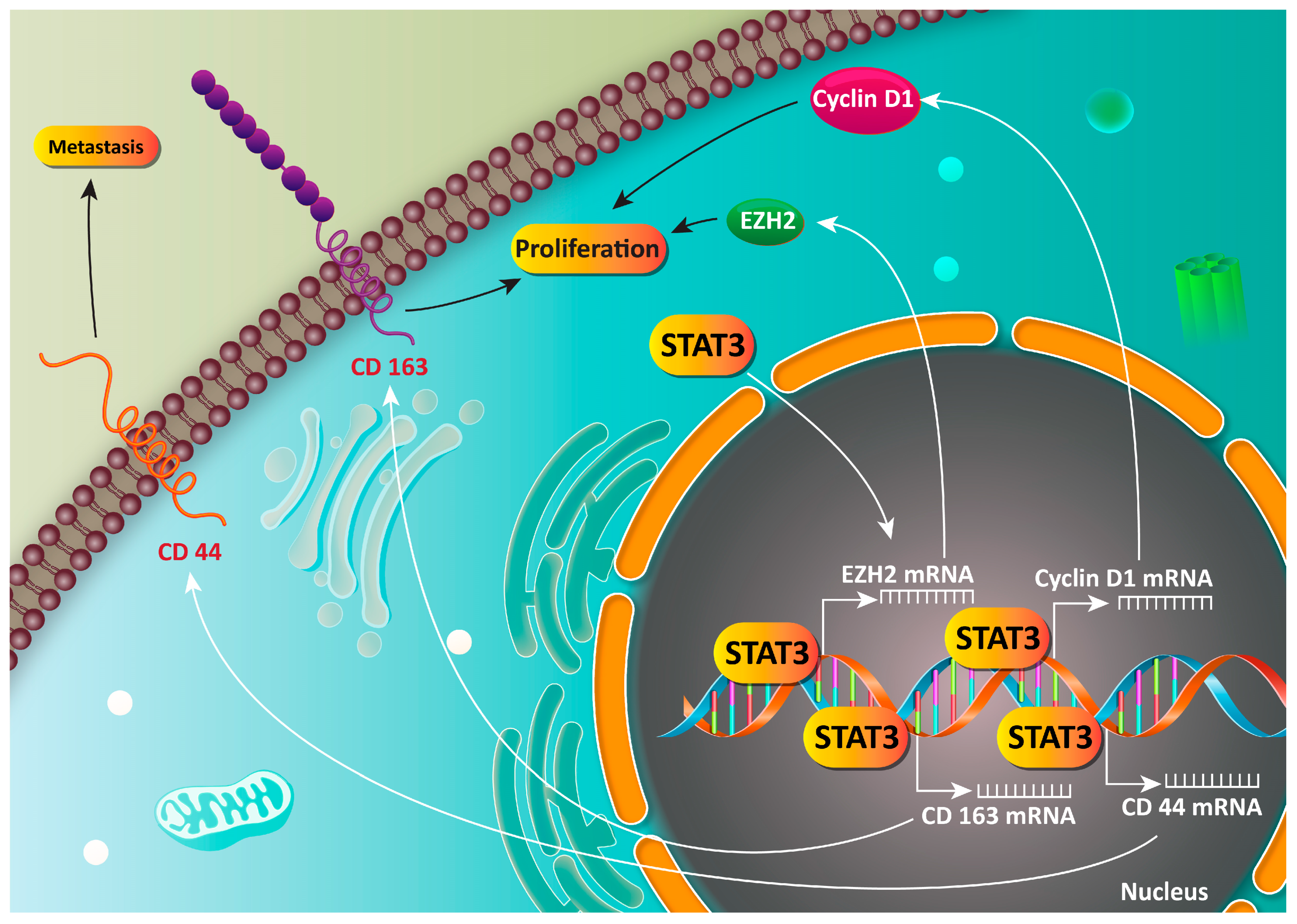
| Cancer Type | Signaling Network | Effect on STAT3 | Results | Refs |
|---|---|---|---|---|
| Breast cancer | BHLH40-AS1/IL-6/STAT3 | Induction | Promoting progression and proliferation | [60] |
| IL-6/STAT3 | Induction | Radiation induces STAT3-mediated inflammation and radio resistance | [61] | |
| MiR-454/VGLL4/STAT3 | Induction | MiR-454 induces the STAT3 signaling pathway via VGLL4 downregulation, leading to cancer malignancy | [62] | |
| PAK1/STAT3 | Induction | Stimulation of the nuclear translocation of STAT3 and enhancing breast cancer stem cell proliferation | [63] | |
| SIRT4/IL-6/STAT3 | Inhibition | Sensitizing cancer cells to tamoxifen chemotherapy | [64] | |
| Prostate cancer | IL-8/STAT3/MALAT1 | Induction | STAT3 upregulates the expression of MALAT1, leading to progression and proliferation | [65] |
| MiR-17/JAK/STAT3 | Inhibition | MiR-17 reduces the expression of pro-survival factors, such as Bcl-2, and induces apoptosis via STAT3 downregulation | [66] | |
| Bladder cancer | MiR-4500/STAT3/CCR7 | Inhibition | Suppressing migration and proliferation | [67] |
| CD44/Akt/ERK/STAT3 | Induction | Inhibition of apoptosis and cell cycle arrest | [68] | |
| Lung cancer | B7-H4/PD-1/STAT3 | Induction | Promoting proliferation and invasion via immune evasion | [69] |
| KCP10043F/STAT3 | Inhibition | Induction of apoptotic cell death | [70] | |
| BIS/STAT3 | Induction | Reducing sensitivity of cancer cells to digoxin-mediated migration and growth inhibition | [71] | |
| Glioblastoma | Annexin-A2/STAT3/oncostatin M receptor | Induction | Promoting the proliferation and invasion of cancer cells | [72] |
| Hsa-miR-181d/STAT3 | Inhibition | Garcinol upregulates the expression of hsa-miR-181d to inhibit STAT3 and the malignancy of cancer cells | [73] | |
| TROP2/JAK2/STAT3 | Induction | Promoting proliferation and migration | [74] | |
| Bradykinin B1 receptor/STAT3/IL-8 | Induction | Enhancing malignant behavior | [75] |
| Drug/Molecular Pathway | Effect on STAT3 | Clinical Trial Phase | Major Outcomes | Refs |
|---|---|---|---|---|
| AZD9150 | Inhibition | Phase I | Anti-tumor activity in pre-clinical models and clinical trial | [76] |
| GRIM19 | Inhibition | - | Sensitizing into radiotherapy | [79] |
| - | - | Phase I | STAT3 provides local progression | [81] |
| Nilotinib | Inhibition | Phase II | Diminution in cancer growth | [82] |
| OPB-31121 | Inhibition | Phase I | High tolerance Inhibition of tumor growth | [83] |
| MiiR | MiR Type | Cancer Cell Line | Effect on STAT3 | Major Outcomes | Refs |
|---|---|---|---|---|---|
| MiR-143 | Onco-suppressor | GC cell lines (AGS, SNU-1, SNU-5, SNU-16, NCIN87 and KATOIII) | Downregulation | Disrupting the proliferation and invasion of cancer cells | [96] |
| MiR-125a | Onco-suppressor | Human GC cell lines MKN45, SGC7901 and NCI-N87 | Downregulation | Reducing the expression of HAS1 and interfering with the migration of cancer cells | [99] |
| MiR-375 | Onco-suppressor | Human GC cell lines BGC-823, AGS, SGC-7901 and MKN-45 | Downregulation | Inhibiting proliferation and migration via STAT3 downregulation | [105] |
| MiR-148a | Onco-suppressor | GC cell lines SNU-1 (ATCC: CRL-5971), SNU-16 (ATCC: CRL-5974), AGS (ATCC: CRL-1739), NCI-N87 (ATCC: CRL-5822) and KATOIII (ATCC: HTB-103) | Downregulation | Suppressing growth and metastasis by the downregulation of CCK-BR via STAT3 downregulation | [108] |
| MiR-874 | Onco-suppressor | Human GC cell lines AGS and BGC823, MKN28 and SGC-7901, as well as the human normal gastric epithelial cell line GES-1 | Downregulation | Disrupting the STAT3/VEGF axis Inhibition of angiogenesis and the malignancy of cancer cells | [117] |
| MiR-216a | Onco-suppressor | Normal human gastric epithelium cell line (GES-1) and GC cell lines (SGC-7901, MGC-803, MKN-28 and BGC-823) | Downregulation | Suppressing the metastasis of cancer cells via disrupting the STAT3/EMT axis | [121] |
| MiR-93-5p | Oncogene | AGS and HEK293 cells | Upregulation | Downregulation of the STAT3/MMP-9 axis Inhibition of metastasis and invasion | [127] |
| MiR-18a | Oncogene | Human GAC cell lines MKN28 and MKN1 | Upregulation | STAT3 induction Promoting the malignant behavior of cancer cells | [131] |
| MiR-155 | Oncogene | Human GC cell lines BGC-823, NCI-N87, SGC-7901, AGS, MKN-45 and immortalized gastric mucosa epithelial cell line GES-1 | Upregulation | Stimulation of STAT3 Activation of MMP-2 and MMP-9 Enhancing invasion and migration of cancer cells | [132] |
| MiR-106a-3p | Oncogene | Human GC cell line including SGC-7901 and BGC-823 | Upregulation | Stimulation of aptinib resistance Activation of JAK2/STAT3 signaling SOCS2, SOCS4 and SOCS5 downregulation | [133] |
| Anti-Tumor Compound | Cell Line | Dose | Duration of Experiment | Results | Refs |
|---|---|---|---|---|---|
| Piperine | TMK-1 human GC cell line | 25, 50 and 100 μM | 1 h | Downregulation of STAT3 Inhibition of IL-1β and IL-6 Decreasing viability and proliferation of cancer cells | [175] |
| Tanshinone IIA | Human GC cell lines (SNU-638, MKN1 and AGS) | 2.5, 5 and 10 μg/mL | 12, 24, 48 and 72 h | Inhibition of STAT3 Reduction in the progression and malignancy of cancer cells | [176] |
| Oxymatrine | Human GC cell lines SGC-7901, MGC-803, BGC-823, HGC-27, AGS and GES-1 | 0.5, 1, 2, 4 and 8 mg/mL | 24, 48 and 72 h | Diminishing proliferation and malignancy of cancer cells Inhibition of IL-21R-mediated STAT3 | [177] |
| Luteolin | Gastric tumor cell lines of SGC7901, SGC7901/DDP, HGC27, MGC803, BGC803 and BGC823 | 10 μM | - | Selective eradication of STAT3 overexpression-GC cells Increasing the binding of STAT3 to SHP-1 | [178] |
| Parthenolide | Human GC drug-resistant SGC-7901/DDP cell line | 1.25, 2.5, 5 and 10 μmol/L | 24, 48 and 72 h | Induction of apoptosis Inhibition of drug resistance via the downregulation of STAT3 | [179] |
| Curcumin analogue | Human GC cell lines (BGC-823, SGC-7901) | 0.5, 1, 5, 10, 20, 50, 80 and 100 μM | 24 and 48 h | Induction of apoptosis and mitotic arrest Downregulation of STAT3 | [180] |
| Nifuratel | Human GC cell lines SGC-7901 and BGC-823 | 75, 150 and 300 μM | 24 h | Inhibition of IL-6-mediated STAT3 activation | [181] |
| Cryptotanshinone | Human GC cell lines SGC-7901 and HGC-27 | 2.5, 5, 7.5, 10, 15 and 20 μM | 4 h | Enhancing anti-tumor activity of doxorubicin Inhibition of STAT3 phosphorylation | [182] |
| Asiatic acid | SGC7901 (metastatic carcinoma of lymph node) | 1, 5, 10, 25 and 50 μM | 12 h | Stimulation of apoptosis Inhibition of proliferation and migration Downregulation of STAT3 | [183] |
| Sulforaphane | Human GC cell lines MGC803 and BGC823 | 2.5, 5 and 10 μM | 72 h | Sensitizing cancer cells to chemotherapy Suppressing cancer stem cell-like properties Upregulation of miR-124 and subsequent downregulation of IL-6/STAT3 | [184] |
| Thymoquinone | Three human GC cells (HGC27, BGC823 and SGC7901) | 25, 50 and 75 μmol/L | 24 h | Suppressing STAT3 phosphorylation Downregulation of survival factors such as Bcl-2 and cyclin-D | [185] |
| Paeoniflorin | Human gastric carcinoma MGC-803 cells and human normal gastric mucosa GES-1 cell lines | 5, 10 and 20 μmol/L | 48 h | Downregulation of STAT3 Interfering with the proliferation and invasion of cancer cells | [186] |
| Eupatilin | Human GC cell line MKN45 | 50 and 100 μM | 16 h | Inhibiting the STAT3 signaling pathway Suppressing VEGF and the growth of cancer cells | [187] |
| Epigallocatechin-6-gallate | Human gastric cancer (AGS) cells | 5, 10, 25 and 50 μmol/L | 24 h | Suppressing IL-6/STAT3/VEGF results in growth inhibition | [188] |
| Cucurbitacin B | GC MKN-45 cells | 0.1, 1 and 10 μM | 12, 24 and 48 h | Sensitizing cancer cells to cell death Downregulation of JAK2/STAT3 | [189] |
| Ponicidin | Human MKN28 cell line | 10, 25 and 50 μmol/L | 48 h | Induction of apoptosis Downregulation of JAK2/STAT3 | [190] |
| Cycloastragenol | Human gastric adenocarcinoma SNU-1 and SNU-16 cells | 1, 5, 10, 30 and 50 μM | 24 h | Inhibition of STAT3 phosphorylation at tyrosine 705 via suppressing Src and JAK1/2 activation Induction of apoptosis | [191] |
| Fucoxanthin | SGC-7901 cells | 25, 50 and 75 μM | 24 h | Downregulation of STAT3 Induction of apoptosis and cell cycle arrest | [192] |
| HJC0152 (niclosamide) | Six GC cell lines (AGS, HGC-27, MKN28, MKN45, SGC7901 and BGC-823) | 5, 10 and 20 μM | 1, 2 and 4 h | Suppressing the STAT3 signaling pathway and subsequent decrease in the expression of survival factors such as Survivin and Mcl-1 | [193] |
| Piceatannol | Human GC SGC-7901 cell line | 10 and 20 μM | - | Inhibiting STAT3 phosphorylation | [194] |
| BP-1-102 | Five human GC cell lines (AGS, HGC-27, MKN28, MGC803 and SGC7901) | 2, 4 and 6 μM | 72 h | Suppressing the invasion and proliferation of cancer cells in a dose- and time-dependent manner Downregulation of STAT3 | [195] |
| lncRNA | Type of lncRNA | Downstream Signaling | Cell Line | Effect on STAT3 | Major Results | Refs |
|---|---|---|---|---|---|---|
| SNHG16 | Oncogene | MiR-135a/JAK2/STAT3 | Four GC cell lines (BGC823, MGC803, MKN45, SGC7901) and normal GC cell line GES-1 | Induction | Promoting colony formation and the proliferation of cancer cells Inhibition of apoptosis | [222] |
| HOTAIR | Oncogene | MiR-454-3p/STAT3/cyclin D1 | AGS and SGC7901 cells | Induction | Knock-down of HOTAIR Sensitizing cancer cells to apoptosis Upregulation of miR-454-3p and the subsequent inhibition of the STAT3/cyclin D1 axis | [216] |
| PVT1 | Oncogene | STAT3/VEGF | GES-1, SGC-7901, BGC-823, MNK-45, AGS, SUN-638, HGC-27 and HUVEC | Induction | Promoting angiogenesis and the growth of cancer cells | [219] |
| NEAT1 | Oncogene | MiR-506/STAT3 | BGC823, SGC-7901, AGS, MGC803, MKN28 cells, GES-1 and HEK-293T cells | Induction | Sponging miR-506 Enhancing expression of STAT3 Increasing the malignancy of cancer cells | [220] |
| GACAT3 | Oncogene | IL-6/STAT3 | Human GC cell lines HGC-27 and SGC-7901 | Induction | Enhancing the proliferation of cancer cells in an inflammatory response behavior | [221] |
| HOXD-AS1 | Oncogene | JAK2/STAT3 | Human GC cell lines (SGC-7901, BGC-823, MGC803 and MKN-45) | Induction | Silencing of HOXD-AS1 is correlated with the downregulation of STAT3 and growth inhibition | [223] |
| Signaling Network | Cell Line | Effect on STAT3 | Results | Refs |
|---|---|---|---|---|
| CXCR4/JAK2/STAT3/VEGF | Human SGC-7901 and MKN45 cells | Induction | Induction of VEGF by CXCR4 Subsequent activation of JAK2/STAT3 Enhancing the migration and proliferation of cancer cells | [285] |
| NOX4/JAK2/STAT3/EMT | Six human GC cell lines (MKN-45, SGC-7901, MGC-803, BGC-823, MKN-28 and AGS) | Induction | Induction of JAK2/STAT3 by NOX4 Stimulation of EMT Promoting invasion | [286] |
| DC-SIGNR | Human GC cell lines, SGC-7901, MGC-803, BGC-823 and AGS, and the control gastric epithelial cell line GES-1 | Induction | Ensuring the growth and viability of cancer cells by the induction of the JAK2/STAT3 pathway | [287] |
| HOXA11 | GC cell lines (KATO III, NCI-N87, SNU-16, AGS and SNU-16) and HEK 293T | Induction | Promoting the stemness and migration of cancer cells by the stimulation of STAT3 | [288] |
| TFF1/STAT3 | AGS cells | Induction | Enhancing the proliferation and migration of cancer cells | [67] |
| DARPP-32/IGF-1R/STAT3 | AGS cells | Induction | Promoting proliferation and invasion | [289] |
| BMX-ARHGAP/STAT3 | Four human GC cell lines (SNU-5, MNK-45, AGS and SGC7901) and the normal gastric epithelial cell line (GES-1) | Induction | Maintaining the carcinogenesis ability of GC stem cells | [279] |
| CXCL16/STAT3/Ror1 | MKN45, MKN45-Luc and KATOIII cells | Induction | Increasing progression and malignancy | [290] |
| Complement C3/JAK2/STAT3 | Human SGC-7901 and MGC-803 cells, normal gastric epithelial cells (GES-1) | Induction | Poor prognosis and enhanced proliferation of cancer cells | [291] |
| BTF3/JAK2/STAT3/EMT | Human gastric epithelial cell line GES-1 and human GC cell lines, including AGS, HGC-27, MKN-28, MGC-803 and SGC-7901 cells | Induction | Induction of JAK2/STAT3 by BTF3 Stimulation of EMT Enhancing the proliferation and migration of cancer cells | [292] |
| IGF1/IGF1R/STAT3/IFITM2 | GC cell lines | Induction | Enhancing the growth and metastasis of cancer cells | [293] |
| IL-6/JAK2/STAT3 | GC cell lines SNU-1, MKN45, SGC7901 and MKN28 | Induction | Secretion of IL-6 by CAMs Stimulation of JAK2/STAT3 by IL-6 | [294] |
| TNF-α/IL-6/STAT3 | SGC7901 cells | Induction | Induction of EMT Ensuring the metastasis of cancer cells | [295] |
| Succinate/STAT3/VEGF | Human gastric mucosal epithelial cell line GES-1 and human GC cell lines AGS (low-differentiated human gastric adenocarcinoma), NCI-N87 (well-differentiated human carcinoma), BGC-823 (low-differentiated human gastric adenocarcinoma) and SGC-7901 (moderate-differentiated human gastric adenocarcinoma) | Induction | Induction of VEGF by succinate via STAT3 overexpression Increasing viability and invasion of cancer cells | [296] |
| Cyclophilin B/STAT3/miR-520d-5p | GC cell lines | Induction | There is feedback consisting of the downregulation of miR-520d-5p and upregulation of cyclophilin B and STAT3, leading to the enhanced growth of cancer cells | [297] |
| CMTM3/STAT3/Twist1/EMT | Human GC cell line SGC-7901 | Inhibition | Downregulation of STAT3 by CMTM3 Suppressing metastasis | [298] |
| HCCR/STAT3 | Human GC cell lines AGS, MKN-45, BGC823, MGC803, HGC27, SGC7901, NCI-N87 | Induction | Triggering chemoresistance | [299] |
| GRIM19/STAT3 | Immortalized normal gastric epithelial cell line GES-1, human embryonic kidney HEK-293 cells, human GC SGC-7901 and BGC-823 cell lines | Inhibition | Suppressing STAT3 Induction of apoptosis | [300] |
| ROS/IL-6/STAT3 | AGS cells | Induction | Enhanced generation of ROS by H. pylori Stimulation of IL-6/STAT3 Enhanced proliferation and invasion of cancer cells | [301] |
| IL-6/STAT3/VEGF | GC cell lines including SGC-7901, MGC, MKN-28 and AGS | Induction | Promoting invasion and angiogenesis Induction of STAT3 and subsequent activation of VEGF | [302] |
| IL-17/STAT3/VEGF | Human GC (AGS) cells and other cells SGC7901, MKN 45 and BGC823 | Induction | Enhancing growth Induction of angiogenesis Activation of STAT3/VEGF | [303] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashrafizadeh, M.; Zarrabi, A.; Orouei, S.; Zarrin, V.; Rahmani Moghadam, E.; Zabolian, A.; Mohammadi, S.; Hushmandi, K.; Gharehaghajlou, Y.; Makvandi, P.; et al. STAT3 Pathway in Gastric Cancer: Signaling, Therapeutic Targeting and Future Prospects. Biology 2020, 9, 126. https://doi.org/10.3390/biology9060126
Ashrafizadeh M, Zarrabi A, Orouei S, Zarrin V, Rahmani Moghadam E, Zabolian A, Mohammadi S, Hushmandi K, Gharehaghajlou Y, Makvandi P, et al. STAT3 Pathway in Gastric Cancer: Signaling, Therapeutic Targeting and Future Prospects. Biology. 2020; 9(6):126. https://doi.org/10.3390/biology9060126
Chicago/Turabian StyleAshrafizadeh, Milad, Ali Zarrabi, Sima Orouei, Vahideh Zarrin, Ebrahim Rahmani Moghadam, Amirhossein Zabolian, Shima Mohammadi, Kiavash Hushmandi, Yashar Gharehaghajlou, Pooyan Makvandi, and et al. 2020. "STAT3 Pathway in Gastric Cancer: Signaling, Therapeutic Targeting and Future Prospects" Biology 9, no. 6: 126. https://doi.org/10.3390/biology9060126
APA StyleAshrafizadeh, M., Zarrabi, A., Orouei, S., Zarrin, V., Rahmani Moghadam, E., Zabolian, A., Mohammadi, S., Hushmandi, K., Gharehaghajlou, Y., Makvandi, P., Najafi, M., & Mohammadinejad, R. (2020). STAT3 Pathway in Gastric Cancer: Signaling, Therapeutic Targeting and Future Prospects. Biology, 9(6), 126. https://doi.org/10.3390/biology9060126







