Mediators of Host–Microbe Circadian Rhythms in Immunity and Metabolism
Abstract
Simple Summary
Abstract
1. Introduction
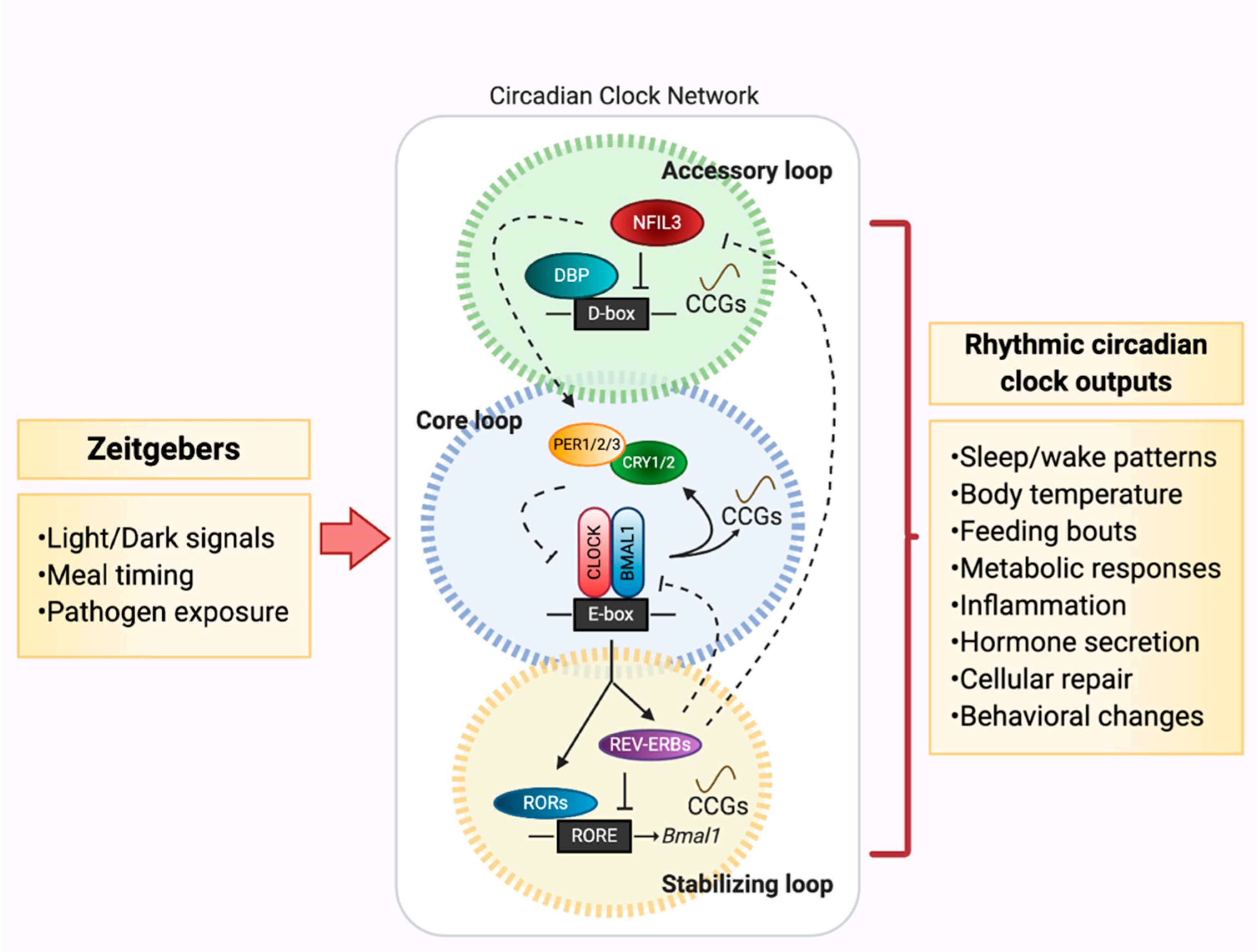
2. Circadian Networks: Key Players that Drive the Metronome of Life
3. Circadian Rhythms Gone Astray: A Defunct Metronome
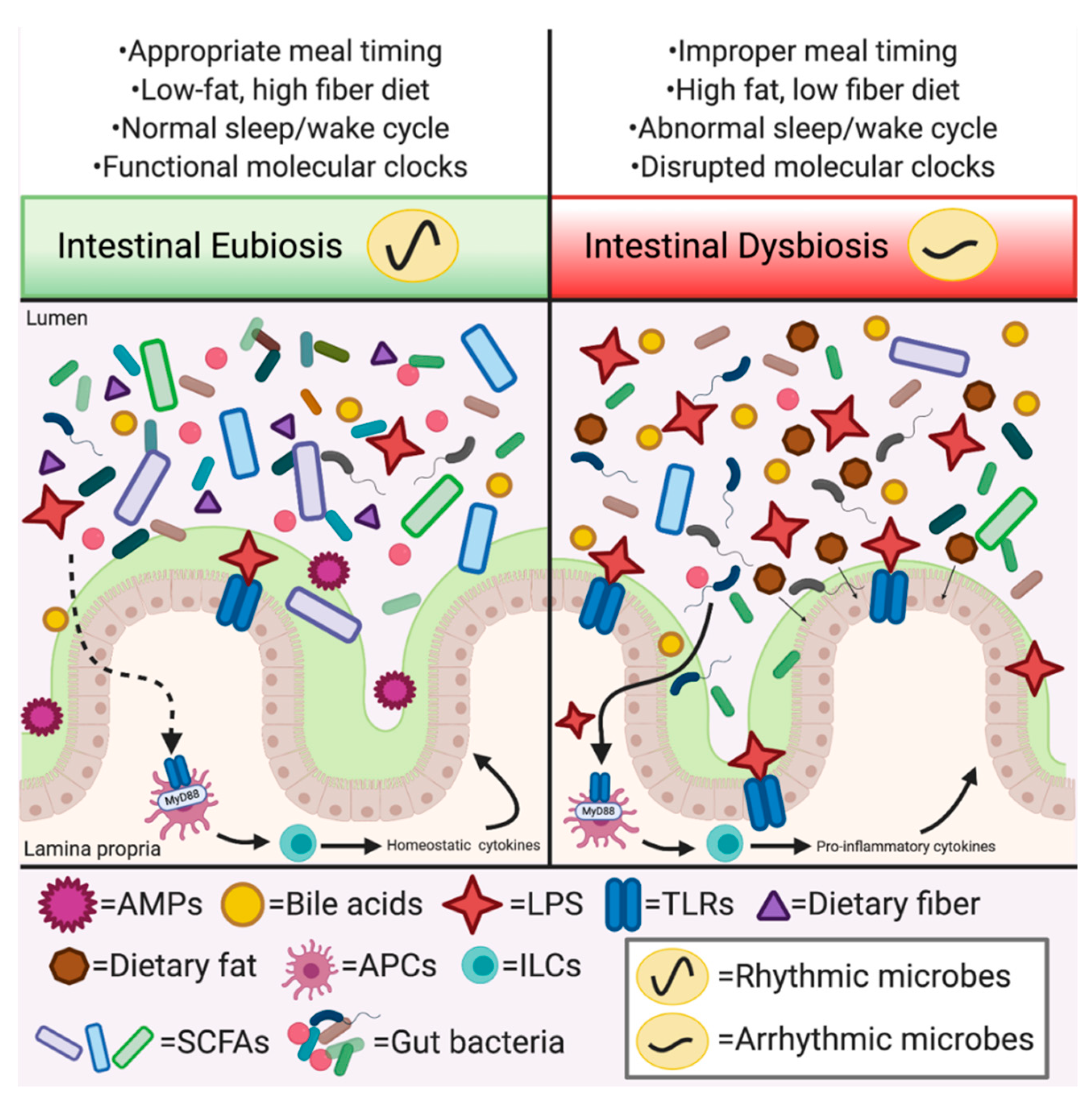
4. Gut Microbes: A Novel Instrument in the Orchestration of Circadian Rhythms
Gut Microbiota, Circadian Rhythmicity, and the Host: A Conserved Orchestration
5. Microbe vs. Host-Mediated Control of Circadian Orchestrations: Who Conducts Who?
5.1. Microbe-Mediated Signals Conduct Host Circadian Rhythms
5.2. Disrupting the Tempo of Circadian Rhythmicity: Host Circadian Network Perturbations and Gut Microbial Oscillations
5.3. Environmental Perturbations Disrupt Orchestration of Circadian Rhythms
5.3.1. Jet Lag and Shiftwork
5.3.2. Time Restricted Feeding
5.3.3. Molecular/Genetic CR Disruption: Circadian Knockout Models
6. Transmitting the Metronome’s Rhythm: Molecular Mediators of Host–Microbe Interactions
6.1. SCFAs
6.2. Bile Acids
6.3. LPS
6.4. Antimicrobial Peptides (AMPs)
6.5. Melatonin
7. Gaps and Challenges in Defining the Role of Host–Microbe Rhythmic Orchestrations
7.1. Defining Additional Orchestrators and Conductors of Microbe-Mediated Circadian Rhythms
7.2. Orchestrations of Host–Microbe Circadian Rhythms in Humans: Association vs. Causation?
7.3. Identifying the Molecular Underpinnings of Host–Microbe Circadian Rhythms
8. Conclusions: The Score Continues in the Orchestration of Host–Microbe Circadian Rhythms
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Partch, C.L.; Green, C.B.; Takahashi, J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014, 24, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Labrecque, N.; Cermakian, N. Circadian Clocks in the Immune System. J. Biol. Rhythm. 2015, 30, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Haspel, J.A.; Anafi, R.; Brown, M.K.; Cermakian, N.; Depner, C.; Desplats, P.; Gelman, A.E.; Haack, M.; Jelic, S.; Kim, B.S.; et al. Perfect timing: Circadian rhythms, sleep, and immunity—An NIH workshop summary. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Frazier, K.; Chang, E.B. Intersection of the Gut Microbiome and Circadian Rhythms in Metabolism. Trends Endocrinol. Metab. 2020, 31, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.H.; Takahashi, J.S. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006, 15, R271–R277. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.M.; Bellet, M.M.; Sassone-Corsi, P.; O’Neill, L. Circadian Clock Proteins and Immunity. Immunity 2014, 40, 178–186. [Google Scholar] [CrossRef]
- Aguilar-López, B.A.; Moreno-Altamirano, M.M.B.; Dockrell, H.M.; Duchen, M.R.; Sánchez-García, F.J. Mitochondria: An Integrative Hub Coordinating Circadian Rhythms, Metabolism, the Microbiome, and Immunity. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef]
- Broussard, J.L.; Van Cauter, E. Disturbances of sleep and circadian rhythms. Curr. Opin. Endocrinol. Diabetes Obes. 2016, 23, 353–359. [Google Scholar] [CrossRef]
- Eckel-Mahan, K.; Sassone-Corsi, P. Metabolism and the Circadian Clock Converge. Physiol. Rev. 2013, 93, 107–135. [Google Scholar] [CrossRef]
- Bunger, M.K.; Wilsbacher, L.D.; Moran, S.M.; Clendenin, C.; Radcliffe, L.A.; HogenEsch, J.B.; Simon, M.C.; Takahashi, J.S.; Bradfield, C.A. Mop3 Is an Essential Component of the Master Circadian Pacemaker in Mammals. Cell 2000, 103, 1009–1017. [Google Scholar] [CrossRef]
- Samsa, W.E.; Vasanji, A.; Midura, R.J.; Kondratov, R.V. Deficiency of circadian clock protein BMAL1 in mice results in a low bone mass phenotype. Bone 2016, 84, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Kondratov, R.V.; Kondratova, A.A.; Gorbacheva, V.Y.; Vykhovanets, O.V.; Antoch, M.P. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev 2006, 20, 1868–1873. [Google Scholar] [CrossRef] [PubMed]
- Rudic, R.D.; McNamara, P.; Curtis, A.-M.; Boston, R.C.; Panda, S.; Hogenesch, J.B.; Fitzgerald, G.A. BMAL1 and CLOCK, Two Essential Components of the Circadian Clock, Are Involved in Glucose Homeostasis. PLoS Biol. 2004, 2, e377. [Google Scholar] [CrossRef] [PubMed]
- Shimba, S.; Ishii, N.; Ohta, Y.; Ohno, T.; Watabe, Y.; Hayashi, M.; Wada, T.; Aoyagi, T.; Tezuka, M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 12071–12076. [Google Scholar] [CrossRef] [PubMed]
- Lamia, K.A.; Storch, K.F.; Weitz, C.J. Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. USA 2008, 105, 15172–151177. [Google Scholar] [CrossRef]
- Sadacca, L.A.; Lamia, K.A.; DeLemos, A.S.; Blum, B.; Weitz, C.J. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia 2011, 54, 120–124. [Google Scholar] [CrossRef]
- Paschos, G.K.; Ibrahim, S.; Song, W.L.; Kunieda, T.; Grant, G.; Reyes, T.M.; Bradfield, A.C.; Vaughan, C.H.; Eiden, M.; Masoodi, M.; et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 2012, 18, 1768–1777. [Google Scholar] [CrossRef]
- Nguyen, K.D.; Fentress, S.J.; Qiu, Y.; Yun, K.; Cox, J.S.; Chawla, A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6Chi inflammatory monocytes. Science 2013, 341, 1483–1488. [Google Scholar] [CrossRef]
- McDearmon, E.L.; Patel, K.N.; Ko, C.H.; Walisser, J.A.; Schook, A.C.; Chong, J.L.; Wilsbacher, L.D.; Song, E.J.; Hong, H.-K.; Bradfield, C.A.; et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science 2006, 314, 1304–1308. [Google Scholar] [CrossRef] [PubMed]
- Kolbe, I.; Oster, H. Chronodisruption, metabolic homeostasis, and the regulation of inflammation in adipose tissues. Yale J. Biol. Med. 2019, 92, 317–325. [Google Scholar]
- Nijhuis, J.; Rensen, S.S.; Slaats, Y.; Van Dielen, F.M.H.; Buurman, W.A.; Greve, J.W.M. Neutrophil activation in morbid obesity, chronic activation of acute inflammation. Obesity 2009, 17, 2014–2018. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S.; Shargill, N.S.; Spiegelman, B.M. Adipose expression of tumor necrosis factor-α: Direct role in obesity-linked insulin resistance. Science 1993, 259, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Feinstein, R.; Kanety, H.; Papa, M.Z.; Lunenfeld, B.; Karasik, A. Tumor necrosis factor-alpha suppresses insulin-induced tyrosine phosphorylation of insulin receptor and its substrates. J. Biol. Chem. 1993, 268, 26055–26058. [Google Scholar] [PubMed]
- Gauthier, M.-S.; Ruderman, N.B. Adipose tissue inflammation and insulin resistance: All obese humans are not created equal. Biochem. J. 2010, 430, e1–e4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Parsons, M.J.; Moffitt, T.E.; Gregory, A.M.; Goldman-Mellor, S.; Nolan, P.M.; Poulton, R.; Caspi, A. Social jetlag, obesity and metabolic disorder: Investigation in a cohort study. Int. J. Obes. 2015, 39, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Guryn, K.; Leone, V.; Chang, E.B. Regional Diversity of the Gastrointestinal Microbiome. Cell Host Microbe 2019, 26, 314–324. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Takeda, K.; Kaisho, T.; Akira, S. Toll-Like Receptors. Annu. Rev. Immunol. 2003, 21, 335–376. [Google Scholar] [CrossRef]
- Soromou, L.W.; Zhang, Z.; Li, R.; Chen, N.; Guo, W.; Huo, M.; Guan, S.; Lu, J.; Deng, X. Regulation of inflammatory cytokines in lipopolysaccharide-stimulated RAW 264.7 murine macrophage by 7-O-methyl-naringenin. Molecules 2012, 17, 3574–3585. [Google Scholar] [CrossRef]
- Heath-Heckman, E.A.C.; Peyer, S.M.; Whistler, C.A.; Apicella, M.A.; Goldman, W.E.; McFall-Ngai, M.J. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-vibrio symbiosis. MBio 2013, 4. [Google Scholar] [CrossRef]
- Kremer, N.; Koch, E.J.; El Filali, A.; Zhou, L.; Heath-Heckman, E.A.C.; Ruby, E.G.; McFall-Ngai, M.J. Persistent Interactions with Bacterial Symbionts Direct Mature-Host Cell Morphology and Gene Expression in the Squid-Vibrio Symbiosis. MSystems 2018, 3, 165–183. [Google Scholar] [CrossRef] [PubMed]
- Nyholm, S.V.; Stewart, J.J.; Ruby, E.G.; McFall-Ngai, M.J. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ. Microbiol. 2009, 11, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Nyholm, S.V.; McFall-Ngai, M.J. The winnowing: Establishing the squid—Vibrios symbiosis. Nat. Rev. Microbiol. 2004, 2, 632–642. [Google Scholar] [CrossRef] [PubMed]
- DeLoney-Marino, C.R.; Wolfe, A.J.; Visick, K.L. Chemoattraction of Vibrio fischeri to Serine, Nucleosides, and N-Acetylneuraminic Acid, a Component of Squid Light-Organ Mucus. Appl. Environ. Microbiol. 2003, 69, 7527–7530. [Google Scholar] [CrossRef] [PubMed]
- McFall-Ngai, M.J. Divining the Essence of Symbiosis: Insights from the Squid-Vibrio Model. PLoS Biol. 2014, 12, e1001783. [Google Scholar] [CrossRef][Green Version]
- Schwartzman, J.A.; Koch, E.; Heath-Heckman, E.A.C.; Zhou, L.; Kremer, N.; McFall-Ngai, M.J.; Ruby, E.G. The chemistry of negotiation: Rhythmic, glycan-driven acidification in a symbiotic conversation. Proc. Natl. Acad. Sci. USA 2015, 112, 566–571. [Google Scholar] [CrossRef]
- Mukherji, A.; Kobiita, A.; Ye, T.; Chambon, P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 2013, 153, 812–827. [Google Scholar] [CrossRef]
- Leone, V.; Gibbons, S.M.; Martinez, K.; Hutchison, A.L.; Huang, E.Y.; Cham, C.M.; Pierre, J.F.; Heneghan, A.F.; Nadimpalli, A.; Hubert, N.; et al. Effects of Diurnal Variation of Gut Microbes and High-Fat Feeding on Host Circadian Clock Function and Metabolism. Cell Host Microbe 2015, 17, 681–689. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Chaix, A.; Yooseph, S.; Panda, S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014, 20, 1006–1017. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zeevi, D.; Levy, M.; Zilberman-Schapira, G.; Suez, J.; Tengeler, A.C.; Abramson, L.; Katz, M.N.; Korem, T.; Zmora, N.; et al. Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell 2014, 159, 514–529. [Google Scholar] [CrossRef]
- Kennedy, E.A.; King, K.Y.; Baldridge, M.T. Mouse microbiota models: Comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front. Physiol. 2018, 9, 1534. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Z.; Wang, Y.; Li, Y.; Ye, C.; Ruhn, K.A.; Behrendt, C.L.; Olson, E.N.; Hooper, L.V. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science 2019, 365, 1428–1434. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.Y.; Ha, C.W.Y.; Campbell, C.R.; Mitchell, A.J.; Dinudom, A.; Oscarsson, J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Increased Gut Permeability and Microbiota Change Associate with Mesenteric Fat Inflammation and Metabolic Dysfunction in Diet-Induced Obese Mice. PLoS ONE 2012, 7, e34233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef]
- Logan, R.W.; Sarkar, D.K. Circadian nature of immune function. Mol. Cell. Endocrinol. 2012, 349, 82–90. [Google Scholar] [CrossRef]
- Silver, A.C.; Arjona, A.; Walker, W.E.; Fikrig, E. The Circadian Clock Controls Toll-like Receptor 9-Mediated Innate and Adaptive Immunity. Immunity 2012, 36, 251–261. [Google Scholar] [CrossRef]
- Voigt, R.M.; Forsyth, C.B.; Green, S.J.; Mutlu, E.; Engen, P.; Vitaterna, M.H.; Turek, F.W.; Keshavarzian, A. Circadian Disorganization Alters Intestinal Microbiota. PLoS ONE 2014, 9, e97500. [Google Scholar] [CrossRef]
- Summa, K.C.; Voigt, R.M.; Forsyth, C.B.; Shaikh, M.; Cavanaugh, K.; Tang, Y.; Vitaterna, M.H.; Song, S.; Turek, F.W.; Keshavarzian, A. Disruption of the Circadian Clock in Mice Increases Intestinal Permeability and Promotes Alcohol-Induced Hepatic Pathology and Inflammation. PLoS ONE 2013, 8, e67102. [Google Scholar] [CrossRef]
- Khalyfa, A.; Poroyko, V.A.; Qiao, Z.; Gileles-Hillel, A.; Khalyfa, A.A.; Akbarpour, M.; Almendros, I.; Farré, R.; Gozal, D. Exosomes and metabolic function in mice exposed to alternating dark-light cycles mimicking night shift work schedules. Front. Physiol. 2017, 8, 882. [Google Scholar] [CrossRef] [PubMed]
- Deaver, J.A.; Eum, S.Y.; Toborek, M. Circadian Disruption Changes Gut Microbiome Taxa and Functional Gene Composition. Front. Microbiol. 2018, 9, 737. [Google Scholar] [CrossRef]
- Casiraghi, L.P.; Alzamendi, A.; Giovambattista, A.; Chiesa, J.J.; Golombek, D.A. Effects of chronic forced circadian desynchronization on body weight and metabolism in male mice. Physiol. Rep. 2016, 4, e12743. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Rollins, D.; Ruhn, K.A.; Stubblefield, J.J.; Green, C.B.; Kashiwada, M.; Rothman, P.B.; Takahashi, J.S.; Hooper, L.V. TH17 cell differentiation is regulated by the circadian clock. Science 2013, 342, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Castanon-Cervantes, O.; Wu, M.; Ehlen, J.C.; Paul, K.; Gamble, K.L.; Johnson, R.L.; Besing, R.C.; Menaker, M.; Gewirtz, A.T.; Davidson, A.J. Dysregulation of Inflammatory Responses by Chronic Circadian Disruption. J. Immunol. 2010, 185, 5796–5805. [Google Scholar] [CrossRef]
- Hatori, M.; Vollmers, C.; Zarrinpar, A.; DiTacchio, L.; Bushong, E.A.; Gill, S.; Leblanc, M.; Chaix, A.; Joens, M.; Fitzpatrick, J.A.J.; et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012, 15, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Delahaye, L.B.; Bloomer, R.J.; Butawan, M.B.; Wyman, J.M.; Hill, J.L.; Lee, H.W.; Liu, A.C.; McAllan, L.; Han, J.C.; Van Der Merwe, M. Time-restricted feeding of a high-fat diet in male C57BL/6 mice reduces adiposity but does not protect against increased systemic inflammation. Appl. Physiol. Nutr. Metab. 2018, 43, 1033–1042. [Google Scholar] [CrossRef]
- Cissé, Y.M.; Borniger, J.C.; Lemanski, E.; Walker, W.H.; Nelson, R.J. Time-Restricted Feeding Alters the Innate Immune Response to Bacterial Endotoxin. J. Immunol. 2018, 200, 681–687. [Google Scholar] [CrossRef]
- Hara, R.; Wan, K.; Wakamatsu, H.; Aida, R.; Moriya, T.; Akiyama, M.; Shibata, S. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells 2001, 6, 269–278. [Google Scholar] [CrossRef]
- Hopwood, T.W.; Hall, S.; Begley, N.; Forman, R.; Brown, S.; Vonslow, R.; Saer, B.; Little, M.C.; Murphy, E.A.; Hurst, R.J.; et al. The circadian regulator BMAL1 programmes responses to parasitic worm infection via a dendritic cell clock. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Liang, X.; Bushman, F.D.; FitzGerald, G.A. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci. USA 2015, 112, 10479–10484. [Google Scholar] [CrossRef] [PubMed]
- Marcheva, B.; Ramsey, K.M.; Buhr, E.D.; Kobayashi, Y.; Su, H.; Ko, C.H.; Ivanova, G.; Omura, C.; Mo, S.; Vitaterna, M.H.; et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nat. Cell Biol. 2010, 466, 627–631. [Google Scholar] [CrossRef]
- Godinho-Silva, C.; Domingues, R.G.; Rendas, M.; Raposo, B.; Ribeiro, H.; da Silva, J.A.; Vieira, A.; Costa, R.M.; Barbosa-Morais, N.L.; Carvalho, T.; et al. Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature 2019, 574, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Druzd, D.; Matveeva, O.; Ince, L.; Harrison, U.; He, W.; Schmal, C.; Herzel, H.; Tsang, A.H.; Kawakami, N.; Leliavski, A.; et al. Lymphocyte Circadian Clocks Control Lymph Node Trafficking and Adaptive Immune Responses. Immunity 2017, 46, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Goc, J.; Zhou, L.; Chu, C.; Shah, M.A.; Eberl, G.; Sonnenberg, G.F. A circadian clock is essential for homeostasis of group 3 innate lymphoid cells in the gut. Sci. Immunol. 2019, 4, 1215. [Google Scholar] [CrossRef] [PubMed]
- Adrover, J.M.; del Fresno, C.; Crainiciuc, G.; Cuartero, M.I.; Casanova-Acebes, M.; Weiss, L.A.; Huerga-Encabo, H.; Silvestre-Roig, C.; Rossaint, J.; Cossío, I.; et al. A Neutrophil Timer Coordinates Immune Defense and Vascular Protection. Immunity 2019, 50, 390.e10–402e10. [Google Scholar] [CrossRef]
- Voigt, R.M.; Summa, K.C.; Forsyth, C.B.; Green, S.J.; Engen, P.; Naqib, A.; Vitaterna, M.H.; Turek, F.W.; Keshavarzian, A. The CircadianClockMutation Promotes Intestinal Dysbiosis. Alcohol. Clin. Exp. Res. 2016, 40, 335–347. [Google Scholar] [CrossRef]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R.; et al. Obesity and Metabolic Syndrome in Circadian Clock Mutant Mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef]
- Ando, N.; Nakamura, Y.; Aoki, R.; Ishimaru, K.; Ogawa, H.; Okumura, K.; Shibata, S.; Shimada, S.; Nakao, A. Circadian Gene Clock Regulates Psoriasis-Like Skin Inflammation in Mice. J. Investig. Dermatol. 2015, 135, 3001–3008. [Google Scholar] [CrossRef]
- Bellet, M.M.; Deriu, E.; Liu, J.Z.; Grimaldi, B.; Blaschitz, C.; Zeller, M.; Edwards, R.A.; Sahar, S.; Dandekar, S.; Baldi, P.; et al. Circadian clock regulates the host response to Salmonella. Proc. Natl. Acad. Sci. USA 2013, 110, 9897–9902. [Google Scholar] [CrossRef]
- Barclay, J.L.; Shostak, A.; Leliavski, A.; Tsang, A.H.K.; Jöhren, O.; Müller-Fielitz, H.; Landgraf, D.; Naujokat, N.; Van Der Horst, G.T.J.; Oster, H. High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in Cry-deficient mice. Am. J. Physiol. Metab. 2013, 304, E1053–E1063. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Zhao, X.; Bai, J.; Gery, S.; Sun, H.; Lin, D.-C.; Chen, Q.; Chen, Z.; Mack, L.; Yang, H.; et al. Circadian clock cryptochrome proteins regulate autoimmunity. Proc. Natl. Acad. Sci. USA 2017, 114, 12548–12553. [Google Scholar] [CrossRef] [PubMed]
- Narasimamurthy, R.; Hatori, M.; Nayak, S.K.; Liu, F.; Panda, S.; Verma, I.M. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc. Natl. Acad. Sci. USA 2012, 109, 12662–12667. [Google Scholar] [CrossRef] [PubMed]
- Hashiramoto, A.; Yamane, T.; Tsumiyama, K.; Yoshida, K.; Komai, K.; Yamada, H.; Yamazaki, F.; Doi, M.; Okamura, H.; Shiozawa, S. Mammalian Clock Gene Cryptochrome Regulates Arthritis via Proinflammatory Cytokine TNF-α. J. Immunol. 2009, 184, 1560–1565. [Google Scholar] [CrossRef]
- Kettner, N.M.; Mayo, S.A.; Hua, J.; Lee, C.; Moore, D.D.; Fu, L. Circadian dysfunction induces leptin resistance in mice. Cell Metab. 2015, 22, 448–459. [Google Scholar] [CrossRef]
- Grimaldi, B.; Bellet, M.M.; Katada, S.; Astarita, G.; Hirayama, J.; Amin, R.H.; Granneman, J.G.; Piomelli, D.; Leff, T.; Sassone-Corsi, P. PER2 Controls Lipid Metabolism by Direct Regulation of PPARγ. Cell Metab. 2010, 12, 509–520. [Google Scholar] [CrossRef]
- Yang, S.; Liu, A.; Weidenhammer, A.; Cooksey, R.C.; McClain, D.; Kim, M.K.; Aguilera, G.; Abel, E.D.; Chung, J.H. The Role of mPer2 Clock Gene in Glucocorticoid and Feeding Rhythms. Endocrinology 2009, 150, 2153–2160. [Google Scholar] [CrossRef]
- Costa, M.J.; So, A.Y.-L.; Kaasik, K.; Krueger, K.C.; Pillsbury, M.L.; Fu, Y.-H.; Ptacek, L.J.; Yamamoto, K.R.; Feldman, B.J. Circadian Rhythm Gene Period 3 Is an Inhibitor of the Adipocyte Cell Fate. J. Biol. Chem. 2011, 286, 9063–9070. [Google Scholar] [CrossRef]
- Arjona, A.; Sarkar, D.K. The circadian gene mPer2 regulates the daily rhythm of IFN-γ. J. Interf. Cytokine Res. 2006, 26, 645–649. [Google Scholar] [CrossRef]
- Logan, R.W.; Wynne, O.; Levitt, D.; Price, D.; Sarkar, D.K. Altered circadian expression of cytokines and cytolytic factors in splenic natural killer cells of per1-/- mutant mice. J. Interf. Cytokine Res. 2013, 33, 108–114. [Google Scholar] [CrossRef]
- Liu, J.; Mankani, G.; Shi, X.; Meyer, M.; Cunningham-Runddles, S.; Malkani, G.; Sun, Z.S. The Circadian Clock Period 2 Gene Regulates Gamma Interferon Production of NK Cells in Host Response to Lipopolysaccharide-Induced Endotoxic Shock. Infect. Immun. 2006, 74, 4750–4756. [Google Scholar] [CrossRef]
- Cho, H.; Zhao, X.; Hatori, M.; Yu, R.T.; Barish, G.D.; Lam, M.T.; Chong, L.-W.; DiTacchio, L.; Atkins, A.R.; Glass, C.K.; et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 2012, 485, 123–127. [Google Scholar] [CrossRef]
- Wang, Y.; Kuang, Z.; Yu, X.; Ruhn, K.A.; Kubo, M.; Hooper, L.V. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 2017, 357, 912–916. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.E.; Blaikley, J.; Beesley, S.; Matthews, L.; Simpson, K.D.; Boyce, S.H.; Farrow, S.N.; Else, K.J.; Singh, D.; Ray, D.W.; et al. The nuclear receptor REV-ERB mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. USA 2012, 109, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Marcheva, B.; Ramsey, K.M.; Peek, C.B.; Affinati, A.; Maury, E.; Bass, J. Circadian Clocks and Metabolism. Handb. Exp. Pharmacol. Handb. Exp. Pharmacol. 2013, 127–155. [Google Scholar] [CrossRef]
- Tahara, Y.; Yamazaki, M.; Sukigara, H.; Motohashi, H.; Sasaki, H.; Miyakawa, H.; Haraguchi, A.; Ikeda, Y.; Fukuda, S.; Shibata, S. Gut Microbiota-Derived Short Chain Fatty Acids Induce Circadian Clock Entrainment in Mouse Peripheral Tissue. Sci. Rep. 2018, 8, 1395. [Google Scholar] [CrossRef] [PubMed]
- Knutsson, A.; Bøggild, H. Gastrointestinal disorders among shift workers. Scand J. Work Environ. Heal. 2010, 36, 85–95. [Google Scholar] [CrossRef]
- Mohren, D.C.L.; Jansen, N.W.H.; Kant Ij Galama, J.M.D.; Van Den Brandt, P.A.; Swaen, G.M.H. Prevalence of common infections among employees in different work schedules. J. Occup. Environ. Med. 2002, 44, 1003–1011. [Google Scholar] [CrossRef]
- Izumo, M.; Pejchal, M.; Schook, A.C.; Lange, R.P.; Walisser, J.A.; Sato, T.R.; Wang, X.; A Bradfield, C.; Takahashi, J.S. Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. Elife 2014, 3. [Google Scholar] [CrossRef]
- Guo, B.; Chatterjee, S.; Li, L.; Kim, J.M.; Lee, J.; Yechoor, V.K.; Minze, L.J.; Hsueh, W.; Ma, K. The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 2012, 26, 3453–3463. [Google Scholar] [CrossRef]
- Shimba, S.; Ogawa, T.; Hitosugi, S.; Ichihashi, Y.; Nakadaira, Y.; Kobayashi, M.; Tezuka, M.; Kosuge, Y.; Ishige, K.; Ito, Y.; et al. Deficient of a Clock Gene, Brain and Muscle Arnt-Like Protein-1 (BMAL1), Induces Dyslipidemia and Ectopic Fat Formation. PLoS ONE 2011, 6, e25231. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, M.-S.; Li, R.; Liu, V.Y.; Fu, L.; Moore, D.D.; Ma, K.; Yechoor, V.K. Loss of Bmal1 leads to uncoupling and impaired glucose-stimulated insulin secretion in β-cells. Islets 2011, 3, 381–388. [Google Scholar] [CrossRef]
- Griebel, G.; Ravinet-Trillou, C.; Beeskã, S.; Avenet, P.; Pichat, P.; Beeské, S. Mice Deficient in Cryptochrome 1 (Cry1−/−) Exhibit Resistance to Obesity Induced by a High-Fat Diet. Front. Endocrinol. 2014, 5, 49. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microb. 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Biancolin, A.D.; Martchenko, A.; Mitova, E.; Gurges, P.; Michalchyshyn, E.; Chalmers, J.A.; Doria, A.; Mychaleckyj, J.C.; Adriaenssens, A.E.; Reimann, F.; et al. The core clock gene, Bmal1, and its downstream target, the SNARE regulatory protein secretagogin, are necessary for circadian secretion of glucagon-like peptide-1. Mol. Metab. 2020, 31, 124–137. [Google Scholar] [CrossRef]
- Beumer, J.; Artegiani, B.; Post, Y.; Reimann, F.; Gribble, F.; Nguyen, T.N.; Zeng, H.; Born, M.V.D.; Van Es, J.H.; Clevers, H. Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nat. Cell Biol. 2018, 20, 909–916. [Google Scholar] [CrossRef]
- Gil-Lozano, M.; Mingomataj, E.L.; Wu, W.K.; Ridout, S.A.; Brubaker, P.L. Circadian Secretion of the Intestinal Hormone GLP-1 by the Rodent L Cell. Diabetes 2014, 63, 3674–3685. [Google Scholar] [CrossRef]
- Gil-Lozano, M.; Wu, W.K.; Martchenko, A.; Brubaker, P.L. High-Fat Diet and Palmitate Alter the Rhythmic Secretion of Glucagon-Like Peptide-1 by the Rodent L-cell. Endocrinology 2016, 157, 586–599. [Google Scholar] [CrossRef]
- Martchenko, A.; Oh, R.H.; Wheeler, S.E.; Gurges, P.; Chalmers, J.A.; Brubaker, P.L. Suppression of circadian secretion of glucagon-like peptide-1 by the saturated fatty acid, palmitate. Acta Physiol. 2018, 222, e13007. [Google Scholar] [CrossRef]
- Larraufie, P.; Martin-Gallausiaux, C.; Lapaque, N.; Dore, J.; Gribble, F.M.; Reimann, F.; Blottiere, H.M. SCFAs strongly stimulate PYY production in human enteroendocrine cells. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Reigstad, C.S.; Salmonson, C.E.; Iii, J.F.R.; Szurszewski, J.H.; Linden, D.R.; Sonnenburg, J.L.; Farrugia, G.; Kashyap, P.C. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015, 29, 1395–1403. [Google Scholar] [CrossRef]
- Bhattarai, Y.; Schmidt, B.A.; Linden, D.R.; Larson, E.D.; Grover, M.; Beyder, A.; Farrugia, G.; Kashyap, P.C. Human-derived gut microbiota modulates colonic secretion in mice by regulating 5-HT3 receptor expression via acetate production. Am. J. Physiol. Liver Physiol. 2017, 313, G80–G87. [Google Scholar] [CrossRef] [PubMed]
- Corrêa-Oliveira, R.; Fachi, J.L.; Vieira, A.; Sato, F.T.; Vinolo, M.A.R. Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 2016, 5, e73. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shimizu, Y.; Kimura, I. Gut microbial metabolite short-chain fatty acids and obesity. Biosci. Microbiota Food Heal. 2017, 36, 135–140. [Google Scholar] [CrossRef]
- Vidrine, K.; Ye, J.; Martin, R.J.; McCutcheon, K.L.; Raggio, A.M.; Pelkman, C.; Durham, H.A.; Zhou, J.; Senevirathne, R.N.; Williams, C.; et al. Resistant starch from high amylose maize (HAM-RS2) and Dietary butyrate reduce abdominal fat by a different apparent mechanism. Obesity 2014, 22, 344–348. [Google Scholar] [CrossRef]
- Chen, G.; Ran, X.; Li, B.; Li, Y.; He, D.; Huang, B.; Fu, S.; Liu, J.; Wang, W. Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-induced Inflammatory Bowel Disease Mice Model. EBioMedicine 2018, 30, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Jiao, N.; Baker, S.S.; Nugent, C.A.; Tsompana, M.; Cai, L.; Wang, Y.; Buck, M.J.; Genco, R.J.; Baker, R.D.; Zhu, R.; et al. Gut microbiome may contribute to insulin resistance and systemic inflammation in obese rodents: A meta-analysis. Physiol. Genom. 2018, 50, 244–254. [Google Scholar] [CrossRef]
- Fukui, H.; Xu, X.; Miwa, H. Role of gut microbiota-gut hormone axis in the pathophysiology of functional gastrointestinal disorders. J. Neurogastroenterol. Motil. 2018, 24, 367–386. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Zhang, R.; Jain, R.; Shi, H.; Zhang, L.; Zhou, G.; Sangwung, P.; Tugal, D.; Atkins, G.B.; Prosdocimo, D.A.; et al. Circadian control of bile acid synthesis by a KLF15-Fgf15 axis. Nat. Commun. 2015, 6, 7231. [Google Scholar] [CrossRef]
- Ma, K.; Xiao, R.; Tseng, H.-T.; Shan, L.; Fu, L.; Moore, D.D. Circadian Dysregulation Disrupts Bile Acid Homeostasis. PLoS ONE 2009, 4, e6843. [Google Scholar] [CrossRef]
- Govindarajan, K.; MacSharry, J.; Casey, P.G.; Shanahan, F.; Joyce, S.A.; Gahan, C.G.M. Unconjugated Bile Acids Influence Expression of Circadian Genes: A Potential Mechanism for Microbe-Host Crosstalk. PLoS ONE 2016, 11, e0167319. [Google Scholar] [CrossRef]
- Liaset, B.; Hao, Q.; Jørgensen, H.; Hallenborg, P.; Du, Z.-Y.; Ma, T.; Marschall, H.-U.; Kruhøffer, M.; Li, R.; Li, Q.; et al. Nutritional Regulation of Bile Acid Metabolism Is Associated with Improved Pathological Characteristics of the Metabolic Syndrome. J. Biol. Chem. 2011, 286, 28382–28395. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.A.; Karpen, S.J. Intestinal transport and metabolism of bile acids. J. Lipid Res. 2015, 56, 1085–1099. [Google Scholar] [CrossRef] [PubMed]
- Kübeck, R.; Bonet-Ripoll, C.; Hoffmann, C.; Walker, A.; Müller, V.M.; Schüppel, V.L.; Lagkouvardos, I.; Scholz, B.; Engel, K.-H.; Daniel, H.; et al. Dietary fat and gut microbiota interactions determine diet-induced obesity in mice. Mol. Metab. 2016, 5, 1162–1174. [Google Scholar] [CrossRef]
- Comas, M.; Gordon, C.J.; Oliver, B.G.G.; Stow, N.W.; King, G.; Sharma, P.; Ammit, A.J.; Grunstein, R.; Phillips, C.L. A circadian based inflammatory response—Implications for respiratory disease and treatment. Sleep Sci. Pract. 2017, 1, 1–19. [Google Scholar] [CrossRef]
- Kheirandish-Gozal, L.; Peris, E.; Wang, Y.; Tamae-Kakazu, M.; Khalyfa, A.; Carreras, A.; Gozal, D. Lipopolysaccharide-Binding Protein Plasma Levels in Children: Effects of Obstructive Sleep Apnea and Obesity. J. Clin. Endocrinol. Metab. 2014, 99, 656–663. [Google Scholar] [CrossRef]
- Salzman, N.H.; Hung, K.; Haribhai, D.; Chu, H.; Karlsson-Sjoeberg, J.; Amir, E.; Teggatz, P.; Barman, M.; Hayward, M.; Eastwood, D.; et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 2010, 11, 76–82. [Google Scholar] [CrossRef]
- Guo, X.; Li, J.; Tang, R.; Zhang, G.; Zeng, H.; Wood, R.J.; Liu, Z. High Fat Diet Alters Gut Microbiota and the Expression of Paneth Cell-Antimicrobial Peptides Preceding Changes of Circulating Inflammatory Cytokines. Mediat. Inflamm. 2017, 2017, 1–9. [Google Scholar] [CrossRef]
- Cash, H.L.; Whitham, C.V.; Behrendt, C.L.; Hooper, L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 2006, 313, 1126–1130. [Google Scholar] [CrossRef]
- Frazier, K.; Kambal, A.; Zale, E.A.; Pierre, J.F.; Hubert, N.; Miyoshi, S.; Miyoshi, J.; Ringus, D.; Harris, D.; Yang, K.; et al. High fat diet disrupts diurnal interactions between REG3γ and small intestinal gut microbes resulting in metabolic dysfunction. bioRxiv 2020. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Levy, M.; Korem, T.; Dohnalová, L.; Shapiro, H.; Jaitin, D.A.; David, E.; Winter, D.R.; Gury-BenAri, M.; Tatirovsky, E.; et al. Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell 2016, 167, 1495–1510.e12. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Q.; Fichna, J.; Bashashati, M.; Li, Y.Y.; Storr, M. Distribution, function and physiological role of melatonin in the lower gut. World J. Gastroenterol. 2011, 17, 3888–3898. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Li, Y.; Han, H.; Ma, J.; Liu, G.; Wu, X.; Huang, X.; Fang, R.; Baba, K.; Bin, P.; et al. Administration of Exogenous Melatonin Improves the Diurnal Rhythms of the Gut Microbiota in Mice Fed a High-Fat Diet. mSystems 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Paulose, J.K.; Wright, J.M.; Patel, A.G.; Cassone, V.M. Human Gut Bacteria Are Sensitive to Melatonin and Express Endogenous Circadian Rhythmicity. PLoS ONE 2016, 11, e0146643. [Google Scholar] [CrossRef]
- Kasahara, T.; Abe, K.; Mekada, K.; Yoshiki, A.; Kato, T. Genetic variation of melatonin productivity in laboratory mice under domestication. Proc. Natl. Acad. Sci. USA 2010, 107, 6412–6417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Silveyra, E.; Jin, N.; Ribelayga, C.P. A congenic line of the C57BL/6J mouse strain that is proficient in melatonin synthesis. J. Pineal. Res. 2018, 65, e12509. [Google Scholar] [CrossRef] [PubMed]
- Froy, O.; Chapnik, N. Circadian oscillation of innate immunity components in mouse small intestine. Mol. Immunol. 2007, 44, 1954–1960. [Google Scholar] [CrossRef] [PubMed]
- Belstrøm, D.; Holmstrup, P.; Bardow, A.; Kokaras, A.; Fiehn, N.-E.; Paster, B.J. Temporal Stability of the Salivary Microbiota in Oral Health. PLoS ONE 2016, 11, e0147472. [Google Scholar] [CrossRef]
- Takayasu, L.; Suda, W.; Takanashi, K.; Iioka, E.; Kurokawa, R.; Shindo, C.; Hattori, Y.; Yamashita, N.; Nishijima, S.; Oshima, K.; et al. Circadian oscillations of microbial and functional composition in the human salivary microbiome. DNA Res. 2017, 24, 261–270. [Google Scholar] [CrossRef]
- Collado, M.C.; A Engen, P.; Bandín, C.; Cabrera-Rubio, R.; Voigt, R.M.; Green, S.J.; Naqib, A.; Keshavarzian, A.; Scheer, F.A.J.L.; Garaulet, M. Timing of food intake impacts daily rhythms of human salivary microbiota: A randomized, crossover study. FASEB J. 2018, 32, 2060–2072. [Google Scholar] [CrossRef]
- Kaczmarek, J.L.; Musaad, S.M.; Holscher, H.D. Time of day and eating behaviors are associated with the composition and function of the human gastrointestinal microbiota. Am. J. Clin. Nutr. 2017, 106, ajcn156380. [Google Scholar] [CrossRef] [PubMed]
- Reitmeier, S.; Kiessling, S.; Clavel, T.; List, M.; Almeida, E.L.; Ghosh, T.S.; Neuhaus, K.; Grallert, H.; Linseisen, J.; Skurk, T.; et al. Arrhythmic Gut Microbiome Signatures Predict Risk of Type 2 Diabetes. Cell Host Microbe 2020, 28, 258–272.e6. [Google Scholar] [CrossRef] [PubMed]
| CRs Disruption | Gut Microbiota | Metabolism | Immune System |
|---|---|---|---|
| Jet lag | - Increased gut permeability [49,50] - Changes in overall fecal community membership and loss of microbial oscillations [40,49,51] - Downregulation of SCFA production [52] | - Increased body weight and adipose tissue content [40,49,51] - Altered food and water intake patterns [40,51,53] | - Increased levels of inflammatory macrophages and decreased T regulatory cells in adipose tissue [51] - Elevated intestinal Th17 cell number [54] - Endotoxemia [55] |
| TRF (active phase) | - Restored gut microbiota community disruption and loss of oscillations induced by HFD [39] | - Restored transcript oscillations in liver and protects against weight gain on HFD [56] | - Did not impact adipose tissue inflammation and macrophage infiltration [57] - Increased expression of serum cytokines [58] |
| TRF (rest phase) | N/A | - Disrupted normal feeding and hepatic transcription patterns [59] | - Disrupted diurnal difference in helminth expulsion rate [60] - Reduced expression of serum cytokines and bacterial killing in response to LPS challenge [58] |
| Bmal1 | - Altered taxonomic composition of fecal microbial community [61] | - Arrhythmic and reduced activity [10] - Impaired glucose and lipid homeostasis [13,14] - Insulin insensitivity [62] | - More susceptible to infection [63] - Loss of rhythmic lymphocyte trafficking [64] - KO in ILCs: reduced ILC numbers in intestine [65] - KO in myeloid cells [18], neutrophils [66], lymphocytes [64]: abolished rhythmic trafficking, reduced infection clearance rate |
| Clock | - Increased gut permeability [50] - Altered fecal microbial community and reduced taxonomic diversity [67] | - Increased body weight and food intake [68] | - Ameliorated induction of dermatitis via IL-23 receptor [69] - Reduced expression of proinflammatory genes; reduced intestinal colonization of Salmonella [70] |
| Cry1/2 | N/A | - Increased weight gain and lipid uptake, and hyperinsulinemia [71] | - Autoimmune response (increased IgG concentration, leukocyte infiltration) [72] - NF-κB activation [73] - Increased activated T cells and TNF-α production; exacerbated joint swelling in arthritis model [74] |
| Per1/2/3 | - Altered fecal microbial community and loss of oscillating taxa [40] | - Leptin resistance [75] - Altered lipid metabolism [76] - Loss of diurnal feeding rhythm [77] - Increased adipose tissue; Decreased muscle tissue [78] | - Loss of daily rhythms of IFN-γ in spleen and serum [79], and cytolytic natural killer cell factors [80] - Protection from LPS-induced mortality [81] |
| Rev-erbα/β | N/A | - Altered lipid homeostatic gene networks and metabolism; altered wheel-running patterns [82] - Reduced body fat and increased lean mass via Nfil3 repression [83] | - Loss of diurnal endotoxin response [84] - Disruption of Th17 cell lineage specification by Nfil3 suppression [54] - Loss of signaling relay between ILCs and Nfil3 [83] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frazier, K.; Frith, M.; Harris, D.; Leone, V.A. Mediators of Host–Microbe Circadian Rhythms in Immunity and Metabolism. Biology 2020, 9, 417. https://doi.org/10.3390/biology9120417
Frazier K, Frith M, Harris D, Leone VA. Mediators of Host–Microbe Circadian Rhythms in Immunity and Metabolism. Biology. 2020; 9(12):417. https://doi.org/10.3390/biology9120417
Chicago/Turabian StyleFrazier, Katya, Mary Frith, Dylan Harris, and Vanessa A. Leone. 2020. "Mediators of Host–Microbe Circadian Rhythms in Immunity and Metabolism" Biology 9, no. 12: 417. https://doi.org/10.3390/biology9120417
APA StyleFrazier, K., Frith, M., Harris, D., & Leone, V. A. (2020). Mediators of Host–Microbe Circadian Rhythms in Immunity and Metabolism. Biology, 9(12), 417. https://doi.org/10.3390/biology9120417






