Next-Generation Sequencing and MALDI Mass Spectrometry in the Study of Multiresistant Processed Meat Vancomycin-Resistant Enterococci (VRE)
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Bacterial Isolation
2.2. Antimicrobial Susceptibility Test
2.3. Whole-Genome Sequencing, Genome Assembly, and Gene Detection
2.4. MALDI-TOF Mass Spectrometry
2.5. Statistics and Bioinformatics Analysis of Spectra
3. Results
3.1. Bacterial Isolation
3.2. Antibiotic Susceptibility Study
3.3. Detection of Antibiotic Resistance Genes
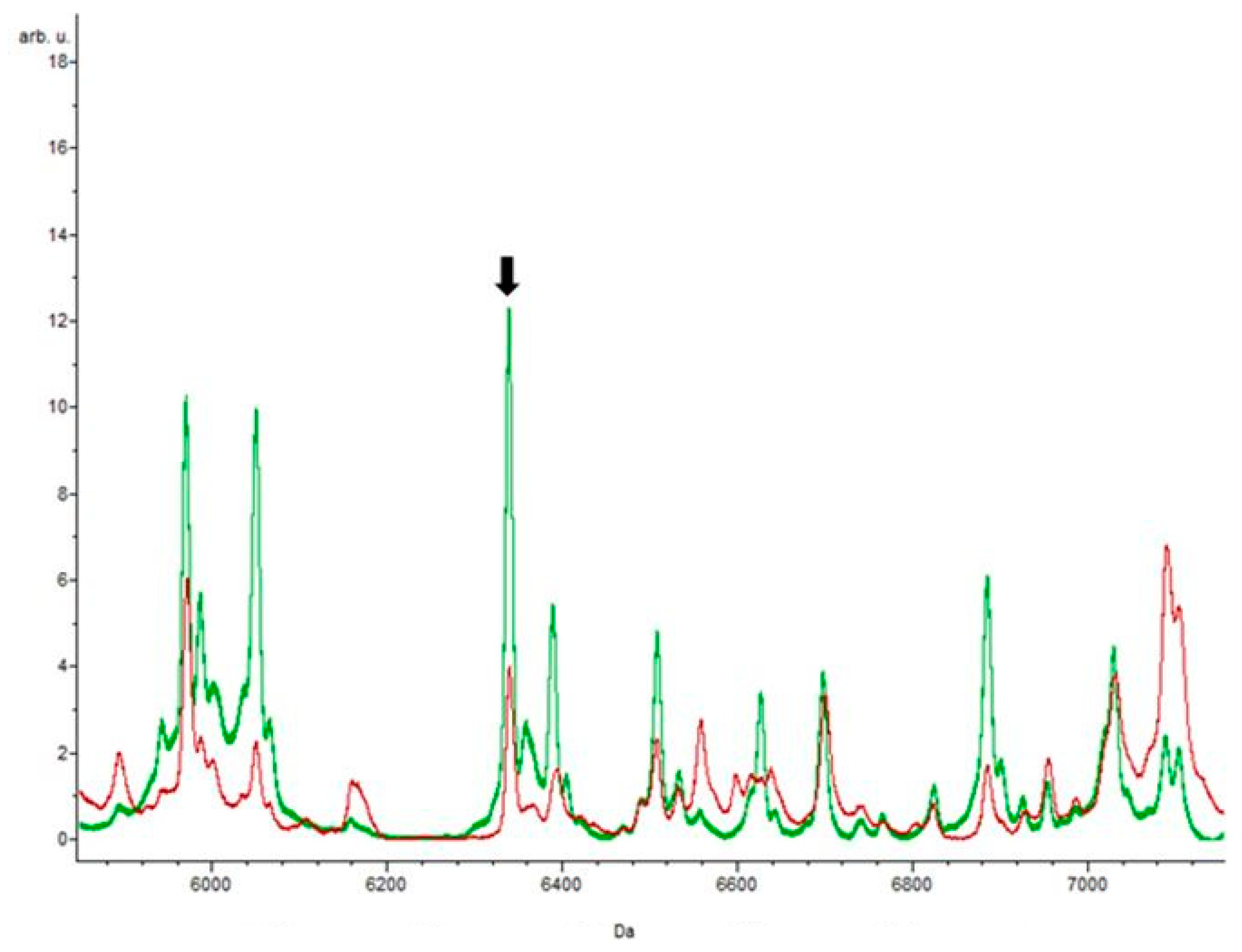
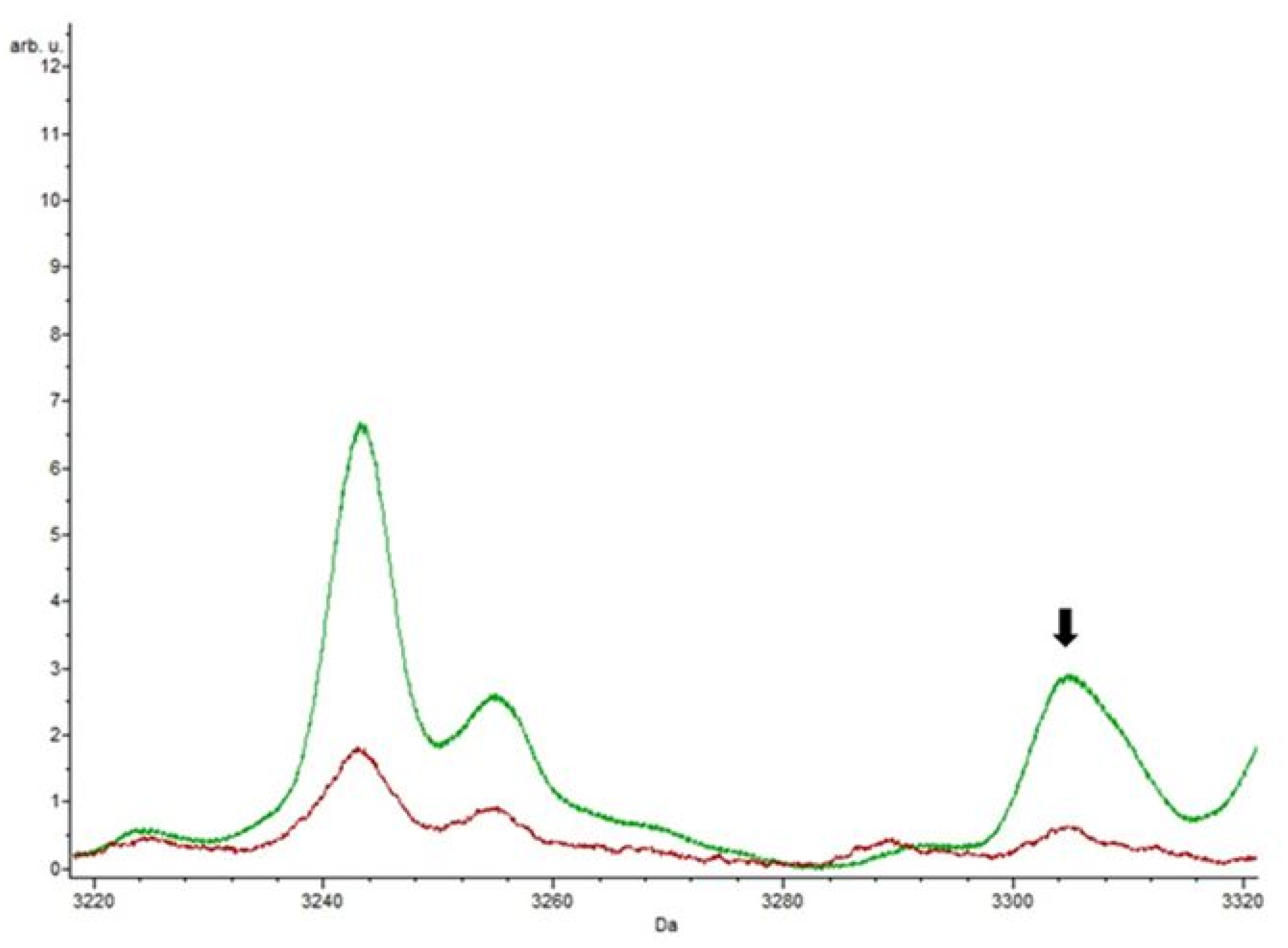
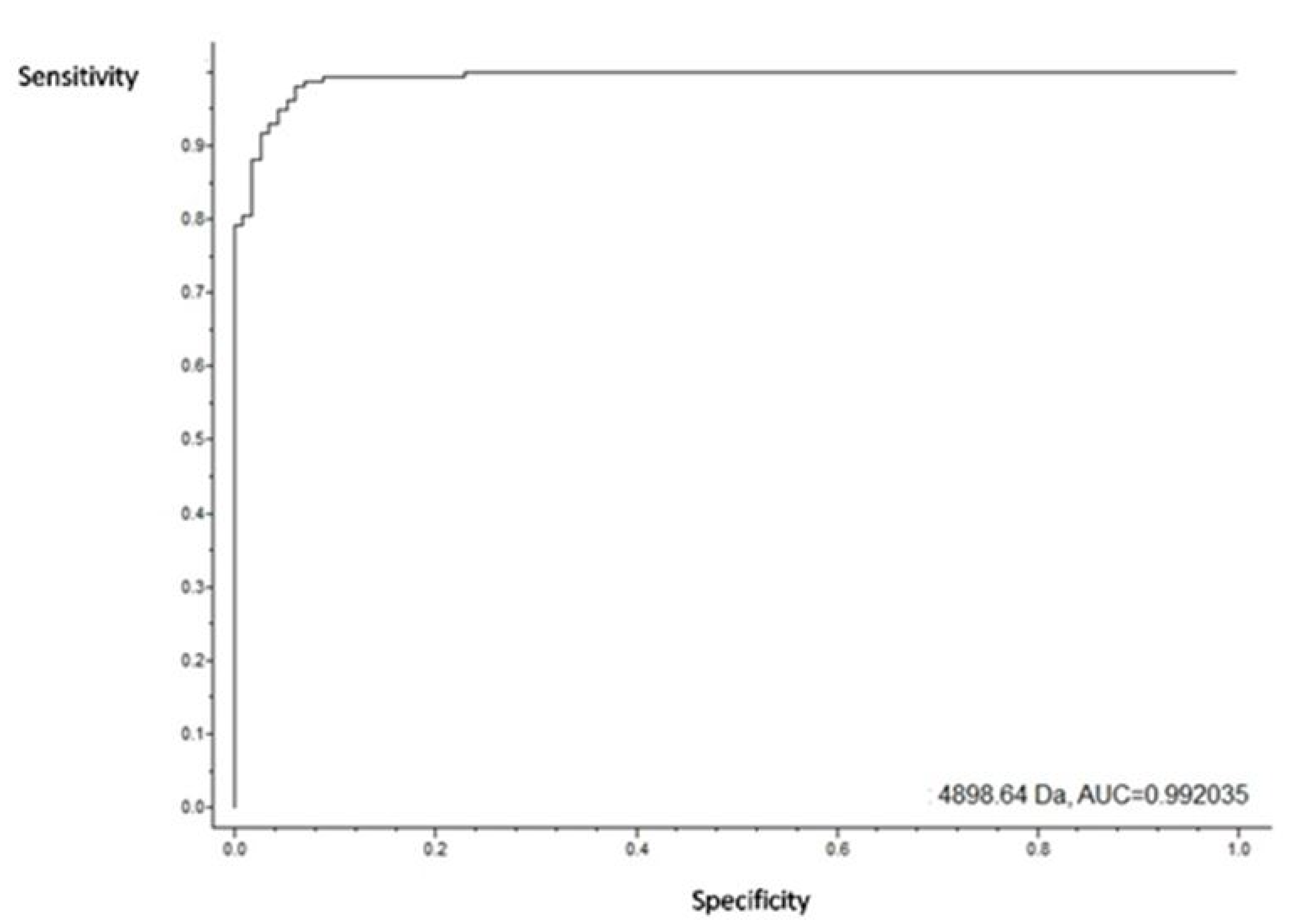
3.4. Putative Biomarkers of Resistance Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lebreton, F.; Willems, R.J.L.; Gilmore, M.S. Enterococcus Diversity, Origins in Nature, and Gut Colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Shepard, B.D.; Gilmore, M.S. Antibiotic-resistant enterococci: the mechanisms and dynamics of drug introduction and resistance. Microbes Infect. 2002, 4, 215–224. [Google Scholar] [CrossRef]
- Gonçalves, A.; Poeta, P.; Silva, N.; Araújo, C.; Lopez, M.; Ruiz, E.; Uliyakina, I.; Direitinho, J.; Igrejas, G.; Torres, C. Characterization of Vancomycin-Resistant Enterococci Isolated from Fecal Samples of Ostriches by Molecular Methods. Foodborne Pathog. Dis. 2010, 7, 1133–1136. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Genet. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Poeta, P.; Costa, D.; Igrejas, G.; Rodrigues, J.; Torres, C. Phenotypic and genotypic characterization of antimicrobial resistance in faecal enterococci from wild boars (Sus scrofa). Vet. Microbiol. 2007, 125, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed]
- Landers, T.; Cohen, B.; Wittum, T.E.; Larson, E.L. A Review of Antibiotic Use in Food Animals: Perspective, Policy, and Potential. Public Heal. Rep. 2012, 127, 4–22. [Google Scholar] [CrossRef]
- Bhavnani, S.M.; Drake, J.A.; Forrest, A.; Deinhart, J.A.; Jones, R.N.; Biedenbach, D.J.; Ballow, C.H. A nationwide, multicenter, case-control study comparing risk factors, treatment, and outcome for vancomycin-resistant and -susceptible enterococcal bacteremia. Diagn. Microbiol. Infect. Dis. 2000, 36, 145–158. [Google Scholar] [CrossRef]
- Taji, A.; Heidari, H.; Ebrahim-Saraie, H.S.; Sarvari, J.; Motamedifar, M. High prevalence of vancomycin and high-level gentamicin resistance in Enterococcus faecalis isolates. Acta Microbiol. et Immunol. Hung. 2019, 66, 203–217. [Google Scholar] [CrossRef]
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-Resistant Enterococci: A Review of Antimicrobial Resistance Mechanisms and Perspectives of Human and Animal Health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef]
- Zalipour, M.; Esfahani, B.N.; Havaei, S.A. Phenotypic and genotypic characterization of glycopeptide, aminoglycoside and macrolide resistance among clinical isolates of Enterococcus faecalis: a multicenter based study. BMC Res. Notes 2019, 12, 292. [Google Scholar] [CrossRef]
- Arthur, M.; Molinas, C.; Depardieu, F.; Courvalin, P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 1993, 175, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Chotinantakul, K.; Chansiw, N.; Okada, S. Antimicrobial resistance of Enterococcus spp. isolated from Thai fermented pork in Chiang Rai Province, Thailand. J. Glob. Antimicrob. Resist. 2018, 12, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Jahan, M.; Zhanel, G.G.; Sparling, R.; Holley, R.A. Horizontal transfer of antibiotic resistance from Enterococcus faecium of fermented meat origin to clinical isolates of E. faecium and Enterococcus faecalis. Int. J. Food Microbiol. 2015, 199, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Vakulenko, S.B.; Donabedian, S.M.; Voskresenskiy, A.M.; Zervos, M.J.; Lerner, S.A.; Chow, J. Multiplex PCR for Detection of Aminoglycoside Resistance Genes in Enterococci. Antimicrob. Agents Chemother. 2003, 47, 1423–1426. [Google Scholar] [CrossRef]
- Abdelkareem, M.Z.; Sayed, M.; Hassuna, N.A.; Mahmoud, M.S.; Abdelwahab, S.F. Multi-drug-resistant Enterococcus faecalisamong Egyptian patients with urinary tract infection. J. Chemother. 2016, 29, 74–82. [Google Scholar] [CrossRef]
- Chouchani, C.; El Salabi, A.; Marrakchi, R.; Ferchichi, L.; Walsh, T.R. First report of mefA and msrA/msrB multidrug efflux pumps associated with blaTEM-1 β-lactamase in Enterococcus faecalis. Int. J. Infect. Dis. 2012, 16, e104–e109. [Google Scholar] [CrossRef]
- Jungblut, P.R.; Holzhütter, H.-G.; Apweiler, R.; Schlüter, H. The speciation of the proteome. Chem. Central J. 2008, 2, 16. [Google Scholar] [CrossRef]
- Radhouani, H.; Pinto, L.; Poeta, P.; Igrejas, G. After genomics, what proteomics tools could help us understand the antimicrobial resistance of Escherichia coli? J. Proteom. 2012, 75, 2773–2789. [Google Scholar] [CrossRef]
- Wolters, M.; Rohde, H.; Maier, T.; Belmar-Campos, C.; Franke, G.; Scherpe, S.; Aepfelbacher, M.; Christner, M. MALDI-TOF MS fingerprinting allows for discrimination of major methicillin-resistant Staphylococcus aureus lineages. Int. J. Med Microbiol. 2011, 301, 64–68. [Google Scholar] [CrossRef]
- Wei, W.; Li-Jun, W.; Sui, W.; Gui, Z.; Xin-Xin, L. Application of Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry in the Screening of VanA-Positive Enterococcus Faecium. Eur. J. Mass Spectrom. 2014, 20, 461–465. [Google Scholar] [CrossRef]
- Croxatto, A.; Prod’Hom, G.; Greub, G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 2012, 36, 380–407. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Sarwal, M.M. The proteogenomic path towards biomarker discovery. Pediatr. Transplant. 2008, 12, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Veenstra, T.D. Global and targeted quantitative proteomics for biomarker discovery. J. Chromatogr. B 2007, 847, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-M.; Han, J.J.; Altwerger, G.; Kohn, E.C. Proteomics and biomarkers in clinical trials for drug development. J. Proteom. 2011, 74, 2632–2641. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2015, 6, 71–79. [Google Scholar] [CrossRef]
- Thornsberry, C. NCCLS Standards for Antimicrobial Susceptibility Tests. Lab. Med. 1983, 14, 549–553. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Single-Cell Genomes and Mini-Metagenomes From Chimeric MDA Products. J. Comput. Boil. 2013, 20, 714–737. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinform. 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLOS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Hunt, M.; Mather, A.E.; Sánchez-Busó, L.; Page, A.J.; Parkhill, J.; Keane, J.A.; Harris, S.R. ARIBA: Rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genomics 2017, 3, e000131. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Freiwald, A.; Sauer, S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 2009, 4, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Reyes, K.; Bardossy, A.C.; Zervos, M. Vancomycin-Resistant Enterococci. Infect. Dis. Clin. North Am. 2016, 30, 953–965. [Google Scholar] [CrossRef]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal Infection-Treatment and Antibiotic Resistance. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Barros, J.; Andrade, M.; Radhouani, H.; Lopez, M.; Igrejas, G.; Poeta, P.; Torres, C. Detection of vanA-Containing Enterococcus Species in Faecal Microbiota of Gilthead Seabream (Sparus aurata). Microbes Environ. 2012, 27, 509–511. [Google Scholar] [CrossRef]
- Gonçalves, A.; Igrejas, G.; Radhouani, H.; Correia, S.; Pacheco, R.; Santos, T.; Monteiro, R.; Guerra, A.; Petrucci-Fonseca, F.; Brito, F.; et al. Antimicrobial resistance in faecal enterococci and Escherichia coli isolates recovered from Iberian wolf. Lett. Appl. Microbiol. 2013, 56, 268–274. [Google Scholar] [CrossRef]
- Marinho, C.M.; Silva, N.; Pombo, S.; Santos, T.; Monteiro, R.; Gonçalves, A.; Micael, J.; Rodrigues, P.; Costa, A.; Igrejas, G.; et al. Echinoderms from Azores islands: An unexpected source of antibiotic resistant Enterococcus spp. and Escherichia coli isolates. Mar. Pollut. Bull. 2013, 69, 122–127. [Google Scholar] [CrossRef]
- Poeta, P.; Radhouani, H.; Igrejas, G.; Gonçalves, A.; Carvalho, C.; Rodrigues, J.; Vinué, L.; Somalo, S.; Torres, C. Seagulls of the Berlengas Natural Reserve of Portugal as Carriers of Fecal Escherichia coli Harboring CTX-M and TEM Extended-Spectrum Beta-Lactamases. Appl. Environ. Microbiol. 2008, 74, 7439–7441. [Google Scholar] [CrossRef]
- Cetinkaya, F.; Mus, T.E.; Soyutemiz, G.E.; Cibik, R. Prevalence and antibiotic resistance of vancomycin-resistant enterococci in animal originated foods. Turk. J. Vet. Anim. Sci. 2013, 37, 588–593. [Google Scholar] [CrossRef]
- Guerrero-Ramos, E.; Molina, D.; Blanco-Morán, S.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C.; Capita, R. Prevalence, Antimicrobial Resistance, and Genotypic Characterization of Vancomycin-Resistant Enterococci in Meat Preparations. J. Food Prot. 2016, 79, 748–756. [Google Scholar] [CrossRef]
- Jung, W.K.; Lim, J.Y.; Kwon, N.H.; Kim, J.M.; Hong, S.K.; Koo, H.C.; Kim, S.H.; Park, Y.H. Vancomycin-resistant enterococci from animal sources in Korea. Int. J. Food Microbiol. 2007, 113, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Klibi, N.; Ben Said, L.; Jouini, A.; Ben Slama, K.; Lopez, M.; Ben Sallem, R.; Boudabous, A.; Torres, C. Species distribution, antibiotic resistance and virulence traits in enterococci from meat in Tunisia. Meat Sci. 2013, 93, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Mus, T.E.; Cetinkaya, F.; Cibik, R.; Soyutemiz, G.E.; Simsek, H.; Coplu, N. Pathogenicity determinants and antibiotic resistance profiles of enterococci from foods of animal origin in Turkey. Acta Veter- Hung. 2017, 65, 461–474. [Google Scholar] [CrossRef]
- Igbinosa, E.O.; Beshiru, A. Antimicrobial Resistance, Virulence Determinants, and Biofilm Formation of Enterococcus Species From Ready-to-Eat Seafood. Front. Microbiol. 2019, 10, 728. [Google Scholar] [CrossRef]
- Zarfel, G.; Galler, H.; Luxner, J.; Petternel, C.; Reinthaler, F.F.; Haas, D.; Kittinger, C.; Grisold, A.; Pless, P.; Feierl, G. Multiresistant Bacteria Isolated from Chicken Meat in Austria. Int. J. Environ. Res. Public Heal. 2014, 11, 12582–12593. [Google Scholar] [CrossRef]
- Hayes, J.R.; English, L.L.; Carter, P.J.; Proescholdt, T.; Lee, K.Y.; Wagner, D.D.; White, D.G. Prevalence and Antimicrobial Resistance of Enterococcus Species Isolated from Retail Meats. Appl. Environ. Microbiol. 2003, 69, 7153–7160. [Google Scholar] [CrossRef]
- Poeta, P.; Costa, D.; Rodrigues, J.; Torres, C. Study of faecal colonization by vanA-containing Enterococcus strains in healthy humans, pets, poultry and wild animals in Portugal. J. Antimicrob. Chemother. 2005, 55, 278–280. [Google Scholar] [CrossRef]
- Fernandez-Guerrero, M.; Rouse, M.S.; Henry, N.K.; E Geraci, J.; Wilson, W.R. In vitro and in vivo activity of ciprofloxacin against enterococci isolated from patients with infective endocarditis. Antimicrob. Agents Chemother. 1987, 31, 430–433. [Google Scholar] [CrossRef]
- Deshpande, L.M.; Fritsche, T.R.; Moet, G.J.; Biedenbach, D.J.; Jones, R.N. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 2007, 58, 163–170. [Google Scholar] [CrossRef]
- Lautenbach, E.; Gould, C.V.; A LaRosa, L.; Marr, A.M.; Nachamkin, I.; Bilker, W.B.; O Fishman, N. Emergence of resistance to chloramphenicol among vancomycin-resistant enterococcal (VRE) bloodstream isolates. Int. J. Antimicrob. Agents 2004, 23, 200–203. [Google Scholar] [CrossRef]
- Lautenbach, E.; Schuster, M.G.; Bilker, W.B.; Brennan, P.J. The role of chloramphenicol in the treatment of bloodstream infection due to vancomycin-resistant Enterococcus. Clin. Infect. Dis. 1998, 27, 1259–1265. [Google Scholar] [CrossRef] [PubMed]
- Patel, G.; Snydman, D.R. Vancomycin-Resistant Enterococcus Infections in Solid Organ Transplantation. Arab. Archaeol. Epigr. 2013, 13, 59–67. [Google Scholar] [CrossRef]
- Patterson, J.E.; Sweeney, A.H.; Simms, M.; Carley, N.; Mangi, R.; Sabetta, J.; Lyons, R.W. An Analysis of 110 Serious Enterococcal Infections Epidemiology, Antibiotic Susceptibility, and Outcome. Med. 1995, 74, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Del Grosso, M.; Caprioli, A.; Chinzari, P.; Fontana, M.C.; Pezzotti, G.; Manfrin, A.; Di Giannatale, E.; Goffredo, E.; Pantosti, A. Detection and Characterization of Vancomycin-Resistant Enterococci in Farm Animals and Raw Meat Products in Italy. Microb. Drug Resist. 2000, 6, 313–318. [Google Scholar] [CrossRef]
- Jackson, C.R.; Fedorka-Cray, P.; Barrett, J.B.; Ladely, S.R. Genetic Relatedness of High-Level Aminoglycoside-Resistant Enterococci Isolated from Poultry Carcasses. Avian Dis. 2004, 48, 100–107. [Google Scholar] [CrossRef]
- Thu, W.P.; Sinwat, N.; Bitrus, A.A.; Angkittitrakul, S.; Prathan, R.; Chuanchuen, R. Prevalence, antimicrobial resistance, virulence gene, and class 1 integrons of Enterococcus faecium and Enterococcus faecalis from pigs, pork and humans in Thai-Laos border provinces. J. Glob. Antimicrob. Resist. 2019, 18, 130–138. [Google Scholar] [CrossRef]
- Beukers, A.G.; Zaheer, R.; Goji, N.; Amoako, K.K.; Chaves, A.V.; Ward, M.; McAllister, T.A. Comparative genomics of Enterococcus spp. isolated from bovine feces. BMC Microbiol. 2017, 17, 52. [Google Scholar] [CrossRef]
- Li, X.-S.; Dong, W.-C.; Wang, X.-M.; Hu, G.-Z.; Cai, B.-Y.; Wu, C.-M.; Du, X.-D. Presence and genetic environment of pleuromutilin-lincosamide-streptogramin A resistance gene lsa(E) in enterococci of human and swine origin. J. Antimicrob. Chemother. 2013, 69, 1424–1426. [Google Scholar] [CrossRef]
- Cavaco, L.; Bernal, J.F.; Zankari, E.; Léon, M.; Hendriksen, R.; Perez-Gutierrez, E.; Aarestrup, F.M.; Donado-Godoy, P. Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia). J. Antimicrob. Chemother. 2016, 72, 678–683. [Google Scholar] [CrossRef]
- Lopez, M.; Kadlec, K.; Schwarz, S.; Torres, C. First Detection of the Staphylococcal Trimethoprim Resistance GenedfrKand thedfrK-Carrying Transposon Tn559in Enterococci. Microb. Drug Resist. 2012, 18, 13–18. [Google Scholar] [CrossRef]
- Lellek, H.; Franke, G.C.; Ruckert, C.; Wolters, M.; Wolschke, C.; Christner, M.; Büttner, H.; Alawi, M.; Kröger, N.; Rohde, H. Emergence of daptomycin non-susceptibility in colonizing vancomycin-resistant Enterococcus faecium isolates during daptomycin therapy. Int. J. Med Microbiol. 2015, 305, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Corona, F.; Martínez, J.L. Phenotypic Resistance to Antibiotics. Antibiotics 2013, 2, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Quintela-Baluja, M.; Böhme, K.; Fernández-No, I.C.; Morandi, S.; Alnakip, M.; Caamaño-Antelo, S.; Velázquez, J.B.; Calo-Mata, P. Characterization of different food-isolated Enterococcus strains by MALDI-TOF mass fingerprinting. Electrophor. 2013, 34, 2240–2250. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.J.B. Proteogenómica de isolados de Enterococcus spp. e Escherichia coli com a utilização do MALDI-TOF MS. Ph.D. Thesis, Universidade de Trás-os-Montes e Alto Douro, Vila Real, Portugal, 2014. (In Spanish). [Google Scholar]
- A Majcherczyk, P.; McKenna, T.; Moreillon, P.; Vaudaux, P. The discriminatory power of MALDI-TOF mass spectrometry to differentiate between isogenic teicoplanin-susceptible and teicoplanin-resistant strains of methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 2006, 255, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Griffin, P.; Price, G.; Schooneveldt, J.M.; Schlebusch, S.; Tilse, M.H.; Urbanski, T.; Hamilton, B.R.; Venter, D. Use of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry To Identify Vancomycin-Resistant Enterococci and Investigate the Epidemiology of an Outbreak. J. Clin. Microbiol. 2012, 50, 2918–2931. [Google Scholar] [CrossRef] [PubMed]
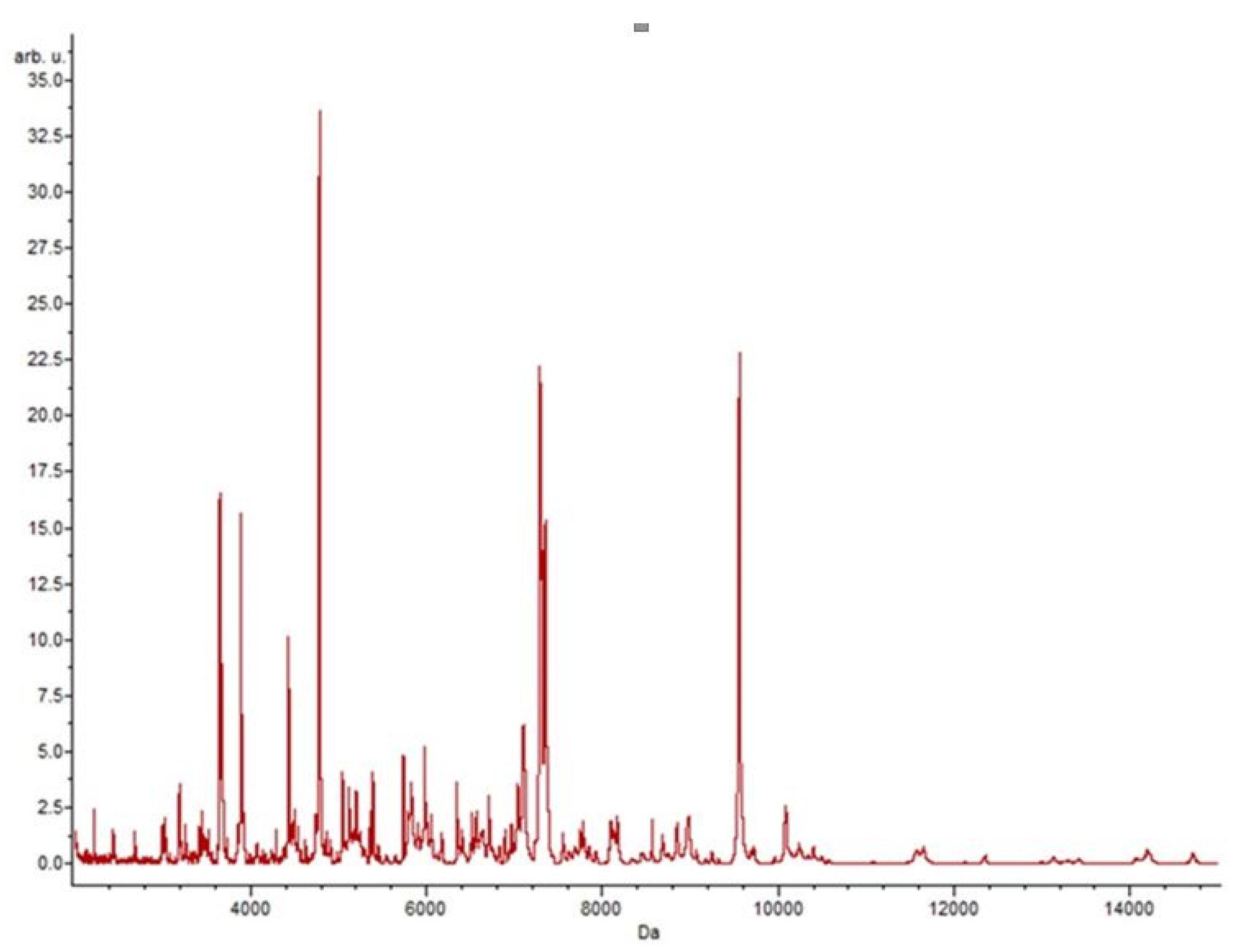
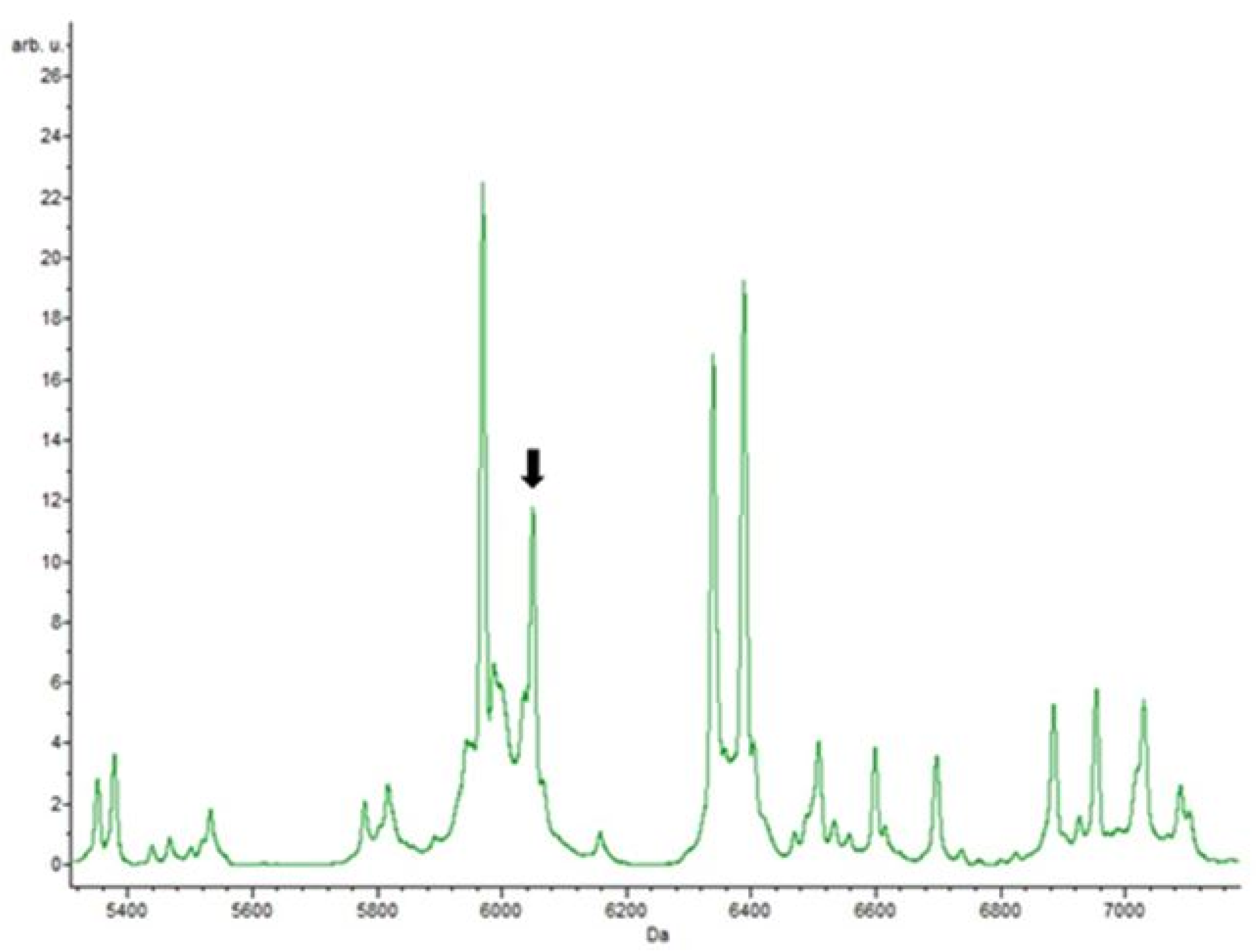
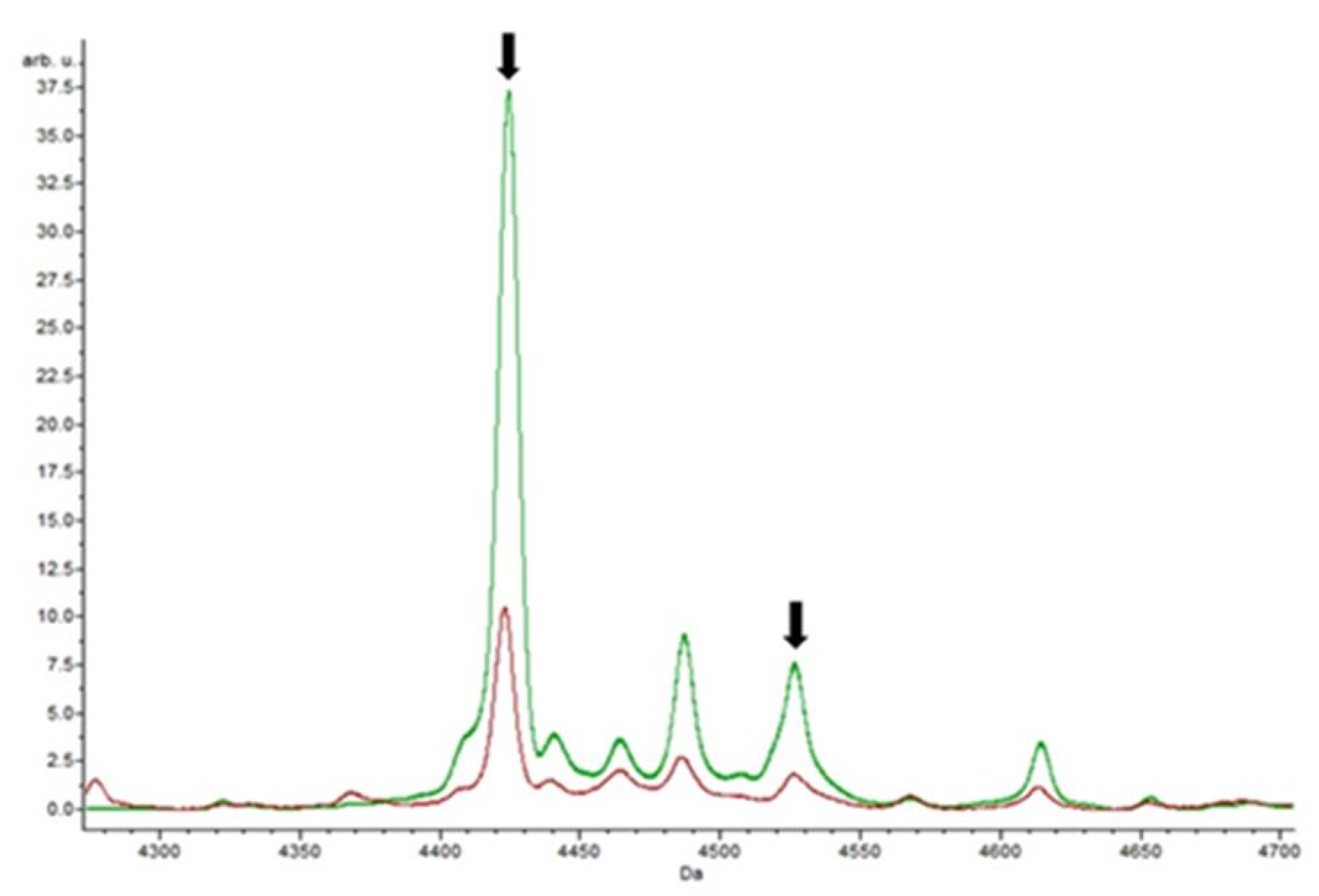
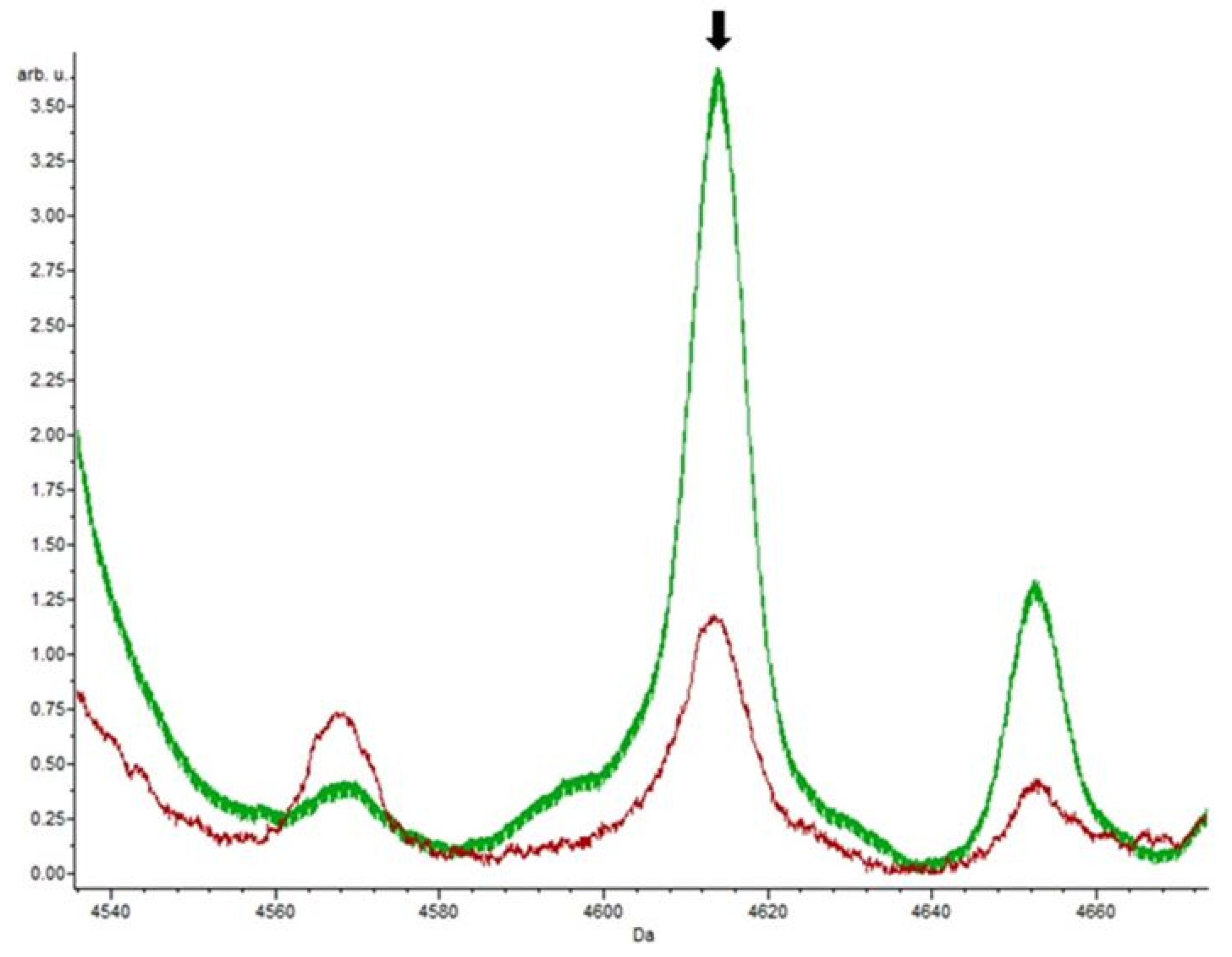
| Isolate | Species | Vancomycin Resistance Gene Detected | Resistance | |
|---|---|---|---|---|
| Phenotype | Genotype | |||
| H1 | Enterococcus faecium | vanA | TET TEI VAN CIP AMP ERY | aac(6’)-Ii, tet(L), tet(M), adeC, efmA, msr(C), cls, erm(T), erm(B), dfrG |
| H2 | E. faecium | vanA | TET TEI VAN AMP | aac(6’)-Ii, aadE, ant(9)-Ia, tet(L), tet(M), adeC, efmA, lnu(B), msr(C), cls, liaS, lsa(E), erm(B) |
| H3 | E. faecium | vanA | TET TEI VAN AMP | aac(6’)-Ii, aadE, ant(9)-Ia, tet(L), tet(M), adeC, efmA, lnu(B), msr(C), cls, liaS, lsa(E), erm(B) |
| H4 | E. faecium | vanA | TET TEI VAN CIP AMP | aac(6’)-Ii, aac(6’)-Ie-aph(2’’)-Ia, adeC, efmA, msr(C), cls, liaS, erm(B), dfrG, dfrK |
| H6 | Enterococcus gallinarum | vanA, vanC1 | TET TEI VAN AMP ERY STR | aac(6’)-Ii, erm(B) |
| H7 | E. faecium | vanA | TET VAN AMP ERY CN K | aac(6’)-Ii, tet(L), tet(M), adeC, efmA, msr(C), cls, liaS, liaR, erm(B) |
| H8 | E. faecium | vanA | TET TEI VAN CIP AMP ERY | aac(6’)-Ii, adeC, efmA, msr(C), cls, erm(B) |
| A10 | E. faecium | vanA | TET TEI VAN AMP ERY | aac(6’)-Ii, aadE, ant(9)-Ia, tet(L), tet(M), adeC, efmA, lnu(B), msr(C), cls, liaS, lsa(E), erm(B) |
| A11 | Enterococcus. durans | vanA | TET TEI VAN AMP ERY | aac(6’)-Iih, tet(L), tet(M), erm(B) |
| A12 | E. gallinarum | vanC1 | TEI VAN AMP ERY | ant(9)-Ia, msr(D), mef(A) |
| A13 | E. faecium | vanA | VAN CIP AMP ERY | aac(6’)-Ii, adeC, efmA, msr(C), cls, erm(B) |
| A14 | E. faecium | vanA | TET TEI VAN ERY | aac(6’)-Ii, aadE, ant(9)-Ia, tet(L), tet(M), adeC, efmA, lnu(B), msr(C), cls, liaS, lsa(E), erm(B) |
| A16 | E. faecium | vanA | TET TEI VAN CIP AMP ERY CN K | aac(6’)-Ii, adeC, efmA, msr(C), cls, erm(B) |
| CP17 | E. faecium | vanA | TET TEI VAN AMP ERY | aac(6’)-Ii, aadE, ant(9)-Ia, tet(L), tet(M), adeC, efmA, lnu(B), msr(C), cls, liaS, lsa(E), erm(B) |
| CP18 | E. faecium | vanA | TET TEI VAN ERY | aac(6’)-Ii, tet(L), tet(M), adeC, efmA, msr(C), cls, liaS, erm(B) |
| CP19 | E. faecium | vanA | TEI VAN CIP AMP ERY | aac(6’)-Ii, adeC, efmA, msr(C), cls, erm(B) |
| CP21 | E. faecium | vanA | TET TEI VAN ERY | aac(6’)-Ii, aadE, ant(9)-Ia, tet(L), tet(M), adeC, efmA, lnu(B), msr(C), cls, liaS, lsa(E), erm(B) |
| CP22 | E. durans | vanA | TET TEI VAN ERY | aac(6’)-Iih, tet(L), tet(M), erm(B) |
| CP23 | E. gallinarum | vanC1 | TET TEI VAN ERY | tet(L), tet(M) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabença, C.; de Sousa, T.; Oliveira, S.; Viala, D.; Théron, L.; Chambon, C.; Hébraud, M.; Beyrouthy, R.; Bonnet, R.; Caniça, M.; et al. Next-Generation Sequencing and MALDI Mass Spectrometry in the Study of Multiresistant Processed Meat Vancomycin-Resistant Enterococci (VRE). Biology 2020, 9, 89. https://doi.org/10.3390/biology9050089
Sabença C, de Sousa T, Oliveira S, Viala D, Théron L, Chambon C, Hébraud M, Beyrouthy R, Bonnet R, Caniça M, et al. Next-Generation Sequencing and MALDI Mass Spectrometry in the Study of Multiresistant Processed Meat Vancomycin-Resistant Enterococci (VRE). Biology. 2020; 9(5):89. https://doi.org/10.3390/biology9050089
Chicago/Turabian StyleSabença, Carolina, Telma de Sousa, Soraia Oliveira, Didier Viala, Laetitia Théron, Christophe Chambon, Michel Hébraud, Racha Beyrouthy, Richard Bonnet, Manuela Caniça, and et al. 2020. "Next-Generation Sequencing and MALDI Mass Spectrometry in the Study of Multiresistant Processed Meat Vancomycin-Resistant Enterococci (VRE)" Biology 9, no. 5: 89. https://doi.org/10.3390/biology9050089
APA StyleSabença, C., de Sousa, T., Oliveira, S., Viala, D., Théron, L., Chambon, C., Hébraud, M., Beyrouthy, R., Bonnet, R., Caniça, M., Poeta, P., & Igrejas, G. (2020). Next-Generation Sequencing and MALDI Mass Spectrometry in the Study of Multiresistant Processed Meat Vancomycin-Resistant Enterococci (VRE). Biology, 9(5), 89. https://doi.org/10.3390/biology9050089









