Cloning of Thalassiosira pseudonana’s Mitochondrial Genome in Saccharomyces cerevisiae and Escherichia coli
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Genomic DNA Isolation by Modified Alkaline Lysis
2.3. DNA Fragment Preparation for Polymerase Chain Reaction (PCR) Cloning
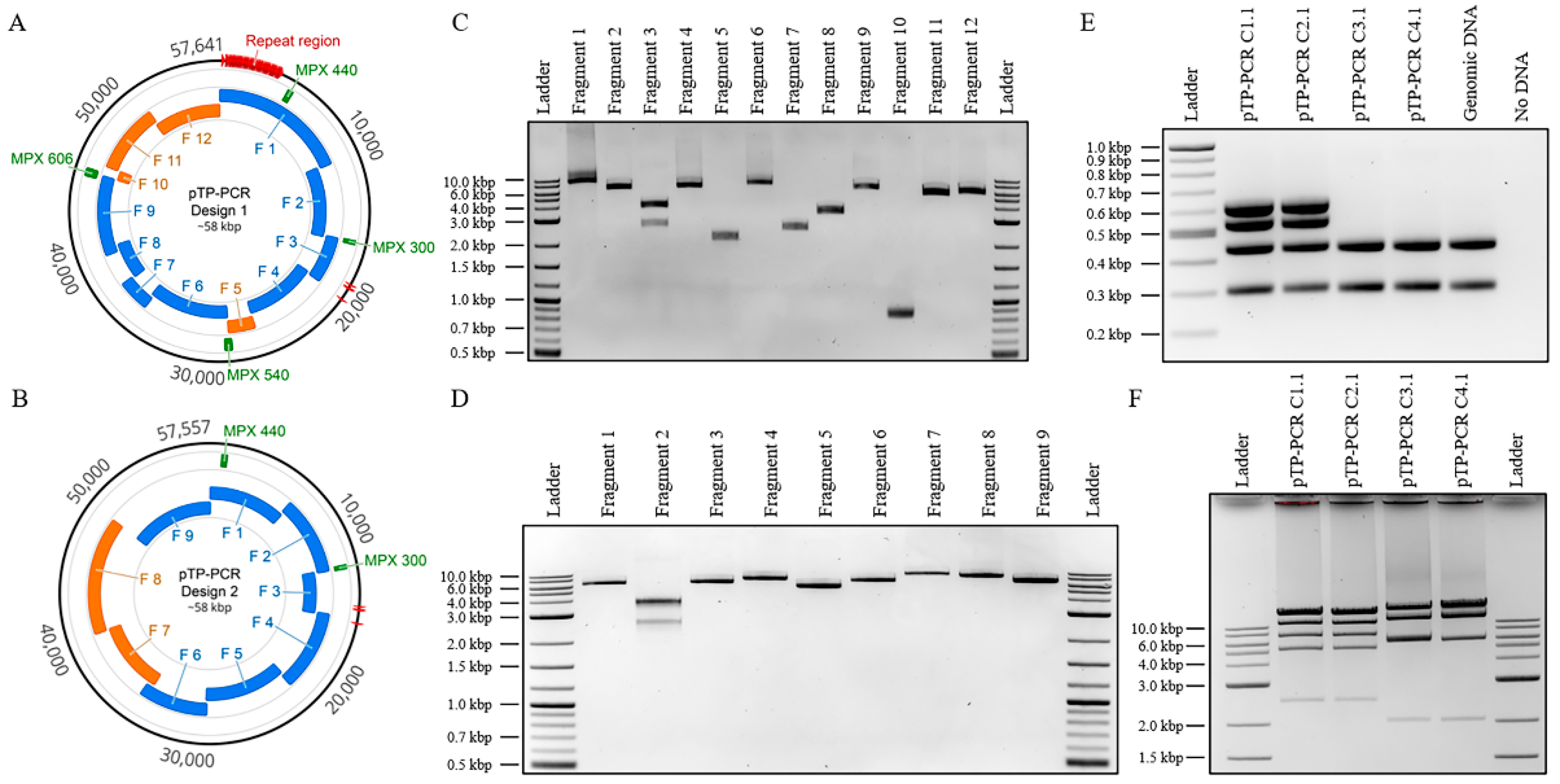
2.3.1. Design 1—Full Genome (pTP-PCR C1 and C2)
2.3.2. Design 2—Reduced Genome Lacking the Repetitive Region (pTP-PCR C3 and C4)
2.4. Yeast Spheroplast Transformation Protocol
2.5. E. coli Transformation
2.6. Screening Strategy
2.6.1. Screening Yeast Colonies
2.6.2. Screening E. coli Colonies
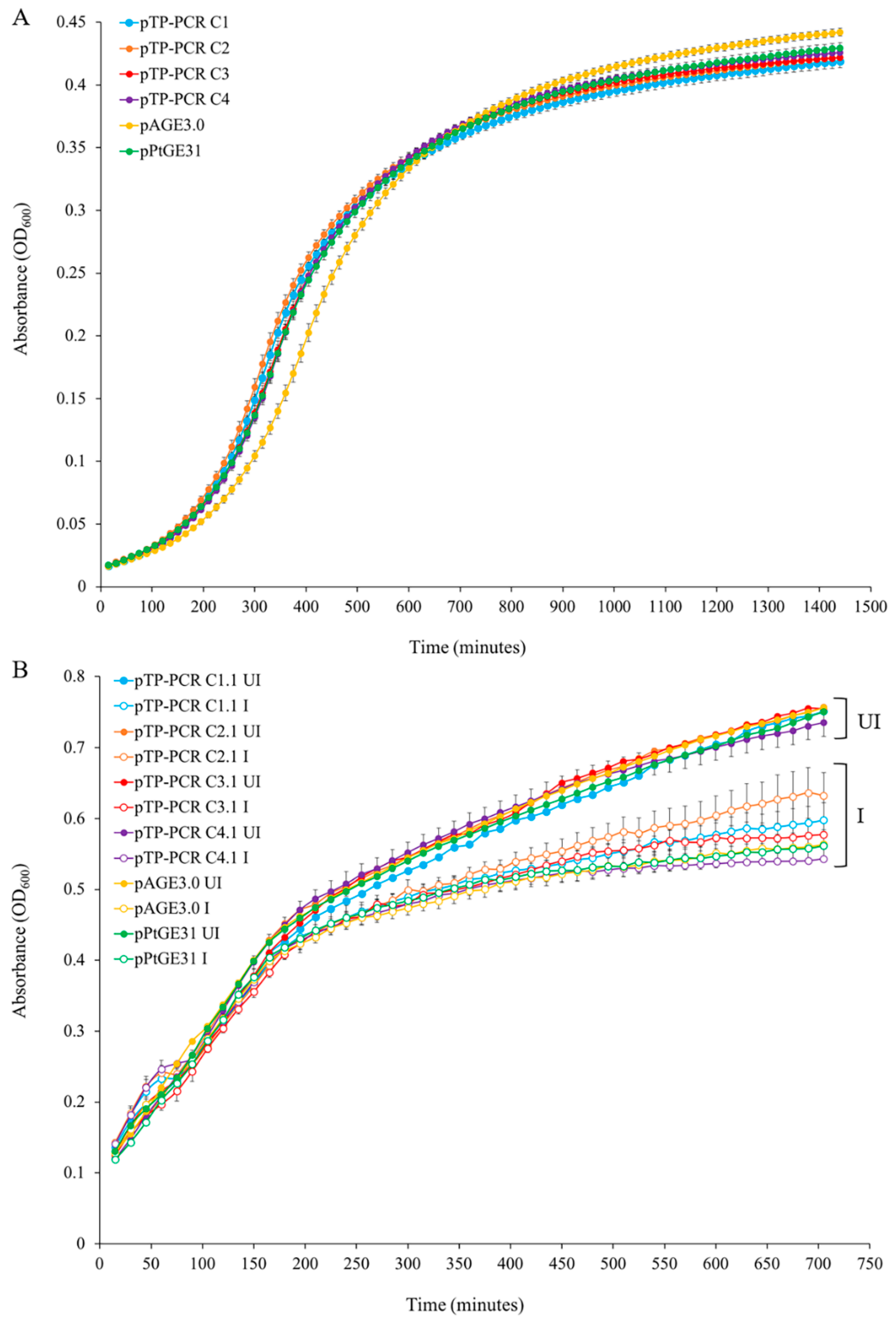
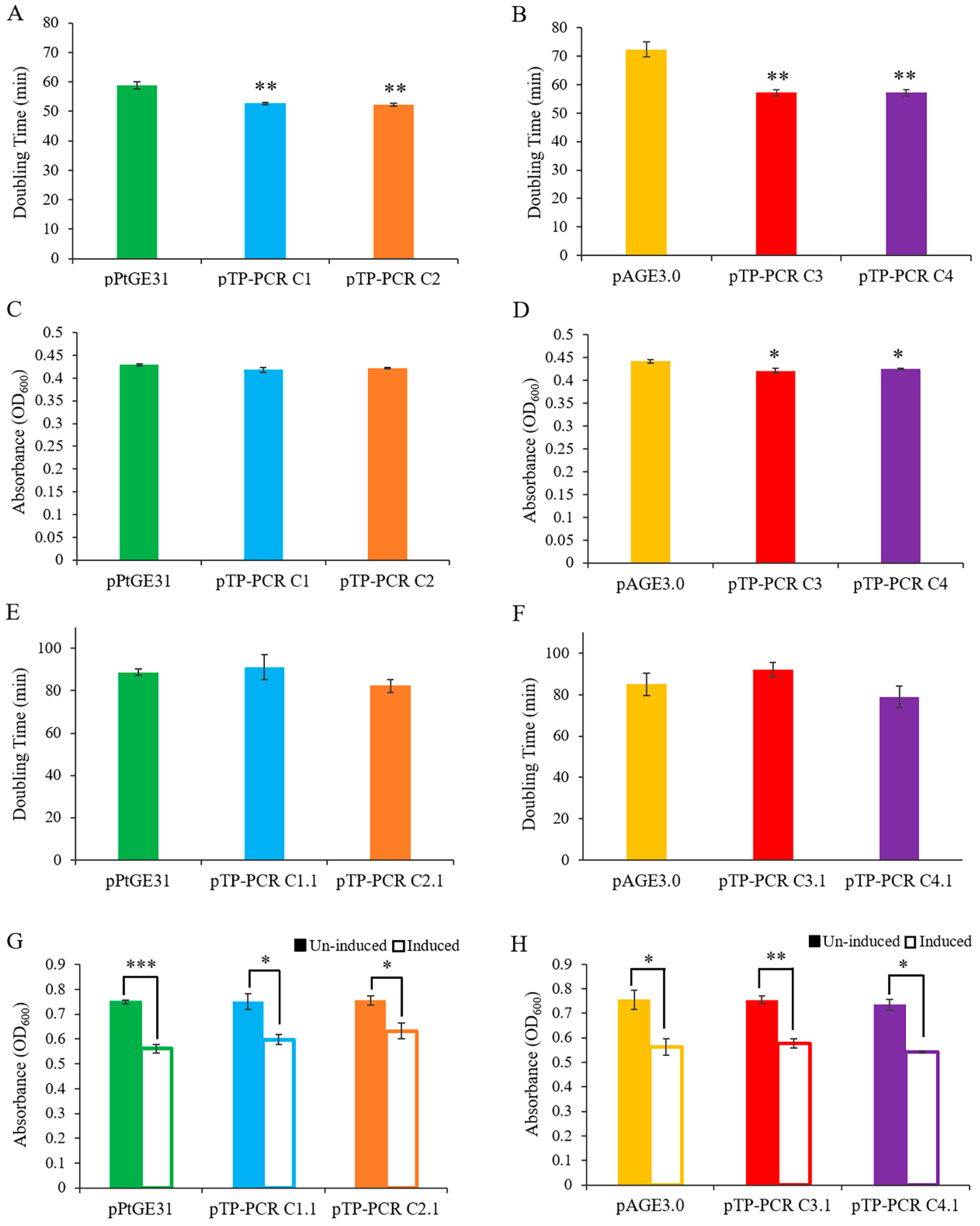
2.7. Evaluation of Growth Phenotypes of Host Strains
2.7.1. S. cerevisiae Growth in Liquid Media
2.7.2. E. coli Growth in Liquid Media
2.8. Bacterial RNA Extraction
2.9. RNA Sequencing
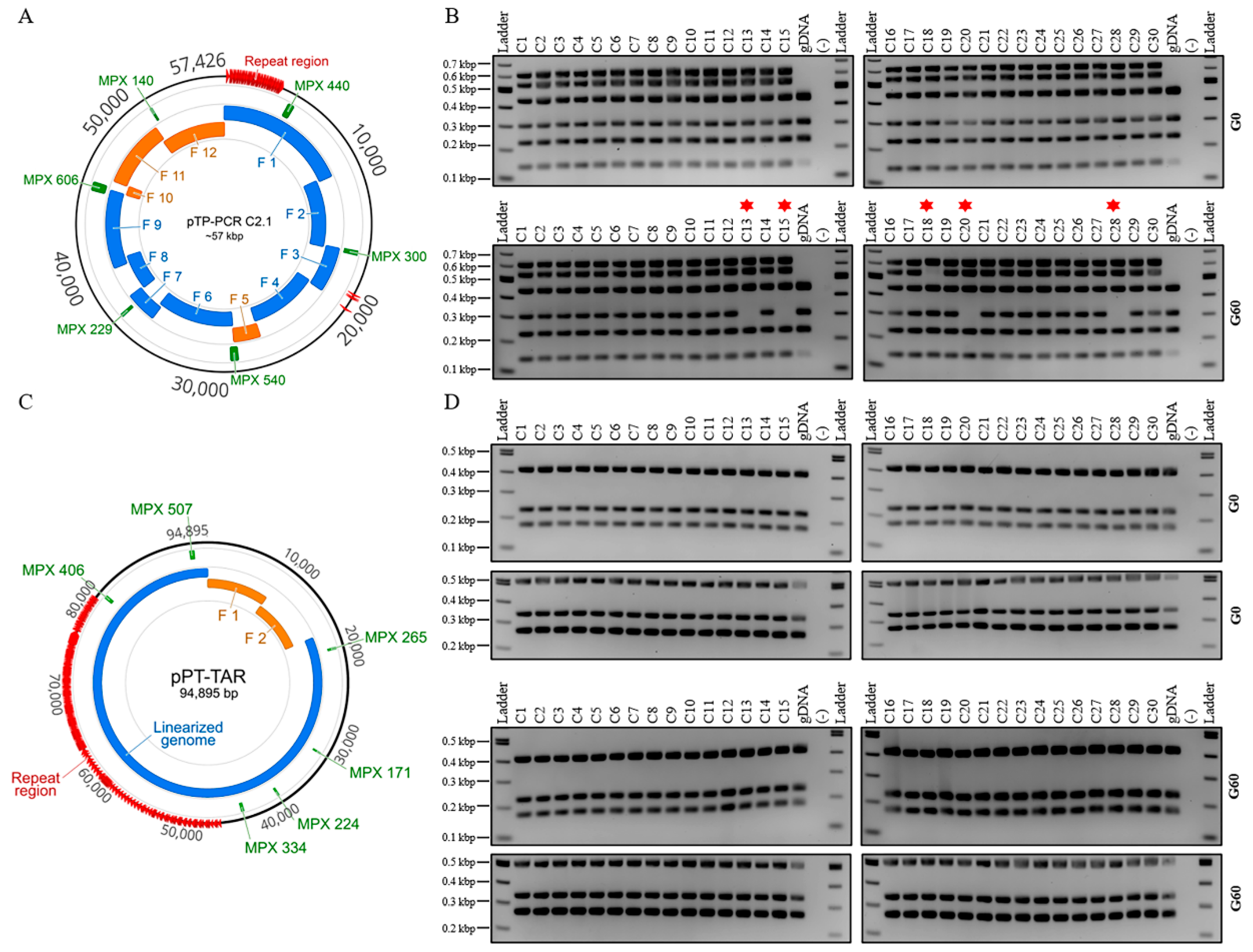
2.10. Plasmid Stability Assay
2.10.1. Propagation of E. coli Strains
2.10.2. Analysis of Descendant E. coli Colonies
2.11. Statistical Analyses
3. Results
3.1. Cloning of T. pseudonana Mitochondrial Genomes
3.2. Sequence Analysis of Cloned T. pseudonana Mitochondrial Genomes
3.3. Maintenance of T. pseudonana Mitochondrial Genomes in Host Organisms
3.4. Assessing the Expression of T. pseudonana and P. tricornutum Mitochondrial Genes in E. coli
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nelson, D.M.; Tréguer, P.; Brzezinski, M.A.; Leynaert, A.; Quéguiner, B. Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem. Cycles 1995, 9, 359–372. [Google Scholar] [CrossRef]
- Tréguer, P.; Nelson, D.M.; Van Bennekom, A.J.; DeMaster, D.J.; Leynaert, A.; Quéguiner, B. The silica balance in the world ocean: A reestimate. Science 1995, 268, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cabero, M.; Puchol, V.; Beltrán, D.; Amorós, P. Thalassiosira pseudonana diatom as biotemplate to produce a macroporous ordered carbon-rich material. Carbon N. Y. 2008, 46, 297–304. [Google Scholar] [CrossRef]
- Delalat, B.; Sheppard, V.C.; Ghaemi, S.R.; Rao, S.; Prestidge, C.A.; McPhee, G.; Rogers, M.-L.; Donoghue, J.F.; Pillay, V.; Johns, T.G.; et al. Targeted drug delivery using genetically engineered diatom biosilica. Nat. Commun. 2015, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Armbrust, E.V.; Berges, J.A.; Bowler, C.; Green, B.R.; Martinez, D.; Putnam, N.H.; Zhou, S.; Allen, A.E.; Apt, K.E.; Bechner, M.; et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 2004, 306, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Oudot-Le Secq, M.P.; Green, B.R. Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene 2011, 476, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Oudot-Le Secq, M.P.; Grimwood, J.; Shapiro, H.; Armbrust, E.V.; Bowler, C.; Green, B.R. Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: Comparison with other plastid genomes of the red lineage. Mol. Genet. Genom. 2007, 277, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Karas, B.J.; Diner, R.E.; Lefebvre, S.C.; McQuaid, J.; Phillips, A.P.R.; Noddings, C.M.; Brunson, J.K.; Valas, R.E.; Deerinck, T.J.; Jablanovic, J.; et al. Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Poulsen, N.; Chesley, P.M.; Kröger, N. Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae). J. Phycol. 2006, 42, 1059–1065. [Google Scholar] [CrossRef]
- Hopes, A.; Nekrasov, V.; Kamoun, S.; Mock, T. Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Methods 2016, 12, 1–12. [Google Scholar] [CrossRef]
- Trentacoste, E.M.; Shrestha, R.P.; Smith, S.R.; Glé, C.; Hartmann, A.C.; Hildebrand, M.; Gerwick, W.H. Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc. Natl. Acad. Sci. USA 2013, 110, 19748–19753. [Google Scholar] [CrossRef]
- Tesson, B.; Lerch, S.J.L.; Hildebrand, M. Characterization of a new protein family associated with the silica deposition vesicle membrane enables genetic manipulation of diatom silica. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Belshaw, N.; Grouneva, I.; Aram, L.; Gal, A.; Hopes, A.; Mock, T. Efficient CRISPR/Cas-mediated homologous recombination in the model diatom Thalassiosira pseudonana. bioRxiv 2017, 215582. [Google Scholar]
- Görlich, S.; Pawolski, D.; Zlotnikov, I.; Kröger, N. Control of biosilica morphology and mechanical performance by the conserved diatom gene Silicanin-1. Commun. Biol. 2019, 2, 1–8. [Google Scholar] [CrossRef]
- Schober, A.F.; Río Bártulos, C.; Bischoff, A.; Lepetit, B.; Gruber, A.; Kroth, P.G. Organelle studies and proteome analyses of mitochondria and plastids fractions from the diatom Thalassiosira pseudonana. Plant Cell Physiol. 2019, 60, 1811–1828. [Google Scholar] [CrossRef] [PubMed]
- Avalos, J.L.; Fink, G.R.; Stephanopoulos, G. Compartmentalization of metabolic pathways in yeast mitochondria improves production of branched chain alcohols. Nat. Biotechnol. 2017, 31, 1–10. [Google Scholar] [CrossRef]
- Bigger, B.W.; Liao, A.-Y.; Sergijenko, A.; Coutelle, C. Trial and error: How the unclonable human mitochondrial genome was cloned in yeast. Pharm. Res. 2011, 28, 2863–2870. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.G.; Koob, M.D. Efficient cloning and engineering of entire mitochondrial genomes in Escherichia coli and transfer into transcriptionally active mitochondria. Nucleic Acids Res. 2003, 31, 1407–1415. [Google Scholar] [CrossRef]
- Gibson, D.G.; Smith, H.O.; Hutchison III, C.A.; Venter, J.C.; Merryman, C. Chemical synthesis of the mouse mitochondrial genome. Nat. Methods 2010, 7, 901–903. [Google Scholar] [CrossRef]
- Gupta, M.; Hoo, B. Entire maize chloroplast genome is stably maintained in a yeast artificial chromosome. Plant Mol. Biol. 1991, 17, 361–369. [Google Scholar] [CrossRef]
- Itaya, M.; Fujita, K.; Kuroki, A.; Tsuge, K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat. Methods 2008, 5, 41–43. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, B.M.; Mikkelson, K.L.; Gutierrez, N.M.; Cunningham, J.L.; Wolff, K.L.; Szyjka, S.J.; Yohn, C.B.; Redding, K.E.; Mendez, M.J. An exogenous chloroplast genome for complex sequence manipulation in algae. Nucleic Acids Res. 2012, 40, 2782–2792. [Google Scholar] [CrossRef]
- Cochrane, R.R.; Brumwell, S.L.; Soltysiak, M.P.M.; Hamadache, S.; Davis, J.G.; Wang, J.; Tholl, S.Q.; Janakirama, P.; Edgell, D.R.; Karas, B.J. Rapid method for generating designer algal mitochondrial genomes. Algal Res. 2020, 50, 1–30. [Google Scholar] [CrossRef]
- Noskov, V.N.; Karas, B.J.; Young, L.; Chuang, R.-Y.; Gibson, D.G.; Lin, Y.-C.; Stam, J.; Yonemoto, I.T.; Suzuki, Y.; Andrews-Pfannkoch, C.; et al. Assembly of large, high G+C bacterial DNA fragments in yeast. ACS Synth. Biol. 2012, 1, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Benders, G.A.; Noskov, V.N.; Denisova, E.A.; Lartigue, C.; Gibson, D.G.; Assad-Garcia, N.; Chuang, R.-Y.; Carrera, W.; Moodie, M.; Algire, M.A.; et al. Cloning whole bacterial genomes in yeast. Nucleic Acids Res. 2010, 38, 2558–2569. [Google Scholar] [CrossRef]
- Karas, B.J.; Suzuki, Y.; Weyman, P.D. Strategies for cloning and manipulating natural and synthetic chromosomes. Chromosom. Res. 2015, 23, 57–68. [Google Scholar] [CrossRef]
- Karas, B.J.; Jablanovic, J.; Sun, L.; Ma, L.; Goldgof, G.M.; Stam, J.; Ramon, A.; Manary, M.J.; Winzeler, E.A.; Venter, J.C.; et al. Direct transfer of whole genomes from bacteria to yeast. Nat. Methods 2013, 10, 410–412. [Google Scholar] [CrossRef]
- Karas, B.J.; Molparia, B.; Jablanovic, J.; Hermann, W.J.; Lin, Y.-C.; Dupont, C.L.; Tagwerker, C.; Yonemoto, I.T.; Noskov, V.N.; Chuang, R.-Y.; et al. Assembly of eukaryotic algal chromosomes in yeast. J. Biol. Eng. 2013, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.G.; Koob, M.D. Toward genetic transformation of mitochondria in mammalian cells using a recoded drug-resistant selection marker. J. Genet. Genom. 2011, 38, 173–179. [Google Scholar] [CrossRef]
- Slattery, S.S.; Diamond, A.; Wang, H.; Therrien, J.A.; Lant, J.T.; Jazey, T.; Lee, K.; Klassen, Z.; Desgagné-Penix, I.; Karas, B.J.; et al. An expanded plasmid-based genetic toolbox enables Cas9 genome editing and stable maintenance of synthetic pathways in Phaeodactylum tricornutum. ACS Synth. Biol. 2018, 7, 328–338. [Google Scholar] [CrossRef]
- Brumwell, S.L.; MacLeod, M.R.; Huang, T.; Cochrane, R.R.; Meaney, R.S.; Zamani, M.; Matysiakiewicz, O.; Dan, K.N.; Janakirama, P.; Edgell, D.R.; et al. Designer Sinorhizobium meliloti strains and multi-functional vectors enable direct inter-kingdom DNA transfer. PLoS ONE 2019, 14, e0206781. [Google Scholar] [CrossRef] [PubMed]
- Sprouffske, K.; Wagner, A. Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinform. 2016, 17, 172. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Godiska, R.; Patterson, M.; Schoenfeld, T.; Mead, D.A. Beyond pUC: Vectors for cloning unstable DNA. Optim. DNA Seq. Process 2005, 1, 55–75. [Google Scholar]
- Bierne, H.; Michel, B. When replication forks stop. Mol. Microbiol. 1994, 13, 17–23. [Google Scholar] [CrossRef]
- Yoon, Y.G.; Koob, M.D. Selection by drug resistance proteins located in the mitochondria of mammalian cells. Mitochondrion 2008, 8, 345–351. [Google Scholar] [CrossRef]
- Rasala, B.A.; Chao, S.-S.; Pier, M.; Barrera, D.J.; Mayfield, S.P. Enhanced genetic tools for engineering multigene traits into green algae. PLoS ONE 2014, 9, e94028. [Google Scholar] [CrossRef]
- Hu, Z.; Fan, Z.; Zhao, Z.; Chen, J.; Li, J. Stable expression of antibiotic-resistant gene ble from Streptoalloteichus hindustanus in the mitochondria of Chlamydomonas reinhardtii. PLoS ONE 2012, 7, e35542. [Google Scholar] [CrossRef]
- Xie, W.H.; Zhu, C.C.; Zhang, N.S.; Li, D.W.; Yang, W.D.; Liu, J.S.; Sathishkumar, R.; Li, H.Y. Construction of novel chloroplast expression vector and development of an efficient transformation system for the diatom Phaeodactylum tricornutum. Mar. Biotechnol. 2014, 16, 538–546. [Google Scholar] [CrossRef] [PubMed]
| Design Assembly Type DNA Source | S. cerevisiae | E. coli | |||||
|---|---|---|---|---|---|---|---|
| Media | Colony Count | Multiplex PCR Screen Positive/Total | Media | Selected Yeast Colony: E. coli Colony Count | Multiplex PCR Screen Positive/Total | Final Genomes Names Selected for Analysis | |
| 1—Full Genome PCR—12 Fragments Genomic DNA | -Histidine -Uracil | 187 | 15/20 | CM | C1: 11 C2: 1137 | C1 = 8/8 C2 = 4/4 | pTP-PCR C1.1 pTP-PCR C2.1 |
| 2—Reduced Genome PCR—9 Fragments Genomic DNA | -Histidine | 680 | 18/20 | CM | C1: 4366 C2: 3530 | C1 = 5/5 C2 = 5/5 | pTP-PCR C3.1 pTP-PCR C4.1 |
| Clone | Point Mutations | Gap Mutations | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Synonymous | Missense | Nonsense | Non-Coding | Non-Coding | Coding | ||||
| Insertion | Deletion | Insertion | Deletion | ||||||
| pTP-PCR C1.1 | 1 | 6 | 0 | 8 | 0 | 6 | 0 | 3 | 24 |
| pTP-PCR C2.1 | 1 | 3 | 0 | 7 | 2 | 8 | 0 | 2 | 23 |
| pTP-PCR C3.1 | 0 | 5 | 0 | 0 | 0 | 1 | 2 | 4 | 12 |
| pTP-PCR C4.1 | 1 | 5 | 0 | 3 | 0 | 1 | 0 | 2 | 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cochrane, R.R.; Brumwell, S.L.; Shrestha, A.; Giguere, D.J.; Hamadache, S.; Gloor, G.B.; Edgell, D.R.; Karas, B.J. Cloning of Thalassiosira pseudonana’s Mitochondrial Genome in Saccharomyces cerevisiae and Escherichia coli. Biology 2020, 9, 358. https://doi.org/10.3390/biology9110358
Cochrane RR, Brumwell SL, Shrestha A, Giguere DJ, Hamadache S, Gloor GB, Edgell DR, Karas BJ. Cloning of Thalassiosira pseudonana’s Mitochondrial Genome in Saccharomyces cerevisiae and Escherichia coli. Biology. 2020; 9(11):358. https://doi.org/10.3390/biology9110358
Chicago/Turabian StyleCochrane, Ryan R., Stephanie L. Brumwell, Arina Shrestha, Daniel J. Giguere, Samir Hamadache, Gregory B. Gloor, David R. Edgell, and Bogumil J. Karas. 2020. "Cloning of Thalassiosira pseudonana’s Mitochondrial Genome in Saccharomyces cerevisiae and Escherichia coli" Biology 9, no. 11: 358. https://doi.org/10.3390/biology9110358
APA StyleCochrane, R. R., Brumwell, S. L., Shrestha, A., Giguere, D. J., Hamadache, S., Gloor, G. B., Edgell, D. R., & Karas, B. J. (2020). Cloning of Thalassiosira pseudonana’s Mitochondrial Genome in Saccharomyces cerevisiae and Escherichia coli. Biology, 9(11), 358. https://doi.org/10.3390/biology9110358





